Abstract
Analyzing sizes of DNA via electrophoresis using a gel has played an important role in the recent, rapid progress of biology and biotechnology. Although analyzing DNA over a wide range of sizes in a short time is desired, no existing electrophoresis methods have been able to fully satisfy these two requirements. Here we propose a novel method using a rigid 3D network structure composed of solid nanowires within a microchannel. This rigid network structure enables analysis of DNA under applied DC electric fields for a large DNA size range (100 bp–166 kbp) within 13 s, which are much wider and faster conditions than those of any existing methods. The network density is readily varied for the targeted DNA size range by tailoring the number of cycles of the nanowire growth only at the desired spatial position within the microchannel. The rigid dense 3D network structure with spatial density control plays an important role in determining the capability for analyzing DNA. Since the present method allows the spatial location and density of the nanostructure within the microchannels to be defined, this unique controllability offers a new strategy to develop an analytical method not only for DNA but also for other biological molecules.
Similar content being viewed by others
Introduction
Since researchers have revealed the critical role played by information stored within DNA molecules in the complex biological activities occurring in living systems1,2,3, various methodologies have been developed to analyze DNA molecules4,5. Among them, gel electrophoresis is the most prominent and commercially available standard method to analyze the size of DNA molecules due to the great controllability of pore sizes and also the inexpensiveness of electrophoresis devices6. On the other hand, the limitations of gel electrophoresis have also been pointed out and they are the difficulty in analyzing relatively long DNA molecules in a short time, the high skill required for appropriately preparing the gel and the requirement of gel concentration control for each targeted DNA size7,8,9. Recently, data for a wide range of DNA size have been obtained using the hydrodynamic chromatography method10. Although the analysis time using this method is faster than that using gel electrophoresis, it still takes several tens of minutes for the fastest analysis. To realize an analysis on the time scale of seconds, nanostructure electrophoresis has attracted much attention as a novel method to overcome the above issues with gel electrophoresis11.
Nanostructure electrophoresis under DC and AC electric fields has been used to analyze DNA molecules by separating differently sized DNA molecules in a short time, which is impossible in gel electrophoresis12. Since Volkmuth and Austin13 first proposed to utilize highly-ordered sieving nanostructures as an alternative for conventional randomly-ordered gel or polymer sieving matrices, many lithography-based nanostructures, such as nanopillar arrays13,14,15,16, nanowall arrays17, nanofilter arrays18,19,20,21 and nanochannels22,23,24, have been applied for analysis of DNA molecules. For example, Kaji et al.15 demonstrated that a nanopillar structure could separate 5 components from a mixture of λ-DNA (48.5 kbp) digested by HindIII under a DC electric field, however it took more than 600 s and the DNA size range had to be over 2 kbp. Huang et al.14 also obtained good separation of large DNA molecules (61–209 kb) in 15 s, however, the nanopillar arrays required AC electric fields and DNA size range was limited to over 60 kb. In a third example from a study by Fu et al.20, continuous separation of DNA molecules was realized in a wide range (0.1–20 kbp), however it was essentially impossible to separate 0.1 and 20 kbp at the same time because of different required separation conditions, which Fu et al. attributed to different separation regimes for 0.1 and 20 kbp. As discussed above, intensive efforts have been devoted to developing new types of nanostructure electrophoresis11,12. Although these artificial nanostructures realize analysis of DNA molecules with fewer time- and labor-intensive operations25, the use of lithography techniques intrinsically limits the available size range (usually over 1 kbp) and also the formation of three-dimensionally (3D) designed spatial nanostructures. Nanostructure electrophoresis needs a relatively long time to analyze short DNA molecules due to the sparseness of geometrical nanostructures compared with 3D gel network structures18,26,27.
A self-assembled nanoparticle array within a microchannel is another candidate methodology, which offers a 3D-ordered nanoporous sieving matrix28,29,30,31,32. Interesting features of nanoparticle arrays have been demonstrated by separating DNA molecules under AC electric fields31. Zeng and Harrison30 reported DNA separations ranging from 100 bp to 2 kbp using the nanoparticle array and the separation time was 10 min and Tabuchi et al.29 showed DNA separations up to 15 kbp within 100 s using nanospheres. Compared with lithography-defined nanostructure electrophoresis, the DNA size range is much wider due to 3D-ordered sieving matrices, however, the spatial controllability of such nanoparticle arrays within microchannels is rather poor due to the intrinsic difficulties in controlling the spatial positions and forming the sparse structures due to the naturally occurring aggregation events of nanoparticles. Additionally, these techniques require special instruments such as AC electric fields generators or a combination of electric fields and pressure application. These limitations, especially as to the spatial controllability, are detrimental to analyze and separate relatively long DNA molecules (over 20 kbp) in a short time; we have seen a general difference between gel electrophoresis and nanostructure electrophoresis. Thus, no existing methods are able to analyze fully a wide range of DNA molecules in a short time.
To overcome these long-standing issues in DNA analysis, here we propose a novel electrophoresis method, which utilizes a rigid 3D network structure composed of solid nanowires only at the desired position within a microchannel. This method allows analysis of DNA molecules under DC electric fields in a much wider range and in a faster time than any other existing methods. Our approach to realize the rigid 3D network structure at the desired spatial position within the microchannel is to use cycling nanowire growth by applying a vapor-liquid-solid (VLS) method with a metal catalyst. Two noteworthy features of this method are: (1) the spatial selectivity of VLS nanowire growth and (2) the density control of the network structures by cycling the supply of metal catalyst.
Results and Discussion
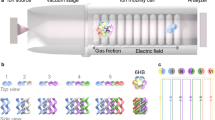
The scheme in Figure 1 summarizes the fabrication procedures; also included are a photo of a fabricated device and SEM and TEM images of the nanowire network structure within the microchannel when varying the number of growth cycles. We fabricated the microchannel on a fused silica substrate by conventional photolithography and utilized Au catalyst for VLS growth of SnO2 nanowires (typical diameter of 10–20 nm)33,34,35. It should be noted that we intentionally did not use electron beam lithography as we wanted to target future commercial use. Other details of the fabrication procedures can be seen in Methods and Supplementary Fig. S1. The nanowire surface can be modified by forming a shell layer on the nanowire sidewall as a core-shell nanowire structure36; and here we utilized the SiO2 shell layer37, which generates a negatively charged surface on the nanowires to avoid the undesired adhesion of negatively charged DNA. The network density is readily controllable by varying the number of growth cycles of the nanowires as shown in Figure 1b, due to the increased number of branched nanowires. Figure 1f shows the pore size distribution data measured from the cross-sectional SEM images. The average pore size decreases with increasing network density via the cycling nanowire growth. It should be noted that the present method allows users to define the spatial location, surface properties, density and pore size of the rigid network nanostructure within the microchannel, which are not attainable by other methods.
3D nanowire network structure.
(a) Schematic illustrations showing nanowire structure after: (I) 1-cycle growth; (II) 3-cycle growth, (III) 5-cycle growth; and (lower row) the overall fabrication procedure. The microchannel and nanowire network structure are formed on fused silica substrate. (b) SEM images (scale bar, 1 μm) of the nanowire network structure showing: (I) 1-cycle growth; (II) 3-cycle growth; and (III) 5-cycle growth. The insets show vertical cross-sectional SEM images; scale bar, 1 μm. (c) Photograph of a fabricated device; scale bar, 1 cm. (d) SEM image of the nanowire network structure embedded in the microchannel; scale bar, 10 μm. (e) TEM images of the nanowire network structure; scale bars, 100 nm (left) and 50 nm (right). Black arrows highlight branched positions. (f) Pore size distribution of the nanowire network structure after (I) 1-cycle growth, (II) 3-cycle growth and (III) 5-cycle growth.
Figure 2 and Supplementary Fig. S2 show the electropherogram data for various DNA mixtures, ranging from 100 bp–166 kbp, obtained using the fabricated rigid 3D network structure when varying the number of nanowire growth cycles. The clear dependence of the electropherogram data on the number of cycles can be seen. In the microchannel with 1-cycle growth nanowires (Type I), the DNA mixtures ranging from 10 kbp to 166 kbp were separable; in particular, the DNA mixture of T4-DNA and λ-DNA was separable within just 3 s with an applied voltage field of 500 V/cm (see Supplementary Fig. S3), which is the fastest time reported to date. The DNA mixtures with DNA sizes smaller than 10 kbp could not be separated within the present experimental range, possibly due to the sparse nanowire density. The network structure with the 3-cycle growth nanowires (Type II) could well separate DNA mixtures down to 5 kbp. In the microchannel with the 5-cycle growth nanowires (Type III), the DNA mixtures down to 100 bp were separable within 20 s. Thus, the nanowire spatial density of the present network structure critically determined the electrophoretic behaviors of different sized DNA molecules. Denser network structures could separate smaller DNA molecules, i.e. there was a correlation between the network density and the separable DNA size range. Since the electrophoretic mobility determined the capability of DNA separation and analysis, we measured the mobility of DNA molecules that ranged from 100 bp to 166 kbp with an applied DC electric field of 500 V/cm.
Separation and mobility data in the network structure.
(a) Separation of 166 kbp DNA (20 ng/μL) and 48.5 kbp DNA (50 ng/μL) in the nanowire network structure with the (I) 1-cycle growth nanowires, (II) 3-cycle growth nanowires and (III) 5-cycle growth nanowires. The electropherogram was obtained at 3 mm from the entrance of the microchannel with the nanowire network structure. The applied DC electric field in the separation channel was 500 V/cm. (b) Separation of (I) 48.5 kbp DNA (20 ng/μL) and 10 kbp DNA (40 ng/μL) in the nanowire network structure with the 1-cycle growth nanowires at the applied DC electric field of 500 V/cm, (II) 10 kbp DNA (30 ng/μL) and 5 kbp DNA (30 ng/μL) in the nanowire network structure with the 3-cycle growth nanowires at the applied DC electric field of 500 V/cm and (III) 1 kbp DNA (30 ng/μL) and 100 bp DNA (30 ng/μL) in the nanowire network structure with the 5-cycle growth nanowires at the applied DC electric field of 500 V/cm. The electropherogram was obtained at 3 mm from the entrance of the microchannel with the nanowire network structure. (c) Dependence of electrophoretic mobility on DNA size in the nanowire network structure with 1-cycle growth nanowires (red circles), 3-cycle growth nanowires (blue circles) and 5-cycle growth nanowires (green circles). Error bars show the standard deviation for a series of measurements (N = 3).
Figure 2c shows the mobility data measured for the network structure with 1-, 3- or 5-cycle growth nanowires (see Supporting Information Movies-1, 2 and 3). These data clearly demonstrate the significant DNA size dependence on the electrophoretic mobility and they also indicate the network density critically affects the size dependence. The shape of the mobility curves for 1- and 3-cycle growth nanowires was identical but shifted in absolute value of mobility (y axis), while the shape of the mobility curve for the 5-cycle growth nanowires was qualitatively different from other two curves and it shifted in absolute value of DNA size (x axis). This seems to indicate that there are two different separation mechanisms as to the change of the pore size in the present network structure. Thus, these experimental data highlight that the present network structure composed of solid nanowires is capable of analyzing a wide range of DNA molecules within a short time.
This DNA size dependence on the electrophoretic mobility is in effect similar to the trend for gel electrophoresis when varying the gel concentration6,7,8,9. It is well known that a denser gel concentration can separate shorter DNA base pairs due to the decreased average pore sizes. In this sense, the spatial density dependence of the present rigid 3D network structure in the DNA separation is quite similar to that of gel electrophoresis. However, there is a significant difference between the present rigid network structure and the soft gel, which is the capability of the former to separate and analyze relatively long DNA molecules within a short time. This is similar to the capability of nanostructure electrophoresis11,12. Thus, the present rigid network structures possess the advantages of both the soft gel and the rigid lithography-defined nanostructures in DNA electrophoresis due to their tailored network structure density, namely the present network structure behaves as a hard gel.
When compared with nanostructure electrophoresis, the present rigid 3D network structure can form 3D confined small pores. The present rigid 3D network structure can enhance the probability of collisions between small DNA molecules and a rigid object more than conventional lithographically defined nanostructures including nanopillars and nanowalls can15,17,38; these collisions might possibly enhance Ogston sieving effects (DNA size < the pore size; mu~ exp(-N), where mu is the mobility and N is the DNA length and mu~ mu0exp(-KC), where mu0 is the free solution mobility, K is the constant of proportionality and C is the concentration of the network-forming matrix) for shorter DNA molecules (see Supplementary Figs. S4a and S4b)39. On the other hand, there would be an argument against Ogston sieving, which stems from the result that the time for transport through the pores (Ttransportation= d/(muE), where d is the pore size and E is the applied DC electric field) in the microchannel with the 5-cycle growth nanowires was shorter than the time for diffusion (Tdiffusion= d2/D, where D is the diffusion coefficient of the DNA molecules) (see Supplementary Fig. S4c)40. In addition, within the dense network structure, the deformed DNA molecules might collide with another rigid post before they contract and this should affect the electrophoretic mobility and also the DNA size dependence on the mobility. Since these results are inconclusive about whether there is Ogston sieving in the matrix, with some evidences in favor and other evidences against the theoretical description, we can say that the presence of 3D confined small rigid pores in the present network structure seems to be responsible for the ability to separate DNA mixtures of wider size ranges than conventional lithographically defined nanostructure electrophoresis.
Based on only the pore sizes and the spatial network structure, the present network structure is quite similar to that of conventional gel structures37,41,42. However, our single molecule imaging experiments revealed a difference between the present network structure and agarose gel regarding the electrophoretic behaviors of 38-kbp DNA, as shown in the video data of the Supporting Information Movie 4. Although their pore sizes are controlled to be similar for both structures, the DNA molecules in the nanowire network structure showed more frequent elongation and contraction by colliding with the rigid nanowire framework and faster migration than those in agarose gel. Thus, a scenario based on the pore size alone cannot explain the present results. One major difference between the present rigid network structure and the conventional gels is the stiffness of the network structures. The present network structure is much more rigid than the conventional gel network structures43; for example, typically the Young's modulus of agarose gel is 0.1–1 MPa, whereas that of the solid nanowires is 100 ± 20 GPa44,45. Since the deformation of DNA molecules must be larger when softer DNA molecules collide with a stiffer object, collision with the rigid nanowire network structure must amplify the DNA deformation more than for the soft gel, considering the stiffness of the DNA molecules. Ou et al.46 have proposed that such mechanical collisions with rigid posts are as important as the large sized DNA separation mechanism. Thus, the different collision behaviors of DNA molecules might cause the different electrophoretic results between the present network structure and the gels, especially for separating mixtures of relatively long DNAs within a short time. The spatial controllability of the present network structure might also be another factor responsible for the faster analytical time due to the spatially concentrated DC electric field. Since the exact separation mechanism of wide-range DNA mixtures in the present network structure must be rather complex, such novel DNA electrophoretic behaviors are an interesting subject for theoreticians and experimentalists alike.
Next we explored the feasibility of using the present network structure to analyze and separate multicomponent DNA mixtures. Figure 3a shows the electropherogram data of a mixture composed of four components including T4-DNA (166 kbp), λ-DNA (48.5 kbp), 10 kbp-DNA and 5 kbp-DNA separated in the microchannel having a network structure formed with 3-cycle growth nanowires. The DC electric field of 500 V/cm was utilized in the electrophoresis. The multicomponent DNA mixtures are clearly separable using the present network structure device and surprisingly within just 4 s, which is the fastest time reported so far15. By using a denser network structure formed with 5-cycle growth nanowires, we can analyze and separate the mixtures composed of even smaller sized DNA molecules 100 bp-DNA, 1 kbp-DNA, 5 kbp-DNA, 20 kbp-DNA and λ-DNA (48.5 kbp), as shown in Figure 3b. Under the DC electric field of 500 V/cm, the five differently sized DNA molecules are readily separated within only 7 s, which is also the fastest time reported so far.
Separation of multicomponent DNA mixtures in the network structure.
(a) Separation of 166 kbp DNA (20 ng/μL), 48.5 kbp DNA (20 ng/μL), 10 kbp DNA (20 ng/μL) and 5 kbp DNA (20 ng/μL) in the nanowire network structure with 3-cycle growth nanowires. The electropherogram was obtained at 1 mm from the entrance of the microchannel with the nanowire network structure. The applied DC electric field in the separation channel was 500 V/cm. (b) Separation of 48.5 kbp DNA (20 ng/μL), 20 kbp DNA (20 ng/μL), 5 kbp DNA (20 ng/μL), 1 kbp DNA (20 ng/μL) and 100 bp DNA (40 ng/μL) in the nanowire network structure with 5-cycle growth nanowires. The electropherogram was obtained at 1 mm from the entrance of the microchannel with the nanowire network structure. The applied DC electric field in the separation channel was 500 V/cm. (c) Separation of lambda DNA digested by HindIII in the nanowire network structure with 5-cycle growth nanowires. The electropherogram was obtained at 3 mm from the entrance of the microchannel with the nanowire network structure. The applied DC electric field in the separation channel was 500 V/cm.
To further demonstrate the capability of the present network structures for analyzing and separating DNA molecules, we separated λ-DNA digested by HindIII, which consists of eight different DNA size fragments, using the microchannel having the network structure formed with 5-cycle growth nanowires (Figure 3c). Measurements were made under an applied DC electric field of 500 V/cm. The DNA fragments were separated as six distinct peaks. Each peak was assigned to a DNA band as indicated in Figure 3c based on peak height and comparison with previous data15. Comparison with the previous top data showed the nanowire network structure had much faster separation time, comparable theoretical plate number and theoretical plate number per column length and lower resolution (Table 1). It should be emphasized that the present DNA separation is superior to that for the previous data in terms of the separation time. The present separation time is 13 s, which is much faster than the previous top time of 680 s obtained using a nanopillar structure15. Thus our experimental data prove that the present rigid 3D network structure can well analyze a wide range (100 bp–166 kbp) of multicomponent DNA mixtures within a short time and the results are superior to those of any existing methods5,6,11,12. Since the present concept based on the rigid 3D network structure in a microchannel has the potential to overcome the long-standing weaknesses in the existing DNA electrophoresis methods, we expect it will open up new research avenues for not only analyzing DNA molecules but also for developing analytical methods for various biological molecules, which are not attainable by existing methods.
In summary, we have demonstrated a novel method based on the rigid 3D network structure comprised of solid branched nanowires within a microchannel, which enables analysis of DNA molecules for a much wider of DNA sizes (100 bp–166 kbp) than any previous methods within a much shorter time (13 s). The network density is readily tunable for the targeted DNA size range by controlling the number of cycles for nanowire growth only at the desired spatial position within a microchannel; this level of flexibility cannot be attained by other methodologies. The rigid 3D network structure with the spatial density control plays an important role in attaining better analysis of DNA molecules. Since the present method allows users to define the spatial location and density of the network nanostructure within the microchannel, such spatial controllability will offer a new strategy to develop analytical methods not only for DNA molecules but also for other biological molecules.
Methods
Fabrication of cycling nanowire growth devices
To fabricate cycling nanowire growth devices, we have utilized a conventional photolithography process, electron beam lithography and vapor-liquid-solid (VLS) growth technique for the nanowires (Supplementary Fig. S1). First, a Cr layer (250 nm thick) was deposited on a substrate by RF sputtering (SVC-700LRF, Sanyu Denshi); this layer was a mask for the dry etching process to fabricate a microchannel. Positive photoresist (TSMR V50, Tokyo Ohka Kogyo Co.) was spin-coated on the Cr layer. Then the microchannel pattern with a width of 25 μm was formed by photolithography. After development of the resist, the patterned Cr area was etched by immersing in Cr etchant (H2O:Ce(NH4)2(NO3)6:HClO4, 85:10:5 by weight percent) for 5 min. The microchannel was formed by etching using reactive ion etching (RIE-10NR, Samco Co.) under CF4 gas ambient. The microchannel depth was controlled to be 2 μm, which is larger than the radius of gyration of T4 DNA (970 nm). The inlet and outlet via holes (1.5 mm diameter) for the microfluidic system were drilled with an ultrasonic driller (SOM-121, Shinoda Co.). We also patterned the metal catalysts to define the spatial position of the nanowires within the microchannel. A Cr layer (10 nm thick) was deposited within the microchannel. Positive resist (ZEP520 A7, Zeon Corp.) was coated on the microchannel by spin-coating and then the pattern was drawn by electron beam lithography (SPG-724, Sanyu Electron Co.). After developing the resist, the Cr layer pattern was removed with Cr etchant. An Au metal catalyst layer (3 nm thick) for nanowire growth was deposited within the microchannel by DC sputtering. And then the resist was lifted off using dimethylformamide, acetone and isopropanol, sequentially. The SnO2 nanowires were fabricated in the microchannel by a pulse laser deposition (PLD) system. Details of the nanowire fabrication conditions can be found elsewhere47. To prepare nanowire branches, the Au catalyst particles were deposited along the nanowire by DC sputtering and the SnO2 nanowires were grown cyclically using the PLD system to increase the density of nanowires in the microchannel. The Cr layer was lifted off with Cr etchant. It should be noted that a 40 nm layer of SiO2 was deposited onto the SnO2 nanowires to avoid charge attraction of the negatively charged DNA molecules. Finally, the microchannel was sealed using a fused silica cover plate (130 μm thick) by the literature method17,38.
DNA observations using the cycling nanowire growth devices
The electrophoretic DNA mobility measurements in the nanowire array structure were done using T4, λ, 20 kbp, 10 kbp, 5 kbp, 2 kbp, 1 kbp, 500 bp and 100 bp DNA molecules (Nippon Gene Co., Ltd.) stained with the dye, YOYO-1 (Invitrogen), at a dye-to-base pair ratio of 1:15 contained in concentrated buffer solution (5 × TBE; 445 mM Tris-Borate and 10 mM EDTA, pH 8.2, Sigma-Aldrich, Inc.). An inverted fluorescent microscope (Eclipse TE-300, Nikon) equipped with a high voltage sequencer (HVS448-1500, Lab Smith) was used to apply the DC electric fields and a Hg lamp was used to observe fluorescently stained DNA molecules. Fluorescence images were captured with an EB-CCD camera (C7190-43, Hamamatsu Photonics K.K.) through a 10 ×/1.40 NA objective lens (Nikon). The images were recorded on a DV tape (DSR-11, Sony) and then analyzed by image-processing software (Cosmos 32, Library, Tokyo, Japan).
DNA observations using the agarose gel
λ-DNA digested Apa I, stained with YOYO-1 at a dye-to-base pair ratio of 1:15, was observed in a polydimethylsiloxane (PDMS) microchannel (650 μm width; 50 μm height), which was filled with 5% agarose gel (Pulsed Field Certified Agarose, Bio-Rad Laboratories, Inc.) in 5× TBE. Under the DC electric field of 300 V/cm, λ-DNA molecules were observed through a 100×/1.40 NA objective lens on an inverted fluorescent microscope (Eclipse TE-300) equipped with a high-voltage sequencer (HVS448-1500), 488 nm wavelength laser (FLS-488-20) and EB-CCD camera (C7190-43). The images were recorded on a DV tape (DSR-11) and then analyzed by image-processing software (Cosmos 32).
References
Watson, J. D. & Crick, F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171, 737–738 (1953).
Ghosh, A. & Bansal, M. A glossary of DNA structures from A to Z. Acta Crystallogr. Sect. D. Biol. Crystallogr. 59, 620–626 (2003).
Gregory, S. G. et al. The DNA sequence and biological annotation of human chromosome 1. Nature 441, 315–321 (2006).
Maxam, A. M. & Gilbert, W. A new method for sequencing DNA. 1977. Biotechnology 24, 99–103 (1992).
Carey, L. & Mitnik, L. Trends in DNA forensic analysis. Electrophoresis 23, 1386–1397 (2002).
Viovy, J. L. Electrophoresis of DNA and other polyelectrolytes: Physical mechanisms. Rev. Mod. Phys. 72, 813–872 (2000).
Duke, T. A., Semenov, A. N. & Viovy, J. L. Mobility of a reptating polymer. Phys. Rev. Lett. 69, 3260–3263 (1992).
Heller, C., Duke, T. & Viovy, J. L. Electrophoretic Mobility of DNA in Gels. 2. Systematic Experimental-Study in Agarose Gels. Biopolymers 34, 249–259 (1994).
Semenov, A. N., Duke, T. A. & Viovy, J. Gel electrophoresis of DNA in moderate fields: The effect of fluctuations. Phys. Rev. E 51, 1520–1537 (1995).
Liu, L. et al. High-resolution hydrodynamic chromatographic separation of large DNA using narrow, bare open capillaries: a rapid and economical alternative technology to pulsed-field gel electrophoresis? Anal. Chem. 86, 729–736 (2014).
Salieb-Beugelaar, G. B., Dorfman, K. D., van den Berg, A. & Eijkel, J. C. Electrophoretic separation of DNA in gels and nanostructures. Lab Chip 9, 2508–2523 (2009).
Dorfman, K. D. DNA electrophoresis in microfabricated devices. Rev. Mod. Phys. 82, 2903–2947 (2010).
Volkmuth, W. D. & Austin, R. H. DNA electrophoresis in microlithographic arrays. Nature 358, 600–602 (1992).
Huang, L. R. et al. A DNA prism for high-speed continuous fractionation of large DNA molecules. Nat. Biotechnol. 20, 1048–1051 (2002).
Kaji, N. et al. Separation of long DNA molecules by quartz nanopillar chips under a direct current electric field. Anal. Chem. 76, 15–22 (2004).
Park, S. G., Olson, D. W. & Dorfman, K. D. DNA electrophoresis in a nanofence array. Lab Chip 12, 1463–1470 (2012).
Yasui, T. et al. DNA separation in nanowall array chips. Anal. Chem. 83, 6635–6640 (2011).
Han, J. & Craighead, H. G. Separation of long DNA molecules in a microfabricated entropic trap array. Science 288, 1026–1029 (2000).
Fu, J., Yoo, J. & Han, J. Molecular sieving in periodic free-energy landscapes created by patterned nanofilter arrays. Phys. Rev. Lett. 97, 018103 (2006).
Fu, J., Schoch, R. B., Stevens, A. L., Tannenbaum, S. R. & Han, J. A patterned anisotropic nanofluidic sieving structure for continuous-flow separation of DNA and proteins. Nat. Nanotech. 2, 121–128 (2007).
Mao, P. & Han, J. Massively-parallel ultra-high-aspect-ratio nanochannels as mesoporous membranes. Lab Chip 9, 586–591 (2009).
Li, W. L. et al. Sacrificial polymers for nanofluidic channels in biological applications. Nanotechnology 14, 578–583 (2003).
Cross, J. D., Strychalski, E. A. & Craighead, H. G. Size-dependent DNA mobility in nanochannels. J. Appl. Phys. 102, 024701 (2007).
Pennathur, S. et al. Free-solution oligonucleotide separation in nanoscale channels. Anal. Chem. 79, 8316–8322 (2007).
Dorfman, K. D., King, S. B., Olson, D. W., Thomas, J. D. P. & Tree, D. R. Beyond Gel Electrophoresis: Microfluidic Separations, Fluorescence Burst Analysis and DNA Stretching. Chem. Rev. 113, 2584–2667 (2013).
Han, J. & Craighead, H. G. Entropic trapping and sieving of long DNA molecules in a nanofluidic channel. J. Vac. Sci. Technol. A 17, 2142–2147 (1999).
Han, J. & Craighead, H. G. Characterization and optimization of an entropic trap for DNA separation. Anal. Chem. 74, 394–401 (2002).
Doyle, P. S., Bibette, J., Bancaud, A. & Viovy, J. L. Self-assembled magnetic matrices for DNA separation chips. Science 295, 2237 (2002).
Tabuchi, M. et al. Nanospheres for DNA separation chips. Nat. Biotechnol. 22, 337–340 (2004).
Zeng, Y. & Harrison, D. J. Self-assembled colloidal arrays as three-dimensional nanofluidic sieves for separation of biomolecules on microchips. Anal. Chem. 79, 2289–2295 (2007).
Zeng, Y., He, M. & Harrison, D. J. Microfluidic self-patterning of large-scale crystalline nanoarrays for high-throughput continuous DNA fractionation. Angew. Chem. Int. Ed. 47, 6388–6391 (2008).
Nazemifard, N., Bhattacharjee, S., Masliyah, J. H. & Harrison, D. J. DNA dynamics in nanoscale confinement under asymmetric pulsed field electrophoresis. Angew. Chem. Int. Ed. 49, 3326–3329 (2010).
Klamchuen, A. et al. Study on transport pathway in oxide nanowire growth by using spacing-controlled regular array. Appl. Phys. Lett. 99, 193105 (2011).
Zhuge, F. et al. Fundamental Strategy for Creating VLS Grown TiO2 Single Crystalline Nanowires. J. Phys. Chem. C 116, 24367–24372 (2012).
Meng, G. et al. Pressure-induced evaporation dynamics of gold nanoparticles on oxide substrate. Phys. Rev. E 87, 012405 (2013).
Chueh, Y. L., Chou, L. J. & Wang, Z. L. SiO2/Ta2O5 core-shell nanowires and nanotubes. Angew Chem Int Edit 45, 7773–7778 (2006).
Yasui, T. et al. DNA Manipulation and Separation in Sublithographic-Scale Nanowire Array. ACS Nano 7, 3029–3035 (2013).
Yasui, T. et al. Electroosmotic flow in microchannels with nanostructures. ACS Nano 5, 7775–7780 (2011).
Grossman, P. D., Menchen, S. & Hershey, D. Quantitative-Analysis of DNA-Sequencing Electrophoresis. Genet. Anal-Biomol. E. 9, 9–16 (1992).
Nkodo, A. E. et al. Diffusion coefficient of DNA molecules during free solution electrophoresis. Electrophoresis 22, 2424–2432 (2001).
Pernodet, N., Maaloum, M. & Tinland, B. Pore size of agarose gels by atomic force microscopy. Electrophoresis 18, 55–58 (1997).
Narayanan, J., Xiong, J.-Y. & Liu, X.-Y. Determination of agarose gel pore size: Absorbance measurements vis a vis other techniques. J. Phys. Conf. Ser. 28, 83–86 (2006).
Ogston, A. G. The Spaces in a Uniform Random Suspension of Fibres. T Faraday Soc 54, 1754–1757 (1958).
Barth, S., Harnagea, C., Mathur, S. & Rosei, F. The elastic moduli of oriented tin oxide nanowires. Nanotechnology 20, 115705 (2009).
Kolahi, K. S. et al. Effect of substrate stiffness on early mouse embryo development. PloS one 7, e41717 (2012).
Ou, J., Cho, J., Olson, D. W. & Dorfman, K. D. DNA electrophoresis in a sparse ordered post array. Phys. Rev. E 79, 061904 (2009).
Klamchuen, A. et al. Crucial role of doping dynamics on transport properties of Sb-doped SnO2 nanowires. Appl. Phys. Lett. 95, 053105 (2009).
Acknowledgements
This research is supported by the Cabinet Office, Government of Japan and the Japan Society for the Promotion of Science (JSPS) through the Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program), JSPS Grant-in-Aid for Scientific Research (A) 24241050, JSPS Grant-in-Aid for Young Scientists (B) 25790028 and Nanotechnology Platform Program (Molecule and Material Synthesis) of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Author information
Authors and Affiliations
Contributions
S.R., T. Yasui, T. Yanagida, T.K. and Y.B. designed the experiments. S.R., T. Yasui, T. Yanagida, K.N., M.K., A.K., M.G., H.Y., F.Z. and N.K. performed experiments. S.R., T. Yasui and T. Yanagida analyzed the data. T. Yasui and T. Yanagida wrote the paper. All authors discussed the results and commented on the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Supplementary Information
SR_SIMovie_1
Supplementary Information
SR_SIMovie_2
Supplementary Information
SR_SIMovie_3
Supplementary Information
SR_SIMovie_4
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Rahong, S., Yasui, T., Yanagida, T. et al. Ultrafast and Wide Range Analysis of DNA Molecules Using Rigid Network Structure of Solid Nanowires. Sci Rep 4, 5252 (2014). https://doi.org/10.1038/srep05252
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep05252
This article is cited by
-
Mechanism of rectification of polymer motion in an asymmetric nano-channel
Journal of Nanostructure in Chemistry (2020)
-
Magnetic and Magnetoresistive Properties of 3D Interconnected NiCo Nanowire Networks
Nanoscale Research Letters (2016)
-
Three-dimensional Nanowire Structures for Ultra-Fast Separation of DNA, Protein and RNA Molecules
Scientific Reports (2015)
-
Influence of elution conditions on DNA transport behavior in free solution by hydrodynamic chromatography
Science China Chemistry (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.