Abstract
We aimed to investigate the expression and clinic significance of Rac GTPase Activating Protein 1 (RACGAP1) in human lung adenocarcinoma (LUAD). Online database analysis revealed a significant increase in RACGAP1 mRNA expression among 26 types of tumor tissues, including LUAD tissues. Online database and tissue microarray analyses indicated that RACGAP1 expression was significantly upregulated in LUAD tissues. Genetic variation analysis identified four different genetic variations of RACGAPs in LUAD. Moreover, online database analysis showed that RACGAP1 upregulation was correlated with shorter survival in patients with LUAD. After silencing RACGAP1 expression in A549 cells using siRNA and assessing its protein levels via Western blotting, we found that RACGAP1 knockdown inhibited cell growth and induced apoptosis determined using the Cell Counting Kit-8 assay, colony formation assay, and flow cytometry. Mechanistically, western blot analysis indicated that Bax expression increased, whereas Bcl-2 expression decreased. Moreover, RACGAP1 knockdown attenuated PI3K/AKT pathway activation in lung cancer cells. Taken together, our findings showed that RACGAP1 was overexpressed in LUAD tissues and played an important role in lung cancer by increasing cell growth through the PI3K/AKT signaling pathway. This study suggests recommends evaluating RACGAP1 in clinical settings as a novel biomarker and potential therapeutic target for lung cancer.
Similar content being viewed by others
Introduction
Lung cancer, including non-small cell adenocarcinoma, squamous cell lung carcinoma, and large cell carcinoma, is the most common type of cancer among men and the third most common type of cancer among women1,2, with lung adenocarcinoma (LUAD) being the most common type of lung cancer. Based on 2020 global cancer statistics, lung cancer still remains the leading cause of cancer death in not only low- and middle-income countries but also most high-income countries3. The treatment of lung cancer depends on the cancer type and stage; however, most patients with lung cancer are diagnosed with advanced-stage disease, and molecular targeted therapies are only limited to specific patients4. Therefore, further exploration of molecular targets are essential for uncovering more innovative diagnostic, preventive, and treatment strategies.
Rac GTPase-activating protein 1 (RACGAP1), also called MgcRACGAP, had been first detected in 1998 from human testis and germ cell tumor extracts and plays an important role in the Rho GTPase activation cycle by regulating Rho GTPase transformation and GTP hydrolysis stimulation5. Studies have shown that RACGAP1 is involved in cytokinesis, induces cell proliferation, and might be related to the well-known proliferative marker Ki676,7,8,9. Moreover, RACGAP1 has been found to play a role in cell transformation, migration, and metastasis10,11,12,13. These reported functions have attracted researcher attention to its potential role in malignancies14. Consequently, an increasing number of studies have reported an the association between RACGAP1 overexpression in several types of cancers, including stomach15, breast16,17, colorectal18, bladder19, liver20, and uterine cancers21, indicating its possible role in tumor proliferation, migration, and recurrence. However, no detailed evidence has been available on the relationship between RACGAP1 and lung cancer diagnosis and prognosis, except for its upregulation in lung cancer22,23.
The present study used bioinformatics analysis of online databases and tissue microarray (TMA) to evaluate the expression profile of RACGAP1 and the relationship between RACGAP1 expression and clinical pathological parameters in lung cancer. Moreover, a series of experiments using representative lung cancer cell line were conducted to examine the functional role of RACGAP1.
Results
Upregulation of RACGAP1 in LUAD
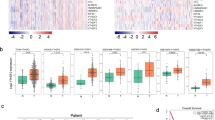
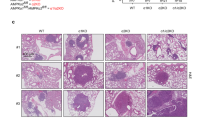
We compared RACGAP1 levels between various types of tumors and adjacent normal tissues using Gene Expression Profiling Interactive Analysis (GEPIA) databases. Our findings showed that although RACGAP1 mRNA expression was downregulated in acute myeloid leukemia, it was dramatically upregulated in most of tumors, including LUAD and lung squamous cell carcinoma (LUSC), compared to adjacent noncancerous tissues (Fig. 1a; P < 0.05). Consistently, data from the University of ALabama at Birmingham CANcer data analysis Portal (UALCAN) website sourced from The Cancer Genome Atlas (TCGA) confirmed that RACGAP1 mRNA expression was upregulated in both LUSC (Fig. 1b) and LUAD (Fig. 1c) tissues compared to that in the normal tissues (P < 0.05). We further investigated the protein levels of RACGAP1 in LUAD using the Human Protein Atlas (HPA) and UALCAN. As shown in Fig. 2a, we found weak or no RACGAP1 expression in normal lung tissues but moderate and high RACGAP1 expression in lung cancer tissues. Consistently, analysis of CPTAC samples using UALCAN also showed that RACGAP1 protein levels were significantly higher in in LUAD tissues than in normal lung tissues (Fig. 2b, P < 0.05). We then examined RACGAP1 expression in surgically resected tumors from 75 patients with lung cancer using immunohistochemistry (IHC). The clinicopathological features of lung adenocarcinoma patients are summarized in Table 1. RACGAP1 expression was significantly greater in tumor tissues than in adjacent normal tissues. A significant difference in RACGAP1 staining pattern was observed between the two groups (Fig. 2c, P < 0.05). The aforementioned findings revealed that RACGAP1 was upregulated in LUAD at both the mRNA and protein levels.
RACGAP1 mRNA expression levels in different human cancers. (a) The mRNA expression of RACGAP1 in different datasets of different cancers was analyzed using the GEPIA website. (b, c) RACGAP1 expression in LUAD and LUSC samples and paired normal samples analyzed through the UALCAN website. *P < 0.05 vs. normal tissues. ACC Adrenocortical Carcinoma, BLCA Bladder Carcinoma, BRCA Breast Invasive Carcinoma, CESC Cervical Carcinoma, CHOL Cholangiocarcinoma, COAD Colon Adenocarcinoma, DLBC Lymphoid Neoplasm Diffuse Large B-cell Lymphoma, ESCA Esophageal Carcinoma, GBM Glioblastoma Multiforme, HNSC Head and Neck Squamous Cell Carcinoma, KICH Kidney Chromophobe, KIRC Kidney Renal Clear Cell Carcinoma, LAML Acute Myeloid Leukemia, LGG Brain Lower Grade Glioma, LIHC Liver Hepatocellular Carcinoma, LUAD Lung Adenocarcinoma, LUSC Lung Squamous Cell Carcinoma, OV Ovarian Serous Cystadenocarcinoma, PAAD Pancreatic Adenocarcinoma, PCPG Pheochromocytoma and Paraganglioma, READ Rectum Adenocarcinoma, SARC Sarcoma, SKCM Skin Cutaneous Melanoma, STAD Stomach Adenocarcinoma, THYM Thymoma, UCEC Uterine Corpus Endometrial Carcinoma, UCS Uterine Carcinosarcoma.
RACGAP1 protein expression in lung cancer tissues and normal tissues. (a) Immunohistochemical staining of RACGAP1 in LUAD tissues and normal lung tissues from the HPA database. (b) RACGAP1 protein expression in LUAD tissues and normal lung tissues from CPTAC samples. (c) RACGAP1 protein levels were determined via IHC using tissue microarrays in 75 pairs of lung adenocarcinoma and adjacent noncancerous lung tissues. The lower right panels show representative images captured at × 4 and × 20 magnification. Each datapoint in the plot represents one tissue sample. Error bars represent the median ± range. T, lung cancer tissue; N, noncancerous lung tissue. *P < 0.05 vs. normal tissues.
Genetic alterations of RACGAP1 in LUAD
Genetic variations of RACGAP1 in 2068 cases retrieved from 7 studies (35 cases from MSKCC; 230 cases from TCGA pub; 566 cases from TCGA, PanCancer; 302 cases from OncoSG, 2020; 516 cases from TCGA; 183 cases from Broad; and 0 cases from TSP) were analyzed using the cBioPortal database (Fig. 3a). We found four different genetic variations of RACGAP1 in LUAD, including missense mutation, truncating mutation, amplification, and deep deletion (Fig. 3b). Most genetic variations of RACGAPS were missense mutations and amplifications (incidence rates of 0.53% and 0.58%, respectively).
Analysis of RACGAP1 alterations in LUAD. (a) CNA and mutation frequency data for RACGAP1 in LUAD were analyzed through the cBioPortal website. (b) OncoPrint visual summary of alterations on a query of RACGAP1. Variations in frequency included missense mutation (green), truncating mutation (gray), amplification (red), and deep deletions (blue).
Relationship between RACGAP1 expression and clinicopathological parameters of LUAD patients
To investigate the correlation between RACGAP1 expression and clinicopathological parameters of patients, the UALCAN database was used to obtain information regarding age, sex, smoking habits, disease stage, nodal metastasis, and p53 mutants, after which their relationship with RACGAP1 levels were determined. Interestingly, all LUAD patients exhibited increased RACGAP1 expression levels (Fig. 4a; 41–60, 61–80, or 81–100 vs. normal, all P < 0.05). Moreover, we found that both male and female patients had increased RACGAP1 mRNA expression compared to normal and that male patients had increased RACGAP1 mRNA expression compared to female patients (Fig. 4b; P < 0.05). Furthermore, we observed higher RACGAP1 mRNA levels in smokers than in nonsmokers, reformed smokers (< 15 years), and reformed smokers (> 15 years) (Fig. 4c; all P < 0.05). Moreover, a comparison between smokers and reformed smokers showed lower levels among the latter (smokers vs. reformed smokers > 15 years or < 15 years; both P < 0.05). In addition, RACGAP1 expression levels were increased across different stages of LUAD (Fig. 4d; compared to normal lung tissues, all P < 0.05), with late-stage tumor tissues exhibiting higher expression levels than did early-stage tumor tissues (Fig. 4d; stage 4 vs. stage 1; P < 0.05). Notably, we also found that RACGAP1 expression was higher in N0-, N1-, or N2-stage LUAD than in normal tissues (Fig. 4e, all P < 0.05). Additionally, TP53 mutants exhibited higher RACGAP1 levels than did normal tissues or TP53 non-mutants (Fig. 4f; all P < 0.05). Differences in the clinicopathological parameters are summarized in Table 2. Our results revealed that high RACGAP1 expression was significantly associated with sex, smoking habit, disease stage, nodal metastasis, and TP53 mutant status.
Upregulation of RACGAP1 was correlated with shorter survival in LUAD patients
The current study further determined whether RACGAP1 was correlated with LUAD patient survival through both GEPIA and Kaplan–Meier survival curves. As shown in Fig. 5a, GEPIA indicated that increased RACGAP1 expression was correlated with decreased overall survival in LUAD patients (P = 1.70E-04). Consistently, analyses using the Kaplan–Meier Plotter revealed that increased RACGAP1 expression was significantly correlated with decreased overall survival (OS; Fig. 5b; hazard ratio [HR] 1.58, log-rank P = 2.0E−12), progression-free survival (PFS; Fig. 5c; HR 1.48, log-rank P = 5.5E−05), and post-progression survival (PPS; Fig. 5d; HR 1.5, log-rank P = 1.7E−03). These findings suggest the potential role of RACGAP1 as a biomarker for predicting prognosis.
Correlation between RACGAP1 expression and survival of LUAD patients. (a) The correlation between RACGAP1 expression and OS of LUAD patients was analyzed using GEPIA. (b–d) The correlation between RACGAP1 expression and OS, PFS, or PPS of LUAD patients was analyzed through the Kaplan–Meier Plotter. OS, overall survival; PFS, free-progression survival; PPS, post-progression survival.
RACGAP1 knockdown inhibits A549 cell growth
To explore the biological function of RACGAP1 on LUAD cell growth, three independent siRNAs targeting RACGAP1 were designed, and knockdown efficiencies were determined using Western blotting. Notably, Western blot analyses revealed that RACGAP1 expression was significantly downregulated at the protein level in A549 cells with all three siRNAs (Fig. 6a). Next, we used transfected with A549 cells to evaluate the effects of RACGAP1 on cellular functions. As shown in Fig. 6b, RACGAP1 knockdown reduced the viability of A549 cells. Moreover, RACGAP1 knockdown inhibited the formation of colonies (Fig. 6c). To further elucidate the possible mechanism underlying cellular growth inhibition induced by RACGAP1 knockdown, the cell cycle assay was performed using flow cytometry. As shown in Fig. 7, RACGAP1 knockdown markedly increased the percentage of cells in the G0/G1 phase but decreased the percentage of cells in the S phase (Fig. 7a), as well as significantly downregulated the expression of G0/G1-related proteins CyclinD1 and CDK4 in A549 cells, excepted for si-RACGAP1-3 in Cyclin D1 and si-RACGAP1 inCDK4 (Fig. 7b). These results suggest that RACGAP1 knockdown may inhibit proliferation by inducing G0/G1 phase arrest.
RACGAP1 knockdown inhibited cell growth in A549 cells. A549 cells were transfected with si-Ctrl or three independent siRNAs for si-RACGAP1. (a) Protein levels of RACGAP1 were determined using Western blot analysis. Representative images of RACGAP1 and β-actin are shown and quantitated using ImageLab software. β-actin was used as an internal control. *P < 0.05 vs. si-Ctrl. (b) Cell viability was determined using the CCK-8 assay. Data were normalized to the viability on day 1 and are represented as fold change. *P < 0.05 vs. si-Ctrl. (c) The colony formation assay was used to determine cell survival. After obtaining the images, colony formation was calculated and normalized to the survival of control cells. *P < 0.05 vs. si-Ctrl.
RACGAP1 knockdown inhibited cell proliferation in A549 cells. A549 cells were transfected with si-Ctrl or three independent siRNAs for si-RACGAP1. (a) Cell cycle progression was determined through PI staining and FACS analysis. Representative plots and percentage of cells at different stages (G0/G1, G2/M, and S phases) were presented. *P < 0.05 vs. si-Ctrl. (b) Protein levels of CDK4 and CyclinD1 after RACGAP1 knockdown were determined using Western blot analysis. Representative images are shown and quantitated using ImageLab software. β-actin was used as an internal control. *P < 0.05 vs. si-Ctrl.
RACGAP1 knockdown induces apoptosis of A549 cells
To investigate the effects of RACGAP1 on cell apoptosis, Annexin V-647 and PI were used to stain apoptotic cells. As shown in Fig. 8, RACGAP1 knockdown significantly increased the percentage of apoptotic cells (Fig. 8a). Mechanistically, Bcl-2 protein levels were downregulated following RACGAP1 knockdown, whereas Bax expression in si-RACGAP1-2 and si-RACGAP1-3 were significantly upregulated (Fig. 8b).
RACGAP1 knockdown induced cell apoptosis in A549 cells. A549 cells were transfected with si-Ctrl or three independent siRNAs for si-RACGAP1. (a) Cell apoptosis was determined using Annexin V/PI staining and FACS analysis. Representative plots and percentage of apoptotic cells are presented. *P < 0.05 vs. si-Ctrl. (b) Protein levels of Bax and Bcl-2 after RACGAP1 knockdown were determined using Western blot analysis. Representative images are shown and quantitated using ImageLab software. β-actin was used as an internal control. *P < 0.05 vs. si-Ctrl.
RACGAP1 knockdown inhibits PI3K/AKT pathway in A549 cells
To investigate whether RACGAP1 knockdown can inhibit the PI3K/AKT pathway, we evaluated the expression of PI3K, AKT and p-AKT after RACGAP1 knockdown (si-RACGAP1-2). As shown in Fig. 9, RACGAP1 knockdown significantly decreased PI3K expression and AKT phosphorylation. These findings suggest that RACGAP1 knockdown inhibited proliferation by suppressing PI3K/AKT pathway activation.
RACGAP1 knockdown inhibited the PI3K/AKT signaling pathway in A549 cells. A549 cells were transfected with si-Ctrl or si-RACGAP1. Protein levels of PI3K, p-AKT, and AKT after RACGAP1 knockdown were determined using Western blot analysis. Representative images are shown and quantitated using ImageLab software. GAPDH was used as an internal control. *P < 0.05 vs. si-Ctrl.
Discussion
Considering its malignant nature, lung cancer has been associated with high morbidity and mortality rates1,3. Diagnosis at an early stage, which can dramatically increase cure rates, is important for LUAD treatment. Owing to the poor effects of molecular targeted therapy in advanced-stage lung cancer, the prognosis of LUAD patients remains poor. Therefore, further studies on novel potential biomarker for early diagnosis and prognostic prediction are urgent needed for lung cancer patients. For this purpose, the current study analyzed multiple databases, with our results showing that RACGAP1 mRNA expression was upregulated in multiple tumor tissues (including lung, liver, and colorectal cancers among others) compared to normal adjacent tissues. These findings confirm that RACGAP1 expression was widely upregulated in several types of cancers, including stomach15, breast16,17 and liver cancers20, suggesting that RACGAP1 upregulation might be a common occurrence and plays an essential role in the development and progression of cancers.
Consistent with our finding on mRNA levels, we also confirmed that RACGAP1 protein expression was significantly upregulated in LUAD tissues compared to normal lung tissues. However, more clinic samples need to be analyzed to further verify RACGAP1 expression in future studies. Furthermore, we used the cBioPortal database to analyze genetic variations of RACGAP1 in LUAD, from which we identified four different genetic variations of RACGAP1, including missense mutation, truncating mutation, amplification, and deep deletion. These results indicate that RACGAP1 could potentially play a role of in LUAD diagnosis. Moreover, we analyzed the relationship between RACGAP1 expression and clinical pathological parameters of LUAD patients. Notably, our findings showed that RACGAP1 was significantly correlated with male sex, smoking, stage, nodal metastasis, and TP53 mutant status in LUAD patients, a finding consistent with those presented in previous study on breast and ovarian cancers24,25. Our survival analysis based on the Kaplan–Meier Plotter online database and GEPIA database revealed that increased RACGAP1 mRNA expression was associated with poor OS and PFS. The current study found that silencing of RACGAP1 expression in A549 cells using three different siRNAs inhibited cell growth as evidenced by decreased cell viability and colony number. Moreover, our analysis revealed that RACGAP1 knockdown blocked G0/G1 progression downregulated the expression of cyclinD1 and CDK4 in A549 cells. Furthermore, the anti-apoptotic protein Bcl-2 and pro-apoptotic protein Bax, which both belong to the Bcl-2 family, have been found to be critical regulators of this pathway26. Notably, we observed that RACGAP1 knockdown increased A549 cell apoptosis. Our analysis of protein expression levels showed that RACGAP1 knockdown downregulated Bcl-2 but upregulated Bax. These findings demonstrated that disruptions in the balance between proliferation and apoptosis in the development of LUAD could be attributed to RACGAP1 overexpression.
The PI3K/AKT signaling pathway is a pivotal cellular signaling cascade that exerts profound regulatory influence over fundamental cellular processes including proliferation, growth, survival, and metabolism27. Activated AKT orchestrates cellular proliferation and growth through multiple pathways. It directly influences cell cycle regulatory proteins, facilitating progression through S and G2/M phases. Additionally, AKT modulates protein synthesis and cell size to promote cellular growth. Furthermore, AKT functions as a crucial regulator of cell survival and apoptosis inhibition. Through modulation of signaling molecules such as the Bcl-2 family and caspases, AKT suppresses apoptosis and promotes cell viability, thereby maintaining cellular homeostasis28. Therefore, we found that RACGAP1 knockdown decreased the protein expression of PI3K and p-AKT, it had only minor effects on the total expression of AKT. The suggests that suppression of PI3K/AKT pathway might be among the underlying mechanisms behind the suppression of LUAD cell growth by silencing of RACGAP1.
In summary, the current study analyzed RACGAP1 mRNA and protein expression in multiple cancers, including lung cancer, and determined their correlation with prognosis and clinicopathological features. Overall, we found that RACGAP1 knockdown exerted anti-tumor activity in vitro, indicating that RACGAP1 might be involved in lung cancer development. Nonetheless, the underlying regulatory mechanisms still need more explored.
Materials and methods
Materials and reagents
The antibody against RACGAP1 (13739-1-AP) was purchased from Proteintech Group (Wuhan, Hubei, China). Antibodies against CDK4, CyclinD1, Bcl-2, Bax, AKT, p-AKT, PI3K, and β-actin were purchased from Cell Signaling Technology (Danvers, MA, United States). Fetal bovine serum, Trypsin–EDTA (0.25%), BCA Protein Assay Kit, and FxCycle PI/RNase Staining Solution were purchased from Thermo Fisher Scientific (Carlsbad, CA, USA). The antibody against GAPDH and the Annexin V 647 Apoptosis Detection Kit and CCK8 kit were obtained from Abbkine (Wuhan, Hubei, China).
Bioinformatics analysis
GEPIA (http://gepia.cancer-pku.cn/) was performed to analyze the expression levels of RACGAP1 in different types of cancers and overall survival analysis of RACGAP1 proteins from TCGA and Genotype-Tissue Expression, especially lung cancer, including LUAD and LUSC, and paired normal tissues. Changes in RACGAP1 mRNA levels in LUAD/LUSC and healthy tissues were further verified using the UALCAN online database. The protein expression of RACGAP1 in LUAD samples was analyzed through HPA and UALCAN databases.
Immunohistochemistry-based TMA
The lung cancer samples and matched noncancerous lung tissues (cat no. HLugA150CS03), containing 75 pairs of lung adenocarcinoma tissues and adjacent noncancerous lung tissues, were used for TMA construction. TMAs were created through a contract service at Shanghai OUTDO Biotech, China. The RACGAP1 antibody (1:3000; cat no. 13739-1-AP; Proteintech Group, Wuhan, China) was used to determine protein expression levels. Human tissues were stained using the EliVision Plus Kit (Maixin, China) according to the manufacturer’s instructions. The RACGAP1 immunostaining score was calculated as the sum of the staining intensity score and positive staining cell rate score. The staining intensity was scored as follows: no staining: 0, weak staining: 1, moderate staining: 2, and strong staining: 3. The positive staining cell rate was scored as follows: 0% to 5%: 0, 5% to 25%: 1, 26% to 50%: 2, 51% to 75%: 3, and > 75%: 4. The Wilcoxon test (raw scores) was applied to determine the significance of RACGAP1 staining in primary lung tumors relative to the matched adjacent tumoral tissues29,30.
Genetic alteration analysis
The cBioPortal database was used to obtain information on the genetic variations of RACGAP1 in 2068 cases retrieved from 7 studies (35 cases from MSKCC; 230 cases from TCGA pub; 566 cases from TCGA, PanCancer; 302 cases from OncoSG, 2020; 516 cases from TCGA; 183 cases from Broad; and 0 cases from TSP).
Clinicopathologic features
The UALCAN database was also used to investigate the relationship between RACGAP1 expression levels and different clinicopathological parameters, which included age, male sex, smoking, stage, nodal metastasis, and TP53 mutant status (summarized in Table 1).
Survival analysis
The Kaplan‑Meier plotter (www.kmplot.com), containing gene expression data and survival information of patients with LUAD, was queried to analyze the association between RACGAP1 mRNA expression and survival of patients with LUAD, including OS, PFS and PPS. The expression of RACGAP1 among the samples was divided into high or low groups according to the median expression, and the association of RACGAP1 expression with the survival of patients with LUAD was analyzed using the log‑rank test method.
Cell line and culture
The lung carcinoma cell line A549 was acquired from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were maintained in RPMI-1640 medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific), penicillin (100,000 U/L), and streptomycin (100 mg/L) at 37 °C in a 5% CO2 atmosphere. The cell lines were tested and authenticated.
Cell transfection
Three different siRNAs, namely anti-RACGAP1 (si-RACGAP1-1/-2/-3) and control siRNA (si-Ctrl), were purchased from Ribobio (Guangzhou, Guangdong, China). Cells were transfected with siRNAs or si-Ctrl at a concentration of 100 nM using Lipofectamine RNAiMax (Thermo Fisher Scientific) for 6–8 h according to manufacturer’s instructions. The cells were then cultured with complete medium at 37℃ in a humidified atmosphere with 5% CO2 for the indicated time points prior to use for experiments.
Western blot analysis
Cells were harvested and lysed in RIPA lysis buffer containing 1 mM phenylmethylsulfonyl fluoride and protease inhibitors. The BCA Protein Assay Kit was used to measure the concentrations of total protein. Thereafter, 50 µg of total protein lysate was separated on 10% SDS polyacrylamide gel and transferred to polyvinylidene fluoride (PVDF) membranes. Next, the membranes were blocked with 5% skim milk in TBST at room temperature for 2 h, incubated overnight at 4 °C with primary antibodies (all 1:1000), followed by incubation with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:5000). Proteins were visualized using an enhanced chemiluminescence (ECL) imager (Thermo Fisher Scientific), and band intensities were quantified using ImageLab software. β-actin expression was used as control. Three independent experiments were performed for each assay.
Cell counting kit-8 (CCK-8) assay
Transfected cells were re-seeded into 96-well plates (2000 cells per well) and cultured at 37 °C and 5% CO2 for the indicated time points. CCK-8 reagent (10 µL; Abbkine, Wuhan, Hubei, China) was added to each well. Thereafter, the plates were incubated for an additional 2 h at 37 °C, and the optical density (OD) was measured at a wave length of 450 nm. Cell viability was calculated based on the OD for each group.
Colony formation assay
Transfected cells were seeded into 12-well plates at a density of 500 cells per well and cultured at 37 °C and 5% CO2 for 10–14 days. Cells were then fixed in 4% paraformaldehyde for 20 min and stained with 0.1% crystal violet for 20 min at room temperature. Colonies were counted manually. Each assay was performed in triplicate.
Cell cycle analysis
Transfected cells were collected and fixed with 70% ethanol at 4 °C overnight. The fixed cells were centrifuged at 2000 rpm for 3 min, washed, and incubated with a mixture of FxCycle PI/ RNase Staining Solution for 30 min at room temperature. Fluorescence-activated cell sorting (FACS; Becton Dickinson, CA, United States) was used to analyze cell cycle progression using ModftLT version 3.0 (Verity Software House Inc., Topsham, ME, USA). The results were the average of three independent experiments.
Cell apoptosis analysis
Transfected cells were collected and washed twice with ice-cold phosphate-buffered saline and then incubated with Annexin V 647 Apoptosis Detection Kit solution (Abbkine, Wuhan, Hubei, China). The apoptosis rate was analyzed using FACS. The results were the average of three independent experiments.
Statistics analysis
Data were analyzed using SPSS 22.0 software. GEPIA was performed to determine the association between RACGAP1 expression and survival of LUAD patients. Kaplan–Meier survival curves were plotted for low- and high-expression groups and then analyzed using the log-rank test. Student’s t-test or the Mann–Whitney U was used for comparisons between two groups. One-way analysis of variance or the Kruskal–Wallis H test was used for multiple group comparisons. All quantitative data were presented as mean ± standard error of the mean. A P value of < 0.05 (two sided) indicated statistical significance. All experiments were performed at least three times.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Mathur, P. et al. Cancer statistics, 2020: Report from national cancer registry programme, India. JCO Glob. Oncol. 6, 1063–1075 (2020).
Bade, B. C. & Dela Cruz, C. S. Lung cancer 2020: Epidemiology, etiology, and prevention. Clin. Chest. Med. 41, 1–24 (2020).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020).
Hirsch, F. R. et al. Lung cancer: Current therapies and new targeted treatments. Lancet. 389, 299–311 (2017).
Toure, A. et al. MgcRacGAP, a new human GTPase-activating protein for Rac and Cdc42 similar to Drosophila rotundRacGAP gene product, is expressed in male germ cells. J. Biol. Chem. 273, 6019–6023 (1998).
Zhao, W. M. & Fang, G. MgcRacGAP controls the assembly of the contractile ring and the initiation of cytokinesis. Proc. Natl. Acad. Sci. USA. 102, 13158–13163 (2005).
Hirose, K., Kawashima, T., Iwamoto, I., Nosaka, T. & Kitamura, T. MgcRacGAP is involved in cytokinesis through associating with mitotic spindle and midbody. J. Biol. Chem. 276, 5821–5828 (2001).
Nishimura, Y. & Yonemura, S. Centralspindlin regulates ECT2 and RhoA accumulation at the equatorial cortex during cytokinesis. J. Cell Sci. 119, 104–114 (2006).
Lee, J. S., Kamijo, K., Ohara, N., Kitamura, T. & Miki, T. MgcRacGAP regulates cortical activity through RhoA during cytokinesis. Exp. Cell Res. 293, 275–282 (2004).
Sahai, E. Mechanisms of cancer cell invasion. Curr. Opin. Genet. Dev. 15, 87–96 (2005).
Lawson, C. D. & Ridley, A. J. Rho GTPase signaling complexes in cell migration and invasion. J. Cell. Biol. 217, 447–457 (2018).
Sanz-Moreno, V. et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 135, 510–523 (2008).
Yamazaki, D., Kurisu, S. & Takenawa, T. Involvement of Rac and Rho signaling in cancer cell motility in 3D substrates. Oncogene. 28, 1570–1583 (2009).
Ke, H. L. et al. Expression of RACGAP1 in high grade meningiomas: a potential role in cancer progression. J. Neurooncol. 113, 327–332 (2013).
Saigusa, S. et al. Clinical significance of RacGAP1 expression at the invasive front of gastric cancer. Gastric Cancer. 18, 84–92 (2015).
Kotoula, V. et al. Sample parameters affecting the clinical relevance of RNA biomarkers in translational breast cancer research. Virchows Arch. 462, 141–154 (2013).
Zhou, D. et al. Long non-coding RNA RACGAP1P promotes breast cancer invasion and metastasis via miR-345-5p/RACGAP1-mediated mitochondrial fission. Mol. Oncol. 15, 543–559 (2021).
Imaoka, H. et al. RacGAP1 expression, increasing tumor malignant potential, as a predictive biomarker for lymph node metastasis and poor prognosis in colorectal cancer. Carcinogenesis. 36, 346–354 (2015).
Stone, R. 2nd. et al. Identification of genes correlated with early-stage bladder cancer progression. Cancer Prev. Res. 3, 776–786 (2010).
Wang, S. M., Ooi, L. L. & Hui, K. M. Upregulation of Rac GTPase-activating protein 1 is significantly associated with the early recurrence of human hepatocellular carcinoma. Clin. Cancer Res. 17, 6040–6051 (2011).
Mi, S. et al. RNA-seq identification of RACGAP1 as a metastatic driver in uterine carcinosarcoma. Clin. Cancer Res. 22, 4676–4686 (2016).
Liang, Y., Liu, M., Wang, P., Ding, X. & Cao, Y. Analysis of 20 genes at chromosome band 12q13: RACGAP1 and MCRS1 overexpression in nonsmall-cell lung cancer. Genes Chromosomes Cancer. 52, 305–315 (2013).
Jin, D., Song, Y., Chen, Y. & Zhang, P. Identification of three lncRNAs as potential predictive biomarkers of lung adenocarcinoma. Biomed. Res. Int. 2020, 7573689 (2020).
Ma, X. J. et al. Gene expression profiles of human breast cancer progression. Proc. Natl. Acad. Sci. USA. 100, 5974–5979 (2003).
Lu, K. H. et al. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin. Cancer Res. 10, 3291–3300 (2004).
Peng, J. et al. Oligomerization of membrane-bound Bcl-2 is involved in its pore formation induced by tBid. Apoptosis. 14, 1145–1153 (2009).
Jin, Y. et al. Activation of PI3K/AKT pathway is a potential mechanism of treatment resistance in small cell lung cancer. Clin. Cancer Res. 28, 526–539 (2022).
Chen, S. et al. PGRN exacerbates the progression of non-small cell lung cancer via PI3K/AKT/Bcl-2 antiapoptotic signaling. Genes Dis. 9, 1650–1661 (2021).
Chen, L. & Su, G. Identification of CTRP1 as a prognostic biomarker and oncogene in human glioblastoma. Biomed. Res. Int. 2019, 2582416 (2019).
Bai, H. et al. Major vault protein suppresses lung cancer cell proliferation by inhibiting STAT3 signaling pathway. BMC Cancer. 19, 454 (2019).
Funding
This study was supported by Fujian Provincial Health Commission for Young and Middle-aged training Program (No. 2020GGB050).
Author information
Authors and Affiliations
Contributions
JX and JG conceived and designed the experiments. ZX, SW and JT conducted bioinformatics analysis and IHC-based TMA analysis. ZX, MW and WL performed cell cultures, CCK8 assays and colony formation assays. ZX, SW and JC conducted cell cycle analysis, Western-blot and data analysis. ZX wrote the manuscript, which JX and JG revised. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, Z., Wu, S., Tu, J. et al. RACGAP1 promotes lung cancer cell proliferation through the PI3K/AKT signaling pathway. Sci Rep 14, 8694 (2024). https://doi.org/10.1038/s41598-024-58539-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-58539-0
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.