Abstract
The vast majority of eukaryotic organisms reproduce sexually, yet the nature of the sexual system and the mechanism of sex determination often vary remarkably, even among closely related species. Some species of animals and plants change sex across their lifespan, some contain hermaphrodites as well as males and females, some determine sex with highly differentiated chromosomes, while others determine sex according to their environment. Testing evolutionary hypotheses regarding the causes and consequences of this diversity requires interspecific data placed in a phylogenetic context. Such comparative studies have been hampered by the lack of accessible data listing sexual systems and sex determination mechanisms across the eukaryotic tree of life. Here, we describe a database developed to facilitate access to sexual system and sex chromosome information, with data on sexual systems from 11,038 plant, 705 fish, 173 amphibian, 593 non-avian reptilian, 195 avian, 479 mammalian, and 11,556 invertebrate species.
Design Type(s) | observation design • species comparison design • data integration |
Measurement Type(s) | phenotypic profiling |
Technology Type(s) | phenotype characterization |
Factor Type(s) | |
Sample Characteristic(s) | Acipenseriformes • Anguilliformes • Anura • Cetartiodactyla • Atheriniformes • Aulopiformes • Beloniformes • Beryciformes • Carcharhiniformes • Carnivora • Caudata • Cetacea • Characiformes • Chelonia • Chiroptera • Clupeiformes • Crocodylia • Cypriniformes • Cyprinodontiformes • Dermoptera • Diprotodontia • Gasterosteiformes • Gobiesociformes • Gymnophiona • Gymnotiformes • Hyracoidea • Insectivora • Lagomorpha • Lamniformes • Marsupialia • Monotremata • Myctophiformes • Myliobatiformes • Osteoglossiformes • Perciformes • Perissodactyla • Petromyzontiformes • Pilosa • Pleuronectiformes • Proboscidea • Primates • Rajiformes • Sphenodontia • Rodentia • Salmoniformes • Scandentia • Scorpaeniformes • Siluriformes • Soricomorpha • Squamata • Squatiniformes • Stomiiformes • Synbranchiformes • Tetraodontiformes • Tubulidentata • Xenarthra • Zeiformes • Acariformes • Blattodea • Coleoptera • Collembola • Dermaptera • Diptera • Embiidina • Ephemeroptera • Grylloblattodea • Hemiptera • Hymenoptera • Isoptera • Lepidoptera • Mantodea • Mecoptera • Megaloptera • Microcoryphia • Neuroptera • Odonata • Orthoptera • Parasitiformes • Phasmatodea • Phthiraptera • Plecoptera • Protura • Psocoptera • Raphidioptera • Siphonaptera • Strepsiptera • Thysanoptera • Trichoptera • Zoraptera • Zygentoma • Apiales • Arecales • Asparagales • Asterales • Austrobaileyales • Brassicales • Caryophyllales • Cucurbitales • Dipsacales • Ericales • Fabales • Gentianales • Lamiales • Laurales • Liliales • Malpighiales • Malvales • Piperales • Poales • Ranunculales • Rhamnales • Rosales • Santalales • Sapindales • Saxifragales • Solanales • Zygophyllales • reproductive system |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Similar content being viewed by others
Background & Summary
Sexual reproduction is a nearly universal feature of eukaryotes, yet a remarkable diversity of sexual systems and sex determining (SD) mechanisms exists. The sexual system of a lineage has important evolutionary and ecological implications, affecting the levels of genetic variation maintained, the degree of inbreeding, the rate of adaptation to novel environments, as well as having longer-term consequences for the formation of new species and the risk of extinction. However, we know little about why and how different sexual systems have evolved. In order to remedy this knowledge gap, the Tree of Sex consortium, a working group of the National Evolutionary Synthesis Center (NESCent), has compiled existing information on sexual systems and sex determination mechanisms, focusing particularly on groups of plants and animals exhibiting variation. In addition to sexual system and mechanism of sex determination (see Tables 1 and 2 for ontology), traits were collated to allow researchers to correlate transitions in mating systems to features of the genome (e.g., chromosome number, ploidy level) and life history (e.g., growth form, life form). To maximize ease of re-use, the data have been deposited in a public repository (see Data Records) and in a trait database custom built by NESCent (TraitDB; http://purl.org/nescent/treeofsex) to which additional data may be uploaded. These data are suitable for in depth comparative analyses of the factors influencing the evolution of sexual systems as well as analyses of the impact of sexual system on, e.g., species ranges, invasiveness, or extinction risk.
To build the database, we surveyed the literature, on-line databases, and expert scientists to obtain species-level descriptions of the traits listed in Tables 1 and 2. For many species, data were obtained from the initial taxonomic description of the species or from subsequent primary literature about the species. While these data are readily available in hard copy in many libraries, it is prohibitively time consuming to track down information species-by-species for use in analyses across broad taxonomic scales. Furthermore, some data are not available in English or require expert interpretation, reducing the usability of the data. For example, botanical knowledge is needed to recognize that plants with ‘perfect flowers’ or ‘monoclinous’ plants are hermaphroditic, with male and female parts contained in the same flowers, or that trioecy and polygamodioecy can both be used to refer to plants with males, females, and hermaphrodites. The Tree of Sex consortium set out to build a database that would make the data accessible and downloadable, using a common ontology describing the traits of interest (Tables 1 and 2).
For some taxa (Coleoptera, Hymenoptera, Diptera, Acari), we have endeavored to make our database comprehensive, representing nearly all known data about the sexual traits compiled. For plants, the database includes 382 genera (348 with data on sexual systems), although we concentrated our data collection on 77 clades (primarily genera), which were known to be variable for sexual system. For these 77 clades, the database has high coverage of the available information, enabling users to address questions about the impact of sexual system on evolutionary and ecological processes. Many genera of plants and invertebrates, however, remain poorly covered in the current database. For vertebrates, our coverage has focused on species with information about the mechanism of sex determination. Figures 1, 2 and 3 provide a summary of data currently available in the database. While the database is not complete, it provides a framework within which additional data can be added by the community. Such a collective effort is needed to fill in details about sexual systems across the Tree of Life.
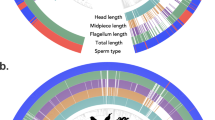
Tree structure is derived from taxonomy, where each tip represents all species in a single genus. Diploid chromosome number is indicated by the height of the innermost ring; all other rings indicate the presence or absence of the trait named at the base of the ring. The ‘Other’ ring includes the states: apomictic, gynomonoecy, andromonoecy, polygamodioecy, and polygamomonoecy. The sexual trait data displayed in the rings is based on 11,038 plant entries.
The XY/ZW ring is colored blue for XY and red for ZW taxa. Complex SCS indicates species with complex sex chromosome karyotypes (e.g., X1X2Y). The sexual trait data displayed in the rings is based on 11,556 invertebrate entries. Remaining features as in Figure 1.
The ‘Other’ ring includes parthenogenesis, gynogenesis, and hybridogenesis. The XY/ZW ring is colored blue for XY and red for ZW taxa. The sexual trait data displayed in the rings is based on 2,145 vertebrate entries. Remaining features as in Figures 1 and 2.
The Tree of Sex database facilitates comparative analyses exploring hypotheses about the evolutionary factors driving transitions among sexual systems. Examples of the type of questions that are being addressed using the database are:
-
Do hermaphrodites diversify more rapidly than species with separate sexes (dioecy)? Information in the database, coupled with phylogenetic information, is being used to determine the impact of dioecy on speciation and extinction rates across multiple genera of plants.
-
Does environmental sex determination place species at heightened risk of extinction, especially in the face of a changing climate? The database allows us to measure the impact of environmental versus genetic sex determination on extinction risk in both turtles and squamates (lizards plus snakes) and to compare extinction risks among groups with different climatic histories.
-
Are some sexual systems more transient than others? By mapping changes in sexual systems to the tree of life, we are assessing whether some transitions are more likely to happen than others (e.g., are ZW systems more likely to transition to XY than vice versa? are gynodioecious species more likely to transition to dioecy than the reverse?).
-
Does the mechanism of sex determination affect genomic evolution? The data are being used to assess which taxa are most likely to undergo fusions between autosomes and sex chromosomes to test ideas about the drivers of fusion events.
-
What factors influence loss of Y chromosomes? In some taxa, Y chromosomes are readily lost, while in others they persist; we have explored the tempo and mode of Y chromosome loss1.
-
Does sociality affect the evolution of chromosome number? Eusocial lifestyles may create selection pressure for increased recombination and indirectly increase the number of chromosomes; we are performing a comparative analysis to test this hypothesis.
By synthesizing existing data on sexual systems, the database will allow biologists to identify the evolutionary and ecological processes that underlie the remarkable diversity in sexual systems across the tree of eukaryotic life. The database will also facilitate rapid identification of suitable taxonomic groups that contain variation in traits of interest for future studies of sexual systems and sex determination.
Methods
Sexual system, karyotype, genome size, ploidy, and life history data were collated from taxonomic accounts, books on flora and fauna, online scientific databases, and the primary scientific literature (sources from the literature were preferred when available). For each species and trait, state values and data sources were entered in the database. In cases where within-species variation in a trait was documented, variants were included in the database, except where noted as ‘rare.’ A notes field in the database for each species allows additional information, such as taxonomic uncertainty or rare variants, to be listed. We avoided using generic information about higher-level taxa (e.g., ‘family X is dioecious’) to inform the trait states of a species, unless the source explicitly listed the species when describing the characteristics of the higher-level taxon.
Vertebrate traits
Data for fish, non-avian reptiles, amphibians, mammals, and birds were compiled from literature searches. Birds appear to be uniform with respect to their sex determination system and were only included if they had specific karyotypic information to distinguish between ZW or complex ZW (e.g., Z1Z2W) systems. Sources included books with karyotype information2–11, online databases12, review papers13,14, and primary research papers. If different values were identified for a particular trait in a given species, multiple entries with the different values are provided for that species.
Invertebrate traits
We performed an extensive literature search using Google Scholar and Web of Science and compiled ~12,000 entries across all orders of hexapods as well as mites. The invertebrate data are drawn from approximately 453 published records including primary research papers, review papers and previous compilations in books. We also incorporated additional data from existing databases (i.e., ScaleNet for scale insects15). For each group in our database we performed literature searches using order and family names in conjunction with the terms: karyotype, cytotaxonomy, cytogenetic, parthenogenesis, haplodiploidy, polyploidy, sex chromosomes and chromosome number. To the extent possible, we reconciled historical karyotype data with currently accepted taxonomy.
Plant traits
The dataset focused on 77 angiosperm clades (primarily at the genus level) exhibiting inter-specific variation in sexual systems to allow species-level analysis of sexual diversity. Genera were chosen from the list containing species with separate sexes compiled by Renner and Ricklefs16, with additional genera from Miller and Venable17, plus genera known to have species with sex chromosomes18, and additional taxa known to the authors to be variable in sexual system. We focused on clades that (a) had at least 15 species, (b) were thought to have at least three dioecious species and three non-dioecious species according to Renner and Ricklefs16, and (c) have sufficient sequence information in GenBank for phylogenetic analyses based on NCBI taxonomy19. Species names were checked against The Plant List (Version 1.1)20. For each of these clades, we gathered data on sexual system, life history, growth form, woodiness and chromosome numbers from four main sources: (1) searching through monographs and local floras (either printed or accessed online via, e.g., eflora.org), (2) detailed search of the primary literature (454 papers), (3) additional online sources (e.g., PLANTS Database21), and (4) consulting with experts with knowledge of the group in question. Finally, we included the extensive information on reproductive traits in parasitic plants from Bellot and Renner22 and on woodiness from Zanne et al.23,24
Data Records
Data record 1
The database files (May 19, 2014 version) in csv format were uploaded to Dryad (Data Citation 1). Separate files are available for the vertebrate, invertebrate, and plant data. Additional notes about the data and pers. comm. files are in this Dryad record as a zip file. Taxonomic information is provided to ensure unique species identity of each record (Order, Family, Genus, species).
Data record 2
The trait data were also uploaded to TraitDB25 within the Tree of Sex project (Data Citation 2). TraitDB is a searchable MySQL-based database, custom built by NESCent, which allows authorized administrators to upload additional data and delete incorrect records. Contributors of additional data should go to treeofsex.org for file configuration information.
Technical Validation
Automated data entries were manually curated to ensure validity. Sub-samples of the records were checked by a different author from the data collector. For the invertebrate and plant data sets, custom scripts were written to identify potentially inconsistent entries (e.g., rows for species with XY karyotypes where the number of chromosomes should be even but was odd, rows specifying that the species is parthenogenetic but describing a male karyotype, species that were said to be both trees and herbs, etc.). All records that appeared to be inconsistent were checked against the original sources to confirm that the entries represent either true trait variation or variation in expert opinion. The on-line database may also be expanded and corrected, as new information becomes available.
Usage Notes
The data are available for download as flat csv files from Dryad (May 19, 2014 version) and from TraitDB. These data may be imported into R26 for use in comparative analyses, e.g., using ape27, diversitree28, GraPhlAn29 (as used to generate Figures 1, 2 and 3), or other packages. The Tree of Sex Consortium places no restrictions on the re-use of the data; we request details of any publications that make substantial use of the database for posting on the website.
Additional information
How to cite this article: The Tree of Sex Consortium. Tree of Sex: A database of sexual systems. Sci. Data 1:140015 doi: 10.1038/sdata.2014.15 (2014).
References
References
Blackmon, H. L. & Demuth, J. P. Estimating tempo and mode of Y chromosome turnover: explaining Y chromosome loss with the fragile Y hypothesis. Genetics 197, 561–572 (2014).
Hsu, T. C. & Benirschke, K. An Atlas of Mammalian Chromosomes Vol. 1 (Springer, 1967).
Hsu, T. C. & Benirschke, K. An Atlas of Mammalian Chromosomes Vol. 2 (Springer, 1968).
Hsu, T. C. & Benirschke, K. An Atlas of Mammalian Chromosomes Vol. 3 (Springer, 1969).
Hsu, T. C. & Benirschke, K. An Atlas of Mammalian Chromosomes Vol. 4 (Springer, 1970).
Hsu, T. C. & Benirschke, K. An Atlas of Mammalian Chromosomes Vol. 5 (Springer, 1971).
Hsu, T. C. & Benirschke, K. An Atlas of Mammalian Chromosomes Vol. 6 (Springer, 1972).
Hsu, T. C. & Benirschke, K. An Atlas of Mammalian Chromosomes Vol. 7 (Springer, 1973).
Hsu, T. C. & Benirschke, K. An Atlas of Mammalian Chromosomes Vol. 8 (Springer, 1974).
Hsu, T. C. & Benirschke, K. An Atlas of Mammalian Chromosomes Vol. 9 (Springer, 1975).
Arai, R. Fish Karyotypes (Springer, 2011).
Olmo, E. & Signorino, G. Chromorep: a reptile chromosomes database, http://chromorep.univpm.it (2005).
Pardo-Manuel de Villena, F. & Sapienza, C. Female meiosis drives karyotype evolution in mammals. Genetics 159, 1179–1189 (2001).
Devlin, R. H. & Nagahama, Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208, 191–364 (2002).
Ben-Dov, Y., Miller, D. R. & Gibson, G. A. P. ScaleNet (http://scalenet.info) (2001).
Renner, S. S. & Ricklefs, R. E. Dioecy and its correlates in the flowering plants. Am. J. Bot. 82, 596–606 (1995).
Miller, J. S. & Venable, D. L. Polyploidy and the evolution of gender dimorphism in plants. Science 289, 2335–2338 (2000).
Ming, R., Bendahmane, A. & Renner, S. S. Sex chromosomes in land plants. Ann. Rev. Plant Biol. 62, 485–514 (2011).
Benson, D. A., et al. GenBank. Nucleic Acids Res. 41, D36–D42 (2013).
The Plant List, v1.1 (http://www.theplantlist.org) (2013).
USDA, NRCS. The PLANTS Database (http://plants.usda.gov) (National Plant Data Team, 2014).
Bellot, S. & Renner, S. S. Pollination and mating systems of Apodanthaceae and the distribution of reproductive traits in parasitic angiosperms. Am. J. Bot. 100, 1083–1094 (2013).
Zanne, A. E. et al. Three keys to the radiation of angiosperms into freezing environments. Nature 506, 89–92 (2014).
Zanne, A. E. et al. Data from: Three keys to the radiation of angiosperms into freezing environments, http://dx.doi.org/10.5061/dryad.63q27.2 (Dryad Digital Repository, 2014b).
Leehr, D., Gosby, M. & Cranston, K. TraitDB: a web application for storing and searching trait data (https://github.com/NESCent/TraitDB) (2014).
R Development Core Team. R: A Language and Environment for Statistical Computing (http://www.R-project.org/) (R foundation for Statistical Computing, 2005).
Paradis, E., Claude, J. & Strimmer, K . APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004).
FitzJohn, R. G. Diversitree: comparative phylogenetic analyses of diversification in R. Methods in Ecology and Evolution 3, 1084–1092 (2012).
Segata, N. GraPhlAn, v0.9 (https://bitbucket.org/nsegata/graphlan) (2012).
Data Citations
The Tree of Sex Consortium Dryad http://dx.doi.org/10.5061/dryad.v1908 (2014)
The Tree of Sex Consortium TreeOfSex in NESCent TraitDB http://purl.org/nescent/treeofsex (2014)
Acknowledgements
We would like to thank the following people for assistance with data collection: Gilad Berman, Marcus Campbell, Anna Johnson, Carlos Larrea, Angelica Lillico-Ouachour, Jason Rae, Nishant Singh, Kohta Yoshida, and Christie Ziegler. We thank Karen Cranston, Mercedes Gosby, Sandra Hall, and Daniel Leehr for developing TraitDB to allow user-designed queries of the database. We are grateful to the following experts in particular taxonomic groups who helped refine the database and/or provide additional data: Rafael Torres Colin, Chris Davies, Bryan Drew, Vladimir Gokhman, Favio Gonzalez, Claes Gustafsson, Peter Heenan, Norbert Holstein, Zhu Hua, Boris Igic, José Luis León de la Luz, Pedro Lorite, Timothy J. Motley, Roy Norton, Rafael Navajas Pérez, Susanne Renner, Jerzy Rzedowski, Stacey Smith, Valerie Souza, Robert Soreng, Daniel Thomas, Henk van der Werff, Amy Zanne. The database development was sponsored by NESCent as part of ‘The Tree of Sex’ working group (NSF #EF-0905606). Funding was also provided by European Research Council Grant #260233 to J.M., National Science Foundation #DEB-1020523 and -1241006 to T.-L.A., #DEB-1120279 to E.E.G., and #MCB-1244355 to N.V., NESCent visiting fellowship to H.B., Marie Curie Reintegration Grant CIG-293878 to I.M., MEXT Grant-in-Aid for Scientific Research on Innovative Areas (#23113007) to J.K., Natural Environment Research Council grant to L.R., and Natural Sciences and Engineering Research Council Discovery grants to S.P.O. and J.C.V.
Author information
Authors and Affiliations
Consortia
Contributions
Working group organizers: D.B., J.E.M., C.L.P. Vertebrate data collectors: J.K., M.K., J.E.M., C.L.P. (fish); N.V. (non-avian reptiles); N.P. (amphibians); J.K. (mammals); J.M. (birds). Invertebrate data collectors: D.B., H.B., M.H., L.R. Plant data collectors: T.-L.A., E.E.G., I.M., R.M., S.P.O., J.C.V. To process and organize the databases, E.E.G., I.M., and S.P.O. wrote custom scripts. S.P.O., C.L.P., H.B., M.W.P., and J.C.V. contributed to the writing of the data descriptor. All authors contributed to the development of the ontology and edited the manuscript. All authors are members of The Tree of Sex Consortium.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
ISA-Tab metadata
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0 Metadata associated with this Data Descriptor is available at http://www.nature.com/sdata/ and is released under the CC0 waiver to maximize reuse.
About this article
Cite this article
The Tree of Sex Consortium. Tree of Sex: A database of sexual systems. Sci Data 1, 140015 (2014). https://doi.org/10.1038/sdata.2014.15
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/sdata.2014.15
This article is cited by
-
HerpSexDet: the herpetological database of sex determination and sex reversal
Scientific Data (2023)
-
Thyroid axis participates in high-temperature-induced male sex reversal through its activation by the stress response
Cellular and Molecular Life Sciences (2023)
-
Identification of sex determination locus in sea cucumber Apostichopus japonicus using genome-wide association study
BMC Genomics (2022)
-
Why not Y naught
Heredity (2022)
-
ROSIE, a database of reptilian offspring sex ratios and sex-determining mechanisms, beginning with Testudines
Scientific Data (2022)