Abstract
Nowadays, Electrocardiogram (ECG) signals can be measured using wearable devices, such as smart watches. Most wearable devices provide only a few details; however, they have the advantage of recording data in real time. In this study, 12-lead ECG signals were generated from lead I and their feasibility was tested to obtain more details. The 12-lead ECG signals were generated using a U-net-based generative adversarial network (GAN) that was trained on ECG data obtained from the Asan Medical Center. Subsequently, unseen PTB-XL PhysioNet data were used to produce real 12-lead ECG signals for classification. The generated and real 12-lead ECG signals were then compared using a ResNet classification model; and the normal, atrial fibrillation (A-fib), left bundle branch block (LBBB), right bundle branch block (RBBB), left ventricular hypertrophy (LVH), and right ventricular hypertrophy (RVH) were classified. The mean precision, recall, and f1-score for the real 12-lead ECG signals are 0.70, 0.72, and 0.70, and that for the generated 12-lead ECG signals are 0.82, 0.80, and 0.81, respectively. In our study, according to the result generated 12-lead ECG signals performed better than real 12-lead ECG.
Similar content being viewed by others
Introduction
Cardiovascular diseases (CVDs) comprise a series of heart blood-vessel abnormalities, which are one of leading reasons for deaths worldwide1. ECG signals are typically used in the early prediction and general diagnosis of abnormal heart rhythms. ECG signals are used to measure the heart’s electrical activity and rhythm through methods, such as resting ECG and Holter monitoring, that are conventional ECG measurement methods. Typically, 12-lead ECG signals are used to diagnose cardiac diseases2. Heart diseases often cause an irregularity in the heart called arrhythmia, wherein A-fib is the most common cardiac arrhythmia. However, real-time ECG measurement is required for early diagnoses.
In this regard, wearable ECG measurement devices are currently in use, with more being developed. Holter ECG devices were developed for long-term ECG monitoring3; however, owing to their limitations such as patient discomfort and limited mobility, ECG monitoring devices, such as patches and watches, were developed. These methods can be used to only measure one of the 12 leads. It is now possible to monitor the patient’s ECG from their home. These ECG monitoring devices are less complex and expensive compared to conventional methods. However, wearable devices, such as patches and smart watches, have a critical limitation; they cannot measure synchronous multiple leads4. Generally, lead I can be representative of limb leads but not precordial leads. Therefore, abnormal cardiac diseases, such as RBBB, LBBB, RVH, and LVH, cannot be diagnosed5. Although single-lead devices are widely used, they are rarely used for diagnosing all cardiac diseases. Due to their lack of information and difficulties mainly A-fib is currently diagnosed by single lead ECG devie6.
Because ECG signals represent a vector of the heart, automated ECG-based classification models have become popular. Most classification models employ 12-lead ECG signals or only lead II signal for learning7. Various deep learning methods, from convolutional neural networks (CNNs) to the more recent transformer encoder models, have been implemented in ECG classification problems and produced promising results8,9,10. Moreover, data generation models, such as the GANs11, transformers12, and diffusion models13, have been implemented on ECG signals for data synthesis14,15.
For related works, there have been attempts to classify A-fib using single-lead ECG signals and the data augmentation method16,17. Hatamian et al.18 proposed a method of classifying A-fib by combining the data augmentation and deep learning methods. However, the GAN input in this method is noises, and the type of GAN used is unconditional GAN; therefore, the generated data does not represent other lead vectors. Moreover, precordial leads were not considered. Lan et al.19 used short-time Fourier transform and GAN to classify and augment data. However, related classification studies mainly focused on augmentation to increase the amount of data. These methods have been verified to increase the performance accuracy; however, the augmented data were not verified.
There have been several studies on lead conversion. Sohn et al. proposed a method for reconstructing a 12-lead ECG from a 3-lead patch device employing an LSTM network20. However, their work requires a device measuring at least three leads, whereas our work only needs lead I. Therefore, it is limited in application to single-lead measurement devices. Afrin et al. proposed a handheld ECG device measuring single-lead ECG, which could measure lead I, lead II, and lead III21. Three different leads are measured asynchronously. Upon the previously measured ECG history, 12-lead ECGs are synchronously reconstructed. Therefore, the proposed method additionally needs previously measured ECG for reconstruction. Huang et al. proposed an ECG system reconstruction method from temporally asynchronous bipolar ECG recordings22. Their reconstruction algorithm is based on multiple ECGs recorded asynchronously from different sites. However, when deriving the optimal weight coefficient, only 11 subjects were recorded, and external data were not tested. SynSigGAN is typically used to generate biomedical signals23 and implement discrete wavelet transformation and Bidirectional Long Short-Term Memory (Bi-LSTM)24 layers for the generation model. However, inputs are typically treated as latent variables; therefore, their use is limited to data augmentation. Shin et al.25 proposed a method for generating a photoplethysmography (PPG) signal from an ECG signal using an LSTM and a CNN. Lee et al.26 proposed the R-peak alignment and time sequence embedding method to transform a one-dimensional time series into a two-dimensional time-series for enhancing the performance of GANs on two-dimensional time series. Chest leads were also converted from limb leads with high accuracy. However, the R-peak, which was used as the median value, resulted in more than one beat during data preprocessing and the model inputs were lead II. Existing methods related to lead conversion performed well. However, the generated ECG signals in these studies were not evaluated based on a classification comparison with real ECG signals. Therefore, the frechet distance (FD) and mean squared error (MSE) scores should be used as evaluation scores, and a comparison with real ECG signals should be considered alongside a Turing test to achieve the usefulness of generated ECG.
In this study, all 12-lead ECG signals were generated using lead I, and their feasibility for usage was determined through a classification performance test. Our study's novelty lies in the analysis of ECG signals generated from Lead I through classification tests, demonstrating their feasibility. In this study, the possibility of generating ECG signals for diagnostic implementation, which reinforces the limitation of single-lead ECG measurement devices, was presented. The proposed method can be applied in out-of-hospital ECG monitoring care without using multiple lead measurement devices.
Methods
In this study, the ECG generation model was based on that in our previous study27. The pix2pix GAN model was trained using MUSE data on patients who had visited the Seoul Asan Medical Center Hospital between January 01, 2001, and February 28, 2022. For classification, the PTB-XL database was used as external data. As illustrated in Fig. 2, six classes were extracted from the PTB-XL database for the classification test. Evaluation was based on the F1-score, precision, recall, and accuracy. The overview of this study is illustrated in Fig. 1.
Datasets and preprocessing
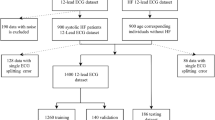
The 12-lead ECG data used in this study were obtained from the MUSE and PTB-XL databases28. The PTB-XL dataset contains 21,837 records obtained from 18,885 patients, and the MUSE database comprises 4 million records obtained from the Asan Medical Center Hospital. The experimental protocols in the data were approved by the Institutional Review Board (IRB) at the Asan Medical Center Hospital, under the approval number IRB No. 2022–0781. All methods were carried out in accordance with relevant guidelines and regulations and informed consent was obtained from all subjects and/or their legal guardian(s).
The duration of both datasets was 10 s and the sampling rate was 500 Hz, wherein the number of sampling points were 5,000. For the generation model, lead I ECG signals obtained from the MUSE database were considered as the input ECG signals, and the remaining leads as reference signals. Additionally, all 12-lead ECG signals from the PTB-XL dataset were used in the classification model. The data descriptions are listed in Supplementary Tables S1, S2 and S3. To generate and classify raw ECG signals using our model, none of the conventional methods, such as filtering or adjusting the baseline, were implemented. Figure 2 illustrates the data preprocess and exclusion criteria. For the MUSE database records, data on patients under 18 years old, unconfirmed data and patient records with sampling rates under 500 Hz were excluded. Furthermore, the 10 s records were segmented into 2.5 s intervals, each containing 1250 sampling points.
GAN architecture
GAN consists of two main networks: a generator and discriminator11. The basis of GAN is a minimax game between the generator and discriminator. In this study, the generator considers lead I as the input and synthesizes the remaining leads; and the discriminator distinguishes the generated signals from the real ones. Figure 1 depicts the overall architecture of the proposed model. The proposed model follows the main objective of conditional GAN, which can be expressed as shown in (1). Conditional GANs29 learn mapping based on the relationship between the signal x and random noise vectors z and y14.
where G tries to minimize the objective of GAN against D, which tries to maximize it (1). Moreover, L1 loss was used (2); thus, the final objective of GAN was represented as follows:
GAN and discriminator network
The generator in this study comprises a U-net based encoder-decoder. The U-net generator is depicted in Supplementary Figure S6. The encoder consists of seven convolution layers, with batch normalization and Leaky ReLU applied in all except the first layer. The decoder is composed of seven up-convolution layers. For all Leaky ReLU functions, the slope was set to 0.2; the kernel size and stride length were 4 and 2, respectively. The discriminator, depicted in Supplementary Figure S7, contains five convolution layers with batch normalization and Leaky ReLU. A convolution layer is added after the last layer to map to a one-dimensional output, followed by a Sigmoid function. The slope for all Leaky ReLU functions is 0.2, with a kernel size of four and a stride length of two. The learning rate is set to 0.0005 for the generator and 0.0001 for the discriminator. Additionally, Adam is employed for hyperparameter optimization, and the batch size is set to 32. A total of 11 models were trained to generate 11 leads. As mentioned in the 'Datasets and Preprocessing' section, the input and output dimensions of the generator, and the input of the discriminator, are all (batch size, 1, 1250).
Evaluation method
The classification of the generated 12-lead ECG signals was performed using the ResNet model. The normal ECG, RBBB, LBBB, LVH, RVH, and A-fib values were then used to evaluate the classification classes. The A-fib and normal ECG were used because most out-of-hospital wearable devices are used to detect AF, and both normal ECG and AF can be classified using single-lead ECG measurement9. By contrast, RBBB, LBBB, LVH, and RVH were diagnosed using the precordial leads (V1, V2, V3, V4, V5, and V6). To test the feasibility of the 12-lead ECG generated from lead I, 5 different methods were compared. First, the classification results of the generated 12-lead ECG and real lead-I ECG signals were compared. To verify the disadvantages of single-lead measurement, the classification performances of the generated 12-lead ECG and real lead-I were compared. Second, the classification results of the generated 12-lead and real 12-lead ECG signals was compared. From the Einthoven triangle formula30 (4–7) was then applied to the generated 12-lead ECG signals to conduct the ablation study. Two different limb leads are required when applying the Einthoven triangle formula. Therefore, three different groups of leads were used in the experiment: input lead I and generated lead II, input lead I and generated lead III, and input lead I and generated lead II, III. The groups were separately evaluated to determine the best outcome results and differences in the number of generated leads.
All the five different sets of methods were evaluated based on their precision, recall and f1-score values (10–12); and PTB-XL external data were used to train and evaluate each classification method.
Results
In this section, the generated signals from the PTB-XL database and their evaluation scores are presented and compared. Figure 3 illustrates the generation and classification processes using lead I. The evaluation of the generation model performance was performed in our previous study27. The evaluation scores for all five results is shown in Table 1 and Table 2.
The precision, recall, and f1-score values of the generated.
12-lead ECG signals and classification performance results of real lead-I are shown in Table 1, where the best results are highlighted in bold. The generated 12-lead ECG signals exhibited the best results followed by the generated lead II. This shows that multi-lead ECG classification is more accurate. Particularly, the classification results of the abnormal.
ECG signals that are typically diagnosed at the precordial lead show a significant difference. The classification performance of all real 12-leads is shown in Table 2, where real lead exhibited poor results using both the 12-lead ECG and single lead I signals.
The confusion matrix results are depicted in Fig. 4. Additionally, the experimented results of the three different lead groups are listed in Table 1 and the confusion matrix is presented in Fig. 5. No significant differences in the results can be observed when the Einthoven formula is used to calculate the other limb leads. Moreover, the ROC and AUC results illustrated in Supplementary Figure S1 exhibit no significant difference. Therefore, generating only Lead II or Lead III and calculating the rest of limb leads using the Einthoven formula reduces both the model complexity and time. The generated 12-lead ECG and reference ECG are illustrated in Supplementary Figure S2. The capability of the generated ECG signals in diagnoses was also tested to verify the classification results of the real ECG signals.
Confusion matrix for the generated ECG. (a) Generated 12-lead ECG signal results, and (b), (c), and (d) are the results of the Einthoven lead conversion. In (b), only lead II was generated using GAN and limb lead and the rest were calculated using the Einthoven formula. In (c), only lead III was generated using GAN and limb lead and the rest were calculated using the Einthoven formula. In (d), lead II and lead III were generated using GAN and limb lead and the rest were calculated using the Einthoven formula.
Discussion
This study demonstrates that generated ECG signals are capable of diagnosing CVDs. Table 3 shows previous studies related to ECG generation by GANs and have evaluated their work with classifications performance that was listed in review paper by Laurenz Berger15. However, as shown in Table 3 most of the previous works’ purpose was focused mainly on solving imbalanced data problems. Also, the input data were noise and simulator where our study focused on lead-to-lead conversion. Previous and related studies have only focused on data augmentations and generated signals were not from single lead ECG. A detailed example of the generated ECG signals is shown in Supplementary Figure S1.
Single-lead ECG signals can be can better classified by implementing the proposed method to classify CVDs, which improves the disadvantages of single-lead ECG signals. This method enables the real-time analysis of ECG signals through single-lead ECG measurement, thereby allowing the use of single-lead ECG measurement devices, such as smart watches, on both patients and the general public. Therefore, the proposed method can be used to alert users and patients of potential danger. Additionally, single-lead measurement, which is a more comfortable method, can be adopted in hospitals instead of 12-lead standard ECG measurement.
The results of the RVH performance were lower than those of the other classes. Moreover, the F1-score difference was up to 0.49 lower than that of the normal class, mainly owing to the low quantity of the RVH data in the PTB-XL database used to train the classification model compared to that used in the other classes. However, its performance was still higher than that of real 12-lead ECG signal classification. 12-lead ECG signals were generated from lead I rather than lead II because general single-lead ECG devices comprise mainly smart watches, which measure lead I.
The classification experiments were performed using an external dataset that was not used to train the generation model. A comparison of the classification of the generated 12-lead and real lead I signals was also performed. This comparison was performed to determine whether the generated ECG signals will have a better classification performance than real lead I. However, the results were more dramatic in precordial-based CVD diagnosis. This result shows that single-lead ECG measurement devices are not capable of diagnosing the various CVD types; however, applying the proposed method improves their classification capability.
Standard ECG signals are used in 12-lead ECG measurement. Multiple electrodes are attached to the surface of the patients’ body, which makes it hard to obtain ECG signals in the long term. However, various devices have been developed with the growth of the single-lead ECG device market. Owing to their real-life ECG measurement capabilities, they are used to detect cardiac diseases, such as A-fib. However, these single-lead measurement devices generally detect or diagnose cardiac diseases based on the rhythmic features; therefore, it is nearly impossible to detect diseases that are diagnosed based on the amplitude or via comparison with other leads.
This study presented the feasibility of generated ECG signals for use in diagnosis. The obtained results were better than those of real ECG signals, which can be implemented in single lead devices. The accuracies, precisions, and F1 scores of the generated 12-lead ECG are shown in Tables 1 and 2. The normal class values are 0.89, 0.92, and 0.91; the A-fib class values are 0.96, 0,76, and 0.84; the LBBB values are 1, 0.96, and 0.98; the RBBB results are 0.87, 0.77, and 0.82; the LVH results are 0.82, 0.94, and 0.87; and the RVH values are 0.38, 0.47, and 0.42, respectively. Among the six classes, A-fib was the only class that was not in sinus rhythm, resulting in higher performance.
The proposed method can also be used to provide insights into various pathological cardiac diagnoses features. This will allow the monitoring of personalized ECG signals during in- and out-of-hospital care, where the cardiologist keeps patient records over a long time. Moreover, further assessment can be made by the cardiologist when a remarkable CVD is detected during the patient’s daily life.
Most of all, the novelty of our study is:
-
1)
A large dataset of over 400 million data is used to train the generative model.
-
2)
No other study has investigated the use of generated ECG signals for diagnosis.
-
3)
Generated ECG classification exhibits a better performance than reference single-lead ECG classification, indicating that the information obtained from the precordial leads are crucial.
As shown in Tables 1 and 2, the proposed method produces a better performance than real ECG classification. Several questions need to be addressed regarding why the generated ECG signal classification results show higher performance. Our proposed model has been trained on a dataset of 4 million samples and has the capability to generate ECG signals that closely resemble real ones. The primary difference between the generated ECG signals and the corresponding reference ECG signals is that the generated ones can fill in missing data and reduce baseline wandering problems shown in Supplementary Figure S4 and S5. However, the crucial outcome of the study is that CVD diagnosis using the entire 12-lead ECG performs better when employing our proposed method.
However, a few limitations exist in this study. First, 6 CVD types containing both precordial and limb leads were classified. Nonetheless, there are various of types of CVDs, such as acute MI (AMI), that are life-threatening. Certain MI, such as ST elevation, are fairly classified using DL (deep learning)43,44,45. However, there are very few AMI record data available owing to its high mortality rate. In the future, more focus should be placed on critical CVDs, which can require out-of-hospital care. Second, for the lead I ECG signals, the input in the proposed method was based on standard 12-lead ECG records. No open data were measured using both the single-lead device and standard 12-lead ECG. However, a few single-lead ECG signals were generated from smart watches, as depicted in Supplementary figure S3, and classified to show the concept of our method. It was important for the proof of concept to show that the ECG signals obtained from single-lead devices can be used to generate 12-lead ECG signals and to detect CVDs.
Conclusion
This study presents a method for generating 12-lead ECG signals that can be used to classify CVDs using DL. ECG data obtained from the Asan medical center and containing 400 million records was used. External data from the PTB-XL database were also used to classify 6 types of cardiac diseases present in the limb and precordial leads. Additionally, the performance of the classification results was compared with those of real and generated ECGs. Consequently, the proposed method exhibited outstanding results during classification, which can be applied in real-life ECG monitoring. Single-lead ECG devices are simple and comfortable to wear; however, owing to the lack of lead information, rhythm features are mainly used to detect abnormal ECG. This approach can be used to solve for the disadvantages of single-lead ECG devices, thereby helping in out-of-hospital CVD detection, which is a crucial step in personalized medicine.
Data availability
The datasets generated or analyzed during the current study are not publicly available due to the restriction policy of the hospital but are available from the corresponding author upon a reasonable request. PTB-XL data base are public data which is available at https://physionet.org/content/ptb-xl/1.0.3/.
Code availability
All algorithms used in this study are available by contacting the corresponding author at any time.
References
Roth, G. A. et al. Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J. Am. Coll. Cardiol. 70, 1–25. https://doi.org/10.1016/j.jacc.2017.04.052 (2017).
Al-Zaiti, S. S., Faramand, Z., Rjoob, K., Finlay, D. & Bond, R. in Cardiovascular and Coronary Artery Imaging (eds Ayman S. El-Baz & Jasjit S. Suri) 45–87 (Academic Press, 2022).
Galli, A., Ambrosini, F. & Lombardi, F. Holter monitoring and loop recorders: From research to clinical practice. Arrhythm. Electrophysiol. Rev. 5, 136–143. https://doi.org/10.15420/aer.2016.17.2 (2016).
Haverkamp, H. T., Fosse, S. O. & Schuster, P. Accuracy and usability of single-lead ECG from smartphones—A clinical study. Indian Pacing Electrophysiol. J. 19, 145–149 (2019).
Miller, J. M., Marchlinski, F. E., Buxton, A. E. & Josephson, M. E. Relationship between the 12-lead electrocardiogram during ventricular tachycardia and endocardial site of origin in patients with coronary artery disease. Circulation 77, 759–766. https://doi.org/10.1161/01.CIR.77.4.759 (1988).
Mathews, S. M., Kambhamettu, C. & Barner, K. E. A novel application of deep learning for single-lead ECG classification. Comput. Biol. Med. 99, 53–62 (2018).
Alday, E. A. P. et al. Classification of 12-lead ecgs: The physionet/computing in cardiology challenge 2020. Physiol. Meas. 41, 124003 (2020).
Yıldırım, Ö., Pławiak, P., Tan, R.-S. & Acharya, U. R. Arrhythmia detection using deep convolutional neural network with long duration ECG signals. Comput. Biol. Med. 102, 411–420 (2018).
Attia, Z. I. et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: A retrospective analysis of outcome prediction. Lancet 394, 861–867 (2019).
Petmezas, G. et al. State-of-the-art deep learning methods on electrocardiogram data: Systematic review. JMIR Med. Inform. 10, e38454 (2022).
Goodfellow, I. et al. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 27, 7966 (2014).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 30, 8796 (2017).
Croitoru, F.-A., Hondru, V., Ionescu, R. T. & Shah, M. Diffusion models in vision: A survey. IEEE Trans. Pattern Anal. Mach. Intell. 25, 896 (2023).
Wang, Y. et al. Transformer-based acoustic modeling for hybrid speech recognition. ICASSP 2020–2020 IEEE International Conf. on Acoustics, Speech and Signal Processing (ICASSP). 6874–6878 (IEEE).
Berger, L., Haberbusch, M. & Moscato, F. Generative adversarial networks in electrocardiogram synthesis: Recent developments and challenges. Artif. Intell. Med. 14, 102632 (2023).
Adib, E., Afghah, F. & Prevost, J. J. Arrhythmia classification using cgan-augmented ecg signals. 2022 IEEE International Conf. on Bioinformatics and Biomedicine (BIBM). 1865–1872 (IEEE).
Zhang, Y.-H. & Babaeizadeh, S. Synthesis of standard 12-lead electrocardiograms using two-dimensional generative adversarial networks. J. Electrocardiol. 69, 6–14 (2021).
Hatamian, F. N. et al. The effect of data augmentation on classification of atrial fibrillation in short single-lead ECG signals using deep neural networks. ICASSP 2020–2020 IEEE International Conf. on Acoustics, Speech and Signal Processing (ICASSP). 1264–1268 (IEEE).
Lan, T., Hu, Q., Liu, X., He, K. & Yang, C. Arrhythmias classification using short-time Fourier transform and GAN based data augmentation. 2020 42nd Annual International Conf. of the IEEE Engineering in Medicine & Biology Society (EMBC). 308–311 (IEEE).
Sohn, J., Yang, S., Lee, J., Ku, Y. & Kim, H. C. Reconstruction of 12-lead electrocardiogram from a three-lead patch-type device using a LSTM network. Sensors 20, 3278 (2020).
Afrin, K., Verma, P., Srivatsa, S. S. & Bukkapatnam, S. T. Simultaneous 12-lead electrocardiogram synthesis using a single-lead ecg signal: Application to handheld ecg devices. (2018) arXiv preprint arXiv:1811.08035.
Huang, C.-Y. et al. Synthesizing the 12-Lead electrocardiograms by the single-lead ECG system-reconstruction of temporal asynchronous bipolar ECG recordings. Circulation 134, A20750–A20750 (2016).
Hazra, D. & Byun, Y.-C. SynSigGAN: Generative adversarial networks for synthetic biomedical signal generation. Biology 9, 441 (2020).
Graves, A., Mohamed, A-R. & Hinton, G. Speech recognition with deep recurrent neural networks. 2013 IEEE International Conf. On Acoustics, Speech And Signal Processing. 6645–6649 (IEEE).
Shin, H., Sun, S., Lee, J. & Kim, H. C. Complementary photoplethysmogram synthesis from electrocardiogram using generative adversarial network. IEEE Access 9, 70639–70649 (2021).
Lee, J., Oh, K., Kim, B. & Yoo, S. K. Synthesis of electrocardiogram V-lead signals from limb-lead measurement using R-peak aligned generative adversarial network. IEEE J. Biomed. Health Inform. 24, 1265–1275 (2019).
Seo, H.-C., Yoon, G.-W., Joo, S. & Nam, G.-B. Multiple electrocardiogram generator with single-lead electrocardiogram. Comput. Methods Programs Biomed. 221, 106858 (2022).
Wagner, P. et al. PTB-XL, a large publicly available electrocardiography dataset. Sci. Data 7, 154. https://doi.org/10.1038/s41597-020-0495-6 (2020).
Mirza, M. & Osindero, S. Conditional generative adversarial nets. (2014) arXiv preprint arXiv:1411.1784.
Butterworth, J. S. & Thorpe, J. J. On evaluating the Einthoven triangle theory. Circulation 3, 923–925 (1951).
Golany, T., Radinsky, K. & Freedman, D. SimGANs: Simulator-based generative adversarial networks for ECG synthesis to improve deep ECG classification. Int. Conf. on Machine Learning 2020. 3597–3606 (PMLR).
Wang, G. et al. Evolving SimGANs to improve abnormal electrocardiogram classification. Proc. of the Genetic and Evolutionary Computation Conf. Companion 2022. 1887–1894.
Golany, T., Freedman, D. & Radinsky, K. Ecg ode-gan: Learning ordinary differential equations of ecg dynamics via generative adversarial learning. Proc. of the AAAI Conf. on Artificial Intelligence 2021. 134–141.
Shaker, A. M., Tantawi, M., Shedeed, H. A. & Tolba, M. F. Generalization of convolutional neural networks for ECG classification using generative adversarial networks. IEEE Access 8, 35592–35605 (2020).
Rath, A., Mishra, D., Panda, G. & Satapathy, S. C. Heart disease detection using deep learning methods from imbalanced ECG samples. Biomed. Signal Process. Control 68, 102820 (2021).
Ma, S., Cui, J., Xiao, W. & Liu, L. Deep learning-based data augmentation and model fusion for automatic arrhythmia identification and classification algorithms. Comput. Intell. Neurosci. 202, 78969 (2022).
Wang, P., Hou, B., Shao, S. & Yan, R. ECG arrhythmias detection using auxiliary classifier generative adversarial network and residual network. IEEE Access 7, 100910–100922 (2019).
Thambawita, V. et al. DeepFake electrocardiograms using generative adversarial networks are the beginning of the end for privacy issues in medicine. Sci. Rep. 11, 21896 (2021).
Yang, H., Liu, J., Zhang, L., Li, Y. & Zhang, H. ProEGAN-MS: A progressive growing generative adversarial networks for electrocardiogram generation. IEEE Access 9, 52089–52100 (2021).
Brophy, E., De Vos, M., Boylan, G. & Ward, T. Multivariate generative adversarial networks and their loss functions for synthesis of multichannel ecgs. IEEE Access 9, 158936–158945 (2021).
Zhou, Z., Zhai, X. & Tin, C. Fully automatic electrocardiogram classification system based on generative adversarial network with auxiliary classifier. Expert Syst. Appl. 174, 114809 (2021).
Li, W., Tang, Y. M., Yu, K. M. & To, S. SLC-GAN: An automated myocardial infarction detection model based on generative adversarial networks and convolutional neural networks with single-lead electrocardiogram synthesis. Inform. Sci. 589, 738–750 (2022).
Cho, Y. et al. Artificial intelligence algorithm for detecting myocardial infarction using six-lead electrocardiography. Sci. Rep. 10, 20495 (2020).
Makimoto, H. et al. Performance of a convolutional neural network derived from an ECG database in recognizing myocardial infarction. Sci. Rep. 10, 8445 (2020).
Tadesse, G. A. et al. DeepMI: Deep multi-lead ECG fusion for identifying myocardial infarction and its occurrence-time. Artif. Intell. Med. 121, 102192 (2021).
Acknowledgements
This work was supported by the Korea Medical Device Development Fund funded by the Korean Government (the Ministry of Science and ICT, the Ministry of Trade, Industry and Energy, the Ministry of Health & Welfare, and the Ministry of Food and Drug Safety) under Project 1711139135 and Project RS-2020-KD000174.
Author information
Authors and Affiliations
Contributions
G.W.Y: Conceptualization, data curation, visualization, investigation, methodology, software, writing—original draft preparation, writing—reviewing and editing. S.J: Supervision, conceptualization, writing—reviewing and editing, and validation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yoon, GW., Joo, S. Classification feasibility test on multi-lead electrocardiography signals generated from single-lead electrocardiography signals. Sci Rep 14, 1888 (2024). https://doi.org/10.1038/s41598-024-52216-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-52216-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.