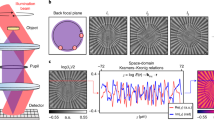
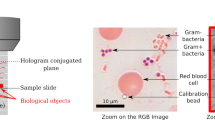
Abstract
An in-line holographic microscope is an optical microscope outfitted with a coherent light source, such as a laser. Light scattered by the specimen interferes with the transmitted beam, and the intensity of that interference pattern constitutes a hologram. Unlike a conventional photograph, a hologram contains information about the phase of the scattered light that is useful for measuring the composition and 3D arrangement of microscopic objects in the specimen. This Primer presents an overview of experimental methods and discusses three recent analysis techniques: fitting scattering models to the hologram; using machine learning to localize and classify the specimen; and a hybrid approach that uses machine learning to initialize fits. The combination of holographic microscopy and model-based analysis is well suited to applications where precise, quantitative results are needed with high acquisition speed. Such applications include studying properties of heterogeneous colloidal dispersions, measuring colloidal interactions, monitoring stresses in soft materials, detecting molecular binding and aggregation, and following the motion of microorganisms in three dimensions. We discuss the reproducibility and current limitations of each method. Finally, we anticipate directions for future development and provide an outlook on the integration between experiment and computational analysis, an emerging paradigm for microscopy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 1 digital issues and online access to articles
$99.00 per year
only $99.00 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sheng, J., Malkiel, E. & Katz, J. Digital holographic microscope for measuring three-dimensional particle distributions and motions. Appl. Opt. 45, 3893–3901 (2006).
Gabor, D. A new microscopic principle. Nature 161, 777–778 (1948).
Gabor, D. & Bragg, W. L. Microscopy by reconstructed wave-fronts. P. Roy. Soc. Lond. A Mat. 197, 454–487 (1949). Together with Gabor (1948), this paper demonstrates that it is possible to optically reconstruct a 3D representation of a specimen from its recorded hologram, a finding that launched the field of holographic microscopy.
Xu, W., Jericho, M. H., Meinertzhagen, I. A. & Kreuzer, H. J. Digital in-line holography for biological applications. Proc. Natl Acad. Sci. USA 98, 11301–11305 (2001).
Xu, W., Jericho, M. H., Kreuzer, H. J. & Meinertzhagen, I. A. Tracking particles in four dimensions with in-line holographic microscopy. Opt. Lett. 28, 164–166 (2003).
Berg, M. J. Tutorial: Aerosol characterization with digital in-line holography. J. Aerosol Sci. 165, 106023 (2022).
Kim, M. K. Principles and techniques of digital holographic microscopy. SPIE Rev. 1, 018005 (2010).
Jericho, S. K., Garcia-Sucerquia, J., Xu, W., Jericho, M. H. & Kreuzer, H. J. Submersible digital in-line holographic microscope. Rev. Sci. Instrum. 77, 043706 (2006).
Garcia-Sucerquia, J. et al. Digital in-line holographic microscopy. Appl. Opt. 45, 836–850 (2006).
Bishara, W., Zhu, H. & Ozcan, A. Holographic opto-fluidic microscopy. Opt. Express 18, 27499–27510 (2010).
Marquet, P. et al. Digital holographic microscopy: a noninvasive contrast imaging technique allowing quantitative visualization of living cells with subwavelength axial accuracy. Opt. Lett. 30, 468–470 (2005).
Mölder, A. et al. Non-invasive, label-free cell counting and quantitative analysis of adherent cells using digital holography. J. Microsc. 232, 240–247 (2008).
Kemper, B. & Bally, G. V. Digital holographic microscopy for live cell applications and technical inspection. Appl. Opt. 47, A52–A61 (2008).
Park, Y., Depeursinge, C. & Popescu, G. Quantitative phase imaging in biomedicine. Nat. Photonics 12, 578–589 (2018).
Barty, A., Nugent, K. A., Roberts, A. & Paganin, D. Quantitative phase tomography. Opt. Comm. 175, 329–336 (2000).
Popescu, G. Quantitative Phase Imaging of Cells and Tissues (McGraw-Hill Education, 2011).
Popescu, G. et al. Imaging red blood cell dynamics by quantitative phase microscopy. Blood Cell Mol. Dis. 41, 10–16 (2008).
Marquet, P., Depeursinge, C. & Magistretti, P. J. Review of quantitative phase-digital holographic microscopy: promising novel imaging technique to resolve neuronal network activity and identify cellular biomarkers of psychiatric disorders. Neurophotonics 1, 020901 (2014).
Mie, G. Beiträge zur Optik trüber Medien, speziell kolloidaler Metallösungen [German]. Ann. Phys. 330, 377–445 (1908).
Ovryn, B. & Izen, S. H. Imaging of transparent spheres through a planar interface using a high-numerical-aperture optical microscope. JOSA 17, 1202–1213 (2000). The authors fit a generative model based on Lorenz–Mie theory to a recorded hologram to determine the properties of a microscopic particle.
Lee, S.-H. et al. Characterizing and tracking single colloidal particles with video holographic microscopy. Opt. Express 15, 18275–18282 (2007). This paper presents a straightforward generative model for hologram formation from a simple sphere, which has become the basis for many later studies on various systems.
Wang, A., Rogers, W. B. & Manoharan, V. N. Effects of contact-line pinning on the adsorption of nonspherical colloids at liquid interfaces. Phys. Rev. Lett. 119, 108004 (2017).
Wang, A. et al. Using the discrete dipole approximation and holographic microscopy to measure rotational dynamics of non-spherical colloidal particles. J. Quant. Spectrosc. Radiat. Transf. 146, 499–509 (2014).
Fung, J. et al. Measuring translational, rotational, and vibrational dynamics in colloids with digital holographic microscopy. Opt. Express 19, 8051 (2011).
Yurkin, M. A. & Hoekstra, A. G. The discrete dipole approximation: an overview and recent developments. J. Quant. Spectrosc. Radiat. Transf. 106, 558–589 (2007).
Yurkin, M. A. & Hoekstra, A. G. The discrete-dipole-approximation code ADDA: capabilities and known limitations. J. Quant. Spectrosc. Radiat. Transf. 112, 2234–2247 (2011).
Pu, Y. & Meng, H. Intrinsic aberrations due to Mie scattering in particle holography. J. Opt. Soc. Am. A 20, 1920 (2003).
Dulin, D., Barland, S., Hachair, X. & Pedaci, F. Efficient illumination for microsecond tracking microscopy. PLoS ONE 9, e107335 (2014).
Giuliano, C. B., Zhang, R. & Wilson, L. G. Digital inline holographic microscopy (DIHM) of weakly-scattering subjects. J. Vis. Exp. 84, e50488 (2014).
Kanka, M., Riesenberg, R., Petruck, P. & Graulig, C. High resolution (NA = 0.8) in lensless in-line holographic microscopy with glass sample carriers. Opt. Lett. 36, 3651–3653 (2011).
Garcia-Sucerquia, J. Noise reduction in digital lensless holographic microscopy by engineering the light from a light-emitting diode. Appl. Opt. 52, A232–A239 (2013).
Hell, S., Reiner, G., Cremer, C. & Stelzer, E. H. K. Aberrations in confocal fluorescence microscopy induced by mismatches in refractive index. J. Microsc. 169, 391–405 (1993).
Wu, Y. & Ozcan, A. Lensless digital holographic microscopy and its applications in biomedicine and environmental monitoring. Methods 136, 4–16 (2018).
Deng, N.-N. et al. Simple and cheap microfluidic devices for the preparation of monodisperse emulsions. Lab. Chip 11, 3963–3969 (2011).
Kaz, D. M., McGorty, R., Mani, M., Brenner, M. P. & Manoharan, V. N. Physical ageing of the contact line on colloidal particles at liquid interfaces. Nat. Mater. 11, 138–142 (2012). This application of a generative modelling approach demonstrates the usefulness of the method: the fast, precise measurements enabled by the approach reveal a previously indiscernible phenomenon.
Moyses, H. W., Krishnatreya, B. J. & Grier, D. G. Robustness of Lorenz–Mie microscopy against defects in illumination. Opt. Express 21, 5968 (2013).
Martin, C., Leahy, B. & Manoharan, V. N. Improving holographic particle characterization by modeling spherical aberration. Opt. Express 29, 18212 (2021).
Fung, J., Perry, R. W., Dimiduk, T. G. & Manoharan, V. N. Imaging multiple colloidal particles by fitting electromagnetic scattering solutions to digital holograms. J. Quant. Spectrosc. Radiat. Transf. 113, 2482–2489 (2012).
Cheong, F. C. et al. Flow visualization and flow cytometry with holographic video microscopy. Opt. Express 17, 13071 (2009).
Dixon, L., Cheong, F. C. & Grier, D. G. Holographic particle-streak velocimetry. Opt. Express 19, 4393–4398 (2011).
Edelstein, A. D. et al. Advanced methods of microscope control using μManager software. J. Biol. Methods 1, e10 (2014).
Vercruysse, D. et al. Three-part differential of unlabeled leukocytes with a compact lens-free imaging flow cytometer. Lab Chip 15, 1123–1132 (2015).
Dimiduk, T. G. et al. A simple, inexpensive holographic microscope. in Biomedical Optics and 3-D Imaging, OSA Technical Digest (CD) JMA38 (Optica, 2010).
Fung, J. Measuring the 3D Dynamics of Multiple Colloidal Particles with Digital Holographic Microscopy. PhD Thesis, Harvard Univ. (2013).
Moreno, D., Santoyo, F. M., Guerrero, J. A. & Funes-Gallanzi, M. Particle positioning from charge-coupled device images by the generalized Lorenz–Mie theory and comparison with experiment. Appl. Opt. 39, 5117–5124 (2000).
Denis, L., Fournier, C., Fournel, T., Ducottet, C. & Jeulin, D. Direct extraction of the mean particle size from a digital hologram. Appl. Opt. 45, 944–952 (2006).
Guerrero-Viramontes, J. A., Moreno-Hernández, D., Mendoza-Santoyo, F. & Funes-Gallanzi, M. 3D particle positioning from CCD images using the generalized Lorenz–Mie and Huygens–Fresnel theories. Meas. Sci. Technol. 17, 2328–2334 (2006).
Yevick, A., Hannel, M. & Grier, D. G. Machine-learning approach to holographic particle characterization. Opt. Express 22, 26884 (2014). This paper is one of the first applications of machine learning to hologram analysis, and demonstrates the increase in speed of analysis that is possible.
Hannel, M. D., Abdulali, A., O’Brien, M. & Grier, D. G. Machine-learning techniques for fast and accurate feature localization in holograms of colloidal particles. Opt. Express 26, 15221 (2018).
Altman, L. E. & Grier, D. G. CATCH: characterizing and tracking colloids holographically using deep neural networks. J. Phys. Chem. B 124, 1602–1610 (2020). This paper demonstrates a fully integrated pipeline for analysis of holograms, with improved automation and precision made possible by combining machine learning with fitting.
Hannel, M., Middleton, C. & Grier, D. G. Holographic characterization of imperfect colloidal spheres. Appl. Phys. Lett. 107, 141905 (2015).
Duda, R. O. & Hart, P. E. Use of the Hough transformation to detect lines and curves in pictures. Commun. ACM 15, 11–15 (1972).
Ballard, D. H. Generalizing the Hough transform to detect arbitrary shapes. Pattern Recogn. 13, 111–122 (1981).
Dimiduk, T. G., Perry, R. W., Fung, J. & Manoharan, V. N. Random-subset fitting of digital holograms for fast three-dimensional particle tracking [invited]. Appl. Opt. 53, G177–G183 (2014).
Dimiduk, T. G. & Manoharan, V. N. Bayesian approach to analyzing holograms of colloidal particles. Opt. Express 24, 24045 (2016). This work demonstrates the use of a Bayesian inference framework for hologram analysis, which has lent several advantages over non-linear least-squares fitting routines, including the formal integration of prior information and MCMC calculation of the posterior over parameters.
Moré, J. J. in Numerical Analysis (ed. Watson, G. A.) 105–116 (Springer, 1978).
Cheong, F. C., Krishnatreya, B. J. & Grier, D. G. Strategies for three-dimensional particle tracking with holographic video microscopy. Opt. Express 18, 13563 (2010).
Krishnatreya, B. J. et al. Measuring Boltzmann’s constant through holographic video microscopy of a single colloidal sphere. Am. J. Phys. 82, 23–31 (2014).
Wang, A., McGorty, R., Kaz, D. M. & Manoharan, V. N. Contact-line pinning controls how quickly colloidal particles equilibrate with liquid interfaces. Soft Matter 12, 8958–8967 (2016).
Wang, A. et al. Before the breach: interactions between colloidal particles and liquid interfaces at nanoscale separations. Phys. Rev. E 100, 042605 (2019).
Roichman, Y., Sun, B., Stolarski, A. & Grier, D. G. Influence of nonconservative optical forces on the dynamics of optically trapped colloidal spheres: the fountain of probability. Phys. Rev. Lett. 101, 128301 (2008).
Sun, B., Lin, J., Darby, E., Grosberg, A. Y. & Grier, D. G. Brownian vortexes. Phys. Rev. E 80, 010401 (2009).
O’Brien, M. J. & Grier, D. G. Above and beyond: holographic tracking of axial displacements in holographic optical tweezers. Opt. Express 27, 25375 (2019).
Xiao, K. & Grier, D. G. Sorting colloidal particles into multiple channels with optical forces: prismatic optical fractionation. Phys. Rev. E 82, 051407 (2010).
Xiao, K. & Grier, D. G. Multidimensional optical fractionation of colloidal particles with holographic verification. Phys. Rev. Lett. 104, 028302 (2010).
Winters, A. et al. Quantitative differentiation of protein aggregates from other subvisible particles in viscous mixtures through holographic characterization. J. Pharm. Sci. 109, 2405–2412 (2020).
Wang, C., Shpaisman, H., Hollingsworth, A. D. & Grier, D. G. Celebrating soft matter’s 10th anniversary: monitoring colloidal growth with holographic microscopy. Soft Matter 11, 1062–1066 (2015).
Shpaisman, H., Jyoti Krishnatreya, B. & Grier, D. G. Holographic microrefractometer. Appl. Phys. Lett. 101, 091102 (2012).
Cheong, F. C., Duarte, S., Lee, S.-H. & Grier, D. G. Holographic microrheology of polysaccharides from Streptococcus mutans biofilms. Rheol. Acta 48, 109–115 (2009).
Wang, C. et al. Holographic characterization of protein aggregates. J. Pharm. Sci. 105, 1074–1085 (2016).
Fung, J. & Hoang, S. Computational assessment of an effective-sphere model for characterizing colloidal fractal aggregates with holographic microscopy. J. Quant. Spectrosc. Radiat. Transf. 236, 106591 (2019). This work demonstrates the range of validity of the effective-sphere model in hologram analysis, used widely in industrial applications.
Wang, C. et al. Holographic characterization of colloidal fractal aggregates. Soft Matter 12, 8774–8780 (2016).
Altman, L. E., Quddus, R., Cheong, F. C. & Grier, D. G. Holographic characterization and tracking of colloidal dimers in the effective-sphere approximation. Soft Matter 17, 2695–2703 (2021).
Philips, L. A. et al. Holographic characterization of contaminants in water: differentiation of suspended particles in heterogeneous dispersions. Water Res. 122, 431–439 (2017).
Cheong, F. C. et al. Holographic characterization of colloidal particles in turbid media. Appl. Phys. Lett. 111, 153702 (2017).
Mackowski, D. W. & Mishchenko, M. I. Calculation of the T matrix and the scattering matrix for ensembles of spheres. J. Opt. Soc. Am. A 13, 2266–2278 (1996).
Leahy, B. et al. Large depth-of-field tracking of colloidal spheres in holographic microscopy by modeling the objective lens. Opt. Express 28, 1061–1075 (2020).
Alexander, R., Leahy, B. & Manoharan, V. N. Precise measurements in digital holographic microscopy by modeling the optical train. J. Appl. Phys. 128, 060902 (2020). This review highlights the historic development of the generative modelling approach to holograms (the only review to our knowledge that does so) and discusses the current abilities and limitations of existing generative models.
Geyer, C. J. in Handbook of Markov Chain Monte Carlo (eds Brooks, S., Gelman, A., Jones, G. L. & Meng, X.-L.) 3–48 (Chapman & Hall/CRC, 2011).
Hansen, N. & Ostermeier, A. Adapting arbitrary normal mutation distributions in evolution strategies: the covariance matrix adaptation. in Proc. IEEE Int. Conf. Evolutionary Computation 312–317 https://doi.org/10.1109/ICEC.1996.542381 (1996).
Neal, R. M. in Handbook of Markov Chain Monte Carlo (eds. Brooks, S., Gelman, A, Jones, G L, & Meng, X L) 113–162 (Chapman & Hall/CRC Handbooks of Modern Statistical Methods, 2011).
Earl, D. J. & Deem, M. W. Parallel tempering: theory, applications, and new perspectives. Phys. Chem. Chem. Phys. 7, 3910–3916 (2005).
Barkley, S. et al. Holographic microscopy with Python and HoloPy. Comput. Sci. Eng. 22, 72–82 (2019).
Crocker, J. & Grier, D. Methods of digital video microscopy for colloidal studies. J. Colloid Interf. Sci. 179, 298–310 (1996).
Krishnatreya, B. J. & Grier, D. G. Fast feature identification for holographic tracking: the orientation alignment transform. Opt. Express 22, 12773 (2014).
Parthasarathy, R. Rapid, accurate particle tracking by calculation of radial symmetry centers. Nat. Methods 9, 724–726 (2012).
Rotskoff, G. M. & Vanden-Eijnden, E. Trainability and accuracy of neural networks: an interacting particle system approach. Preprint at https://doi.org/10.48550/arXiv.1805.00915 (2018).
Newby, J. M., Schaefer, A. M., Lee, P. T., Forest, M. G. & Lai, S. K. Convolutional neural networks automate detection for tracking of submicron-scale particles in 2D and 3D. Proc. Natl Acad. Sci. USA 115, 9026–9031 (2018).
Schneider, B., Dambre, J. & Bienstman, P. Fast particle characterization using digital holography and neural networks. Appl. Opt. 55, 133 (2016).
Klein, E. Structure and Dynamics of Colloidal Clusters. PhD Thesis, Harvard Univ. (2019).
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. Preprint at https://doi.org/10.48550/arXiv.1412.6980 (2014).
Bottou, L. in Proc. COMPSTAT’2010 (eds Lechevallier, Y. & Saporta, G.) 177–186 (Physica-Verlag HD, 2010).
Glorot, X., Bordes, A. & Bengio, Y. in Proc. Fourteenth Int. Conf. Artificial Intelligence and Statistics Vol. 15 (eds Gordon, G., Dunson, D. & Dudík, M.) 315–323 (PMLR, 2011).
Redmon, J. & Farhadi, A. YOLOv3: an incremental improvement. Preprint at https://doi.org/10.48550/arXiv.1804.02767 (2018).
Meng, G., Arkus, N., Brenner, M. P. & Manoharan, V. N. The free-energy landscape of clusters of attractive hard spheres. Science 327, 560–563 (2010).
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the inception architecture for computer vision. in Proc. IEEE Conf. Computer Vision and Pattern Recognition (CVPR) 2818–2826 (IEEE, 2016).
Pickering, S. U. Emulsions. J. Chem. Soc. Trans. 91, 2001–2021 (1907).
Xiao, J., Li, Y. & Huang, Q. Recent advances on food-grade particles stabilized Pickering emulsions: fabrication, characterization and research trends. Trends Food Sci. Tech. 55, 48–60 (2016).
Yoon, K. Y. et al. Core flooding of complex nanoscale colloidal dispersions for enhanced oil recovery by in situ formation of stable oil-in-water Pickering emulsions. Energ. Fuels 30, 2628–2635 (2016).
Bhargava, A., Francis, A. V. & Biswas, A. K. Interfacial studies related to the recovery of mineral slimes in a water–hydrocarbon liquid-collector system. J. Colloid Interf. Sci. 64, 214–227 (1978).
Aveyard, R., Binks, B. P. & Clint, J. H. Emulsions stabilised solely by colloidal particles. Adv. Colloid Interfac. 100–102, 503–546 (2003).
Dinsmore, A. D. et al. Colloidosomes: selectively permeable capsules composed of colloidal particles. Science 298, 1006–1009 (2002).
Rahmani, A. M., Wang, A., Manoharan, V. N. & Colosqui, C. E. Colloidal particle adsorption at liquid interfaces: capillary driven dynamics and thermally activated kinetics. Soft Matter 12, 6365–6372 (2016).
Fung, J. & Manoharan, V. N. Holographic measurements of anisotropic three-dimensional diffusion of colloidal clusters. Phys. Rev. E 88, 020302 (2013).
Perry, R. W., Meng, G., Dimiduk, T. G., Fung, J. & Manoharan, V. N. Real-space studies of the structure and dynamics of self-assembled colloidal clusters. Faraday Discuss. 159, 211–234 (2013).
Zia, R. N. Active and passive microrheology: theory and simulation. Annu. Rev. Fluid Mech. 50, 371–405 (2018).
Style, R. W. et al. Traction force microscopy in physics and biology. Soft Matter 10, 4047–4055 (2014).
Cheong, F. C. & Grier, D. G. Rotational and translational diffusion of copper oxide nanorods measured with holographic video microscopy. Opt. Express 18, 6555 (2010).
Makarchuk, S., Beyer, N., Gaiddon, C., Grange, W. & Hébraud, P. Holographic traction force microscopy. Sci. Rep. 8, 3038 (2018).
Moerner, W. E. & Fromm, D. P. Methods of single-molecule fluorescence spectroscopy and microscopy. Rev. Sci. Instrum. 74, 3597–3619 (2003).
Steelman, Z. A., Eldridge, W. J., Weintraub, J. B. & Wax, A. Is the nuclear refractive index lower than cytoplasm? Validation of phase measurements and implications for light scattering technologies. J. Biophotonics 10, 1714–1722 (2017).
Liu, P. Y. et al. Real-time measurement of single bacterium’s refractive index using optofluidic immersion refractometry. Procedia Eng. 87, 356–359 (2014).
Molaei, M. & Sheng, J. Imaging bacterial 3D motion using digital in-line holographic microscopy and correlation-based de-noising algorithm. Opt. Express 22, 32119 (2014).
Wang, A., Garmann, R. F. & Manoharan, V. N. Tracking E. coli runs and tumbles with scattering solutions and digital holographic microscopy. Opt. Express 24, 23719–23725 (2016).
Bozzuto, G. & Molinari, A. Liposomes as nanomedical devices. Int. J. Nanomed. 10, 975–999 (2015).
Deamer, D. The role of lipid membranes in life’s origin. Life 7, 5 (2017).
Schwille, P. & Diez, S. Synthetic biology of minimal systems. Crit. Rev. Biochem. Mol. 44, 223–242 (2009).
Spustova, K., Köksal, E. S., Ainla, A. & Gözen, I. Subcompartmentalization and pseudo-division of model protocells. Small 17, 2005320 (2021).
Wang, A., Chan Miller, C. & Szostak, J. W. Core-shell modeling of light scattering by vesicles: effect of size, contents, and lamellarity. Biophys. J. 116, 659–669 (2019).
Tran, L. H. A. et al. Measuring vesicle loading with holographic microscopy. Preprint at https://doi.org/10.48550/arXiv.2204.13068 (2022).
Quinn, M. K. et al. How fluorescent labelling alters the solution behaviour of proteins. Phys. Chem. Chem. Phys. 17, 31177–31187 (2015).
Hughes, L. D., Rawle, R. J. & Boxer, S. G. Choose your label wisely: water-soluble fluorophores often interact with lipid bilayers. PLoS ONE 9, e87649 (2014).
Markel, V. Introduction to the Maxwell Garnett approximation: tutorial. J. Opt. Soc. Am. A 33, 1244–1256 (2016).
Zagzag, Y., Soddu, M. F., Hollingsworth, A. D. & Grier, D. G. Holographic molecular binding assays. Sci. Rep. 10, 1932 (2020).
Altman, L. E. & Grier, D. G. Interpreting holographic molecular binding assays with effective medium theory. Biomed. Opt. Express 11, 5225 (2020).
Snyder, K., Quddus, R., Hollingsworth, A. D., Kirshenbaum, K. & Grier, D. G. Holographic immunoassays: direct detection of antibodies binding to colloidal spheres. Soft Matter 16, 10180–10186 (2020).
Draine, B. T. The discrete-dipole approximation and its application to interstellar graphite grains. Astrophys. J. 333, 848–872 (1988).
Ruffner, D. B., Cheong, F. C., Blusewicz, J. M. & Philips, L. A. Lifting degeneracy in holographic characterization of colloidal particles using multi-color imaging. Opt. Express 26, 13239–13251 (2018).
Rawat, S., Wendoloski, J. & Wang, A. cGAN-assisted imaging through stationary scattering media. Opt. Express 30, 18145–18155 (2022).
Abadi, M. et al. TensorFlow: large-scale machine learning on heterogeneous distributed systems. Preprint at https://doi.org/10.48550/arXiv.1603.04467 (2015).
Bradbury, J. et al. JAX: composable transformations of Python + NumPy programs. GitHub http://github.com/google/jax (2018).
Kucukelbir, A., Tran, D., Ranganath, R., Gelman, A. & Blei, D. M. Automatic differentiation variational inference. J. Mach. Learn. Res. 18, 1–45 (2017).
Jouppi, N. P. et al. In-datacenter performance analysis of a Tensor Processing Unit. in Proc. 44th Annual Int. Symp. Computer Architecture 1–12 (Association for Computing Machinery, 2017).
Leith, E. N., Upatnieks, J. & Haines, K. A. Microscopy by wavefront reconstruction. J. Opt. Soc. Am. 55, 981–986 (1965).
Schnars, U. & Jüptner, W. Direct recording of holograms by a CCD target and numerical reconstruction. Appl. Opt. 33, 179–181 (1994). This paper represents another major development in the field of holographic microscopy: the application of the digital camera, which allows holograms to be reconstructed numerically rather than optically.
Hickling, R. Holography of liquid droplets. J. Opt. Soc. Am. 59, 1334–1339 (1969).
Slimani, F., Grehan, G., Gouesbet, G. & Allano, D. Near-field Lorenz–Mie theory and its application to microholography. Appl. Opt. 23, 4140–4148 (1984).
Trujillo, C., Castañeda, R., Piedrahita-Quintero, P. & Garcia-Sucerquia, J. Automatic full compensation of quantitative phase imaging in off-axis digital holographic microscopy. Appl. Opt. 55, 10299–10306 (2016).
Popescu, G. et al. Fourier phase microscopy for investigation of biological structures and dynamics. Opt. Lett. 29, 2503–2505 (2004).
Joo, C., Akkin, T., Cense, B., Park, B. H. & de. Boer, J. F. Spectral-domain optical coherence phase microscopy for quantitative phase-contrast imaging. Opt. Lett. 30, 2131–2133 (2005).
Piliarik, M. & Sandoghdar, V. Direct optical sensing of single unlabelled proteins and super-resolution imaging of their binding sites. Nat. Commun. 5, 1–8 (2014).
Young, G. et al. Quantitative mass imaging of single biological macromolecules. Science 360, 423–427 (2018).
Mahmoodabadi, R. G. et al. Point spread function in interferometric scattering microscopy (iSCAT). Part I: aberrations in defocusing and axial localization. Opt. Express 28, 25969–25988 (2020).
Acknowledgements
Work at Harvard is supported by the National Science Foundation under grant DMR-2011754 and by the Department of Defense through the National Defense Science & Engineering Graduate Fellowship Program. Work at UNSW Sydney was supported by the Human Frontier of Science Program Grant (RPG0029/2020 to A.W.), and A.W. was supported by the Australian Research Council Discovery Early Career Award (DE210100291). Work at NYU was supported by the National Science Foundation under grants DMR-2104837, DMR-1420073 and DMR-0922680, and by the National Institutes of Health under grant R44TR001590.
Author information
Authors and Affiliations
Contributions
Introduction (C.M., L.E.A., S.R., A.W., D.G.G. and V.N.M.); Experimentation (C.M., L.E.A., S.R., A.W., D.G.G. and V.N.M.); Results (C.M., L.E.A., S.R., A.W., D.G.G. and V.N.M.); Applications (C.M., L.E.A., S.R., A.W., D.G.G. and V.N.M.); Reproducibility and data deposition (C.M., L.E.A., S.R., A.W., D.G.G. and V.N.M.); Limitations and optimizations (C.M., L.E.A., S.R., A.W., D.G.G. and V.N.M.); Outlook (C.M., L.E.A., S.R., A.W., D.G.G. and V.N.M.); Overview of the Primer (C.M., L.E.A., S.R., A.W., D.G.G. and V.N.M.).
Corresponding author
Ethics declarations
Competing interests
D.G.G. is a founder of Spheryx, Inc., which manufactures instruments for holographic particle characterization. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Methods Primers thanks Radim Chmelik; Laurence Wilson, who co-reviewed with Sam Matthews; and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
CATCH: https://github.com/laltman2/CATCH
HDF5: https://www.hdfgroup.org/solutions/hdf5/
HoloPy: http://holopy.readthedocs.io/
pylorenzmie: https://github.com/davidgrier/pylorenzmie
Glossary
- Holograms
-
2D intensity patterns resulting from the interference between light scattered from an object and a reference beam.
- Fringes
-
The bright or dark bands in an image that are produced by the interference of light.
- Holographic microscopy
-
The use of a microscope with a coherent or semi-coherent light source to record holograms of microscopic objects.
- Reconstruction
-
The process of illuminating a hologram with a beam such that the hologram acts as a diffraction grating.
- Tomograms
-
Images recorded by a penetrating wave that represents a cross section of a 3D object.
- Colloidal particles
-
Nanoparticles or microparticles suspended in a fluid or other medium.
- Scattering
-
The interaction of electromagnetic radiation with an object resulting in a change in the direction of the light.
- Coherent
-
Light that has a narrow distribution of frequencies and a well-defined phase, such that interference can be observed.
- Collimated
-
A beam of light with parallel rays.
- Objective lens
-
A lens or collection of lenses that focus and magnify light to form an image.
- Tube lens
-
A lens or series of lenses that focus parallel rays to form an image on a sensor or eyepiece.
- Speckle
-
Fluctuating bright and dark regions in an image that arise from extraneous scattering and interference.
- Spherical aberration
-
A type of optical aberration in which rays nearer the edge of a lens are deflected more than those near its axis.
- Capillary action
-
Flow driven by interfacial tension.
- Syringe pumps
-
Volumetrically controlled pumps that deliver fluid by moving a syringe piston, typically resulting in a constant flow rate.
- Pressure pumps
-
Pumps that are pressure-driven and have controllers to maintain constant pressure.
- Dynamic range
-
The range of intensities that a sensor can record.
- Quantum efficiency
-
A measure of the sensitivity of a detector, determined by how many incident photons are converted into electrons.
- Dark count
-
The intensity recorded by a sensor or camera in the absence of a signal.
- Bayesian parameter estimation
-
A statistical inference technique yielding the probability distribution of the parameters of a model given the data.
- Marginalized uncertainties
-
The uncertainties in a model parameter determined by accounting for correlations with other parameters.
- Markov-chain Monte Carlo
-
(MCMC). A numerical method that uses a biased random walk through the parameter space to both estimate a probability distribution and integrate it.
- Convolutional neural networks
-
(CNNs). Machine-learning algorithms that use convolution layers to process higher-dimensional image data.
- Support vector machines
-
Machine-learning algorithms that distinguish data points using hyperplanes in a high-dimensional parameter space.
- ReLU
-
The rectified linear activation function, a piecewise function that returns zero for negative inputs and returns the input for positive inputs.
- Brownian motion
-
The random motion of particles suspended in a medium due to collisions with the surrounding molecules.
- Polydispersity
-
The distribution of sizes within a sample.
- Graticule
-
A set of parallel lines with known spacing used for measuring scale.
- Lossy encoding
-
Methods of compressing or transferring data that approximate or down-sample the data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Martin, C., Altman, L.E., Rawat, S. et al. In-line holographic microscopy with model-based analysis. Nat Rev Methods Primers 2, 83 (2022). https://doi.org/10.1038/s43586-022-00165-z
Accepted:
Published:
DOI: https://doi.org/10.1038/s43586-022-00165-z