Abstract
Free amino acids in potato tubers contribute to their nutritional value and processing quality. Exploring the natural variation in their accumulation in tubers across diverse genetic backgrounds is critical to potato breeding programs aiming to enhance or partition their distribution effectively. This study assessed variation in the tuber-bound free amino acids in a diversity panel of tetraploid potato clones developed and maintained by the Texas A&M Potato Breeding Program to explore their genetic basis and to obtain genomic-estimated breeding values for applied breeding purposes. Free amino acids content was evaluated in tubers of 217 tetraploid potato clones collected from Dalhart, Texas in 2019 and 2020, and Springlake, Texas in 2020. Most tuber amino acids were not affected by growing location, except histidine and proline, which were significantly lower (− 59.0%) and higher (+ 129.0%), respectively, at Springlake, Texas (a location that regularly suffers from abiotic stresses, mainly high-temperature stress). Single nucleotide polymorphism markers were used for genome-wide association studies and genomic selection of clones based on amino acid content. Most amino acids showed significant variations among potato clones and moderate to high heritabilities. Principal component analysis separated fresh from processing potato market classes based on amino acids distribution patterns. Genome-wide association studies discovered 33 QTL associated with 13 free amino acids. Genomic-estimated breeding values were calculated and are recommended for practical potato breeding applications to select parents and advance clones with the desired free amino acid content.
Similar content being viewed by others
Introduction
Potato is a major dietary staple crop consumed worldwide. Potato tubers are a rich source of carbohydrates (mainly starch), dietary fiber, high-quality protein, vitamins, antioxidants, and minerals1,2. The total protein content of potato tubers is relatively low (1.0–3.0% FW or 5.0–14.0% DW) but comparable to cereals on a dry basis and better than commonly consumed vegetables3. Potato protein has high biological activity due to its high free amino acid score (69.0–76.0%)4,5 and higher nutritional value than other commonly consumed plant sources due to its high content of essential amino acids6. Potato contains all nine essential amino acids3, qualitatively superior to many other plant-based proteins7. Nearly half of the total amino acids in potatoes are in the free form8,9. Asparagine and glutamine amides dominate the free amino acids (34.0–90.0%)10,11, whereas lysine, tyrosine, and the sulfur-containing amino acids methionine and cysteine are still limited in potatoes3,12. Nevertheless, potato tubers contain higher amounts of lysine than cereals13. Enhancing the protein quality of potato tubers by traditional breeding or genetic manipulation of pathways involved in the metabolism of limiting amino acids holds the potential to improve potato’s biological activity and nutritional value and thus enhance human health and nutrition.
Free amino acids react with reducing sugars during frying, resulting in a non-enzymatic browning reaction (Maillard) which affects the organoleptic and other quality traits of processed potato products14,15. Asparagine reacts with reducing sugars to form a carcinogenic compound called acrylamide16,17, a major concern in processed potato products18,19,20,21,22. Genetically modified (GM) potatoes produced by manipulating the expression of the asparagine synthetase gene using RNA interference have low asparagine content and reduced acrylamide levels after processing23,24. Free asparagine and reducing sugars are precursors for acrylamide formation via deamination and decarboxylation processes16,17,25. In potatoes, the precursor concentration plays a significant role in determining the acrylamide-forming potential18,20,21,22,26,27,28. A study reported a significant correlation between free asparagine concentration and acrylamide-forming potential in French fry varieties18,28. Despite the need, polyploidy and high heterozygosity make breeding and selection of low acrylamide-forming potential potato cultivars for healthy fries and chips challenging. Possibilities of using the Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) system to manipulate reducing sugars and aspargine metabolism in potatoes are already being considered29. Another amino acid affecting flavor is methionine, a precursor of methional, a major volatile associated with the aroma of potato chips and french fries30. Enhancing free methionine in tubers for nutritional and aroma benefits is of interest to the food industry and consumers. Transgenic approaches to improve free methionine levels in potato tubers have been used but had, in several cases, undesired effects, including morphological changes in tubers or plants31,32,33,34,35,36,37. Given the uncertainty about the acceptance of GM potatoes, selecting low asparagine and high methionine in potato tubers through conventional breeding could be the way to develop healthy and flavorful potato varieties. Most studies addressing quality traits focus on increasing starch content38,39,40, diminishing reducing sugars41,42, and possessing glycoalkaloid levels below toxic limits43,44 in potato tubers. Increasing the protein content in potato tubers is also desirable. However, breeding for more free amino acids in potato tubers may favor indirect selection for low carbohydrate levels because of their inverse relationship with protein content45.
Significant genetic variation in the relative amounts of free proteinogenic and non-proteinogenic amino acids has been observed in cultivated potatoes and landraces46,47,48. Screening of a diploid potato population using targeted49 and untargeted46 metabolite analysis showed significant variation in amino acid content, explaining the underlying genetic variation and identifying metabolic quantitative trait loci (QTL). Due to technological advancements in genotyping, genome-wide association studies (GWAS) have evolved as a powerful tool for exploring the genetic basis of various traits in several crop species, including potatoes50,51,52,53,54,55,56,57,58. A GWAS study performed using the metabolome of cooked potatoes of the Solanaceae Coordinated Agricultural Project (SolCAP) diversity panel59 identified several metabolites, including five free amino acids, significantly associated with SNP markers60. Evaluating phenotypic variation for free amino acid levels in diversity panels used by breeding programs should provide a baseline to plan selection strategies to improve potatoes’ nutritional properties. Diversity panels can be used to understand the genetic basis of the traits using GWAS and generate genomic-estimated breeding values (GEBVs) of traits using genomic selection (GS) for practical marker-assisted selection in breeding programs. Losses in nitrogen and amino acids in potato tubers differ significantly according to cooking techniques; chips and canned potatoes show the maximum loss, followed by drum-dried and french-fried potatoes61,62,63,64. Field production practices, growing environmental conditions, nitrogen-fertilization rate and application time, residual soil nitrogen, soil-moisture availability, and storage conditions also influence the amino acid content65,66,67,68. The broader goal of this research would be to assist the breeding program in developing healthier and more nutritious potatoes using conventional breeding and guide the development of strategies for metabolic engineering. The specific objectives were to i) evaluate the phenotypic variation for free amino acids of raw potato tubers of a diversity panel of tetraploid clones and to assess the effect of field location and year ii) understand the genetic basis of free amino acids using GWAS, and iii) generate GEBVs via GS to facilitate the selection of parents and advancement of clones through the breeding pipeline.
Results
Phenotypic variation for free amino acids in potato
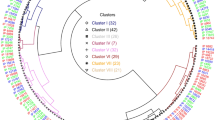
The diversity panel (217) of cultivated potato clones evaluated in Dalhart, Texas in 2019 and 2020, and Springlake, Texas in 2020 showed phenotypic variation for the relative amounts of all 19 free amino acids in potato tubers. The phenotypic distributions of amino acid content were normal or close to normal in most cases (Supplementary Fig. 1). Histidine, arginine, asparagine, aspartic acid, alanine, proline, valine, leucine, and phenylalanine distributions differed significantly from normal per Shapiro–Wilk test69 (p < 0.05) (Supplementary Fig. 1). Hierarchical cluster analysis of free amino acids reflected the metabolic relationship between amino acids (Fig. 1).
For all traits evaluated, the analysis of variance indicated significant differences between clones (Supplementary Table 1). The interactions between clones and environments were significant for most amino acids, except arginine, asparagine, glutamine, threonine, glycine, and lysine. Analysis of variance from the two Texas locations (Dalhart and Springlake) in 2020 indicated that most tuber amino acids were not affected by growing location, except histidine and proline (Fig. 2 and Supplementary Table 2), which were significantly lower (− 59.0%) and higher (+ 129.0%), respectively, at Springlake, TX (a location that regularly suffers from abiotic stresses, mainly high-temperature stress). The average histidine content in tubers at Springlake was 0.5%, whereas tubers harvested from the Dalhart location had an average of 1.2%. In relation to proline, the average in Springlake was 1.6%, whereas the value was 3.7% in Dalhart (Fig. 2). In Springlake, the lowest amount of histidine was observed in Russet Norkotah 112 (0.1%), and the highest was found in ATTX98453-3R (1.4%). In Dalhart, the lowest amount of histidine was observed in ATX08181-5Y/Y (0.1%) the highest was found in ATX9332-8Ru (2.2%) (Supplementary Table 2). Regarding proline, in Springlake, the lowest value was obtained in TX05249-11W (1.3%), and the highest value was obtained in clone NDTX7590-3R (8.1%) (Supplementary Table 2). In Dalhart, the lowest value of proline was found in ATX9332-8Ru (0.5%), and the highest value was observed in AORTX09037-1W/Y (4.6%) (Supplementary Table 2). Analysis of variance with data from 2 years (2019, 2020, Dalhart location) revealed no significant differences between years, but the interaction between clone and year was significant for histidine, serine, and proline (Supplementary Table 1).
Box-plots for histidine (A) and proline (B) content in raw potato tubers diversity panel from the two locations (Springlake and Dalhart, TX). The amino acid means of tubers from the diversity panel (217 clones) at each location are represented by bold horizontal lines through the box-plot. Different letters above the boxplots denote significant differences between the means of the two locations based on Student t-test (p < 0.05).
Across the diversity panel, asparagine and glutamine were the most abundant amino acids (33.0%, and 21.0%, respectively). In contrast, histidine and glycine were the least abundant, with average relative compositions of (0.8%, and 0.9%, respectively) (Table 1). Asparagine was highest in the clone NDTX060700C-1W (55.0%) and lowest in the clone COTX94216-1R (21.0%) (Supplementary Table 3). The clone ATTX01178-1R had the highest concentration of glutamine (32.0%), whereas COTX08365F-3P/P had the highest concentration of valine (10.7%) (Supplementary Table 3). The highest proportion of methionine (2.5%) was found in the clone ATX9130-1Ru, whereas COTX03134-1Y/W had the highest concentration of lysine (5.6%) (Supplementary Table 3).
Significant variation was observed between potato market groups for various free amino acids (Supplementary Table 4). Histidine was significantly higher in purples than in chippings and russets potatoes. The reds had a significantly lower asparagine (30.0%) than chippers (38.9%) and yellows (35.0%). Glutamine was significantly higher in chippers and phenylalanine in reds than in other market groups. Methionine was significantly higher in russets (1.6%) than in chippers (1.4%), yellows (1.4%), and purples (1.3%) (Supplementary Table 4). No significant differences were found between market groups for aspartic acid and lysine contents.
The diversity panel of clones was subjected to principal component analysis (PCA) based on free amino acid percentages and separation between fresh and processed (mainly chippers, since most of the russets included in the diversity panel belong to the fresh or dual-purpose) potato market classes. The first two principal components accounted for 47.9% of the total variance in the data, of which first principal component (PC1) explained 37.0% and second principal component (PC2) explained 10.9% of this variance (Fig. 3). Asparagine, isoleucine, valine, serine, alanine, phenylalanine, tyrosine, and glycine contributed the most to the PC1 and PC2 as observed by the intense red color in the contributions scale (Fig. 3, Supplementary Fig. 2).
Principal component analysis separating fresh (specific gravity < 1.069) and processing (specific gravity > 1.070) potato market classes based on the free amino acid (His = histidine, Arg = arginine, Asn = asparagine, Gln = glutamine, Ser = serine, Glu = glutamic acid, Asp = aspartic acid, Thr = threonine, Gly = glycine, Ala = alanine, Pro = proline, Lys = lysine, Tyr = tyrosine, Met = methionine, Val = valine, Ile = isoleucine, Leu = leucine, Phe = phenylalanine) composition of 217 potato clones. Each point represents the projection of an individual potato clone in the PC1 and PC2 axes, and the dot colors represent the market group (purple = fresh and cyan = processing). The ellipses represent 95% confidence intervals around the centroid of each data cluster. The contributions (%) of free amino acids to PC1 and PC2 from low to high are indicated by blue to red color gradient, respectively.
The relationships among free amino acids in potato tubers were examined through a correlation network (Fig. 4). The correlation coefficient threshold for the network was set at 0.5, with p-values less than 0.05. The network consisted of 12 connected amino acids, and correlation coefficients were positive except with asparagine. Asparagine had the highest number of connections (nodal degree), followed by valine, tyrosine, valine, and serine, indicating its centrality to the network. The most abundant amino acid, glutamine, had a low level of connection. The connection between lysine, phenylalanine, and threonine was also poor. Valine had the highest positive and significant correlation with isoleucine (r = 0.80). Significant negative correlations were found between asparagine and alanine (r = − 0.71), valine (r = − 0.68), and glycine (r = − 0.67). Broad sense heritability values of the relative levels of free amino acids ranged from 0.58 to 0.89 for most amino acids except histidine, 0.15 (Table 1).
Correlation network of the free amino acid composition based on Pearson’s correlation matrix obtained based on the evaluation of 217 potato clones in Dalhart, Texas in 2019 and 2020, and Springlake, Texas in 2020. The correlation coefficient threshold was set at 0.5 with a p-value of less than 0.05.
Average tuber weight per plant and average weight per tuber were significantly lower at Springlake, Texas compared to Dalhart, Texas. The correlations were examined and weak and/or moderate correlations were obtained. Interestingly, proline was negatively correlated with average tuber weight per plant and average weight per tuber (Table 2). Histidine was positively correlated with average tuber weight per plant and average weight per tuber (Table 2). There was no significant correlation between proline and average tuber number per plant. Also, no significant correlation was found between histidine and average tuber number per plant. The correlations of other amino acids with yield components were also examined. Negative correlation was observed for aspartic acid and proline with tuber weight (Supplementary Table 5). Likewise, positive correlations were observed for lysine, tyrosine, and methionine with tuber weight (Supplementary Table 5).
Linkage disequilibrium (LD)
Using 10,106 SNPs for the full panel, pairwise LD was assessed using the squared-allele frequency correlations (r2), and LD was found to be low even at zero distance (Supplementary Fig. 3). The function LD.plot was used to illustrate the amount of LD in the panel. The 5–10 Mb window size seems appropriate to filter the output such that just the most significant marker inside a given window is returned based on the curve’s shape.
Genome-wide association mapping—quantitative trait loci identification
We examined the inflation of the − log10(p) using quantile–quantile (QQ) plots of the observed vs. expected values under the null hypothesis shown in Supplementary Fig. 4. Strongly associated SNPs deviated from the plot’s diagonal at the upper-right end. A plot of the − log10(p-value) of the association statistic on the y-axis versus the chromosomal position of the SNP on the x-axis, commonly known as Manhattan plots, gave the visualization of the GWAS results (Fig. 5). Along the plot, regions with many highly linked SNPs in linkage disequilibrium emerged as the “Manhattan skyline”.
Manhattan plots displaying the marker–trait associations for Histidine (A), Arginine (B), Asparagine (C), Glutamine (D), Serine (E), Aspartic Acid (F), Threonine (G), Glycine (H), Alanine (I), Valine (J), Isoleucine (K), Leucine (L), Phenylalanine (M) by GWASpoly using the additive and dominant model in three combined environments. The horizontal axis indicates the chromosome number and the position of each SNP. The vertical axis indicates the negative logarithm of the p-value for each SNP. Each dot signifies an SNP. The broken line indicates the Bonferroni threshold level of 0.05.
Allele dosage and different inheritance models were considered in GWASpoly58. The additive inheritance model identified nine SNPs associated with eight free amino acids. The simplex dominant models identified seven (1-dom ref) and eight (1-dom alt) SNPs associated with eight and nine free amino acids, respectively (Table 3). The duplex dominant models identified six (2-dom ref) and five (2-dom alt) SNPs associated with six and five free amino acids, respectively (Table 3).
A total of 33 QTL were significantly associated with 13 free amino acids on chromosomes 1, 2, 3, 5, 6, 7, and 10 (Table 3). However, no QTL were detected above the threshold for five amino acids, which are glutamic acid, proline, lysine, tyrosine, and methionine. A QTL on chromosome 6 was identified for histidine in the additive and the dominant model. GWAS analysis of arginine content identified four QTL in chromosome 2. Five QTL were identified for asparagine content in chromosomes 2 and 7. Two QTL were identified for glutamine content in chromosomes 3 and 6. Four QTL were identified for serine content in chromosomes 2, 5, and 7. For aspartic acid content, the only QTL identified was in chromosome 1. For threonine content, two QTL were identified in chromosome 1. Three QTL were identified for glycine content in chromosome 2. Three QTL were identified for alanine content in chromosome 2. Four QTL were identified for valine content in chromosomes 2 and 3. One QTL was identified for isoleucine content in chromosome 2. A QTL was observed in chromosome 5 for leucine content, which explained 3.6% of the phenotypic variance. Two QTL were identified for phenylalanine content on chromosomes 2 and 10 under additive and dominant genetic models, respectively. QTL co-localization was discovered for a few amino acids regardless of the inheritance model tested. Asparagine, glycine, and serine showed two colocalizing QTL on chromosome 2 (SNP peak positions at 25.6 Mb and 26.6 Mb), and isoleucine and valine shared a QTL in another region of chromosome 2 (SNPs peak position at 27.9 Mb). The position of the QTL, SNP at the peak and the phenotypic variance explained by each QTL are reported in Table 3.
The genes located in the 100 kbp (kilobase pair) flanking genomic region of each peak SNP were retrieved from the potato reference genome (Supplementary Table 6). The selection of a 100 kbp interval is a commonly used size range when investigating candidate genes. Gene annotation of DM version 4.03 and UniProt (www.uniprot.org) were used to evaluate these candidate genes’ putative function. Several of the genes (around 150 candidate genes) had unknown functions (Supplementary Table 6). However, a few potential genes directly linked to primary metabolic pathways were found in our investigation. A proenzyme, S-adenosylmethionine decarboxylase proenzyme (SAMDC) (PGSC0003DMG400010051), was identified as part of the arginine metabolism. Another potential gene, proline oxidase/dehydrogenase (PGSC0003DMG400010050), was found for proline metabolism (Supplementary Table 6). Given the degree of linkage disequilibrium in potatoes, it cannot be ruled out that genes regulating amino acids could be located megabases away from important SNPs.
Genomic selection – genomic-estimated breeding values for traits
Genomic estimated breeding values (GEBVs) can help guide parental selection and the advancement of superior clones in breeding programs. Box plot of the predicted reliabilities (the squared correlation between the true and predicted values) showed that most of the predicted reliabilities are higher than 0.5 for most of the free amino acids content except histidine (Fig. 6). The GEBVs (Supplementary Table 7) showed that clone TX12474-1P/R ranked topmost for histidine. NDTX060700C-1W ranked topmost for asparagine and aspartic acid. Considering the GEBVs, the topmost was BTX1544-2W/Y for arginine, NDTX050169-1R for glutamine, and COTX00104-7R for serine content. BTX1544-2W/Y came out on top for arginine, whereas NDTX050169-1R came out on top for glutamine. COTX00104-7R had the highest GEBV for serine content, and AOTX97213-1Ru had the highest GEBV for glutamic acid. ATTX96746-1R came out on top for threonine, and Krantz came on top for glycine. COTX05211-4R ranked topmost for alanine, and NDTX081648CB-4W ranked topmost for proline. COTX04015-3W/Y had the highest GEBVs for lysine. COTX05261-1R/Y ranked topmost for tyrosine, and ATX9130-1Ru ranked topmost for methionine. Based on the GEBVs, ATTX98500-3P/Y came on top for valine. The topmost clone for isoleucine was ATTX98510-1R/Y, leucine was ATTX03516-2R, and phenylalanine was ATTX10265-4R/Y.
Box plot of the predicted reliabilities (the squared correlation between the true and predicted values) for free amino acids content (His = histidine, Arg = arginine, Asn = asparagine, Gln = glutamine, Ser = serine, Glu = glutamic acid, Asp = aspartic acid, Thr = threonine, Gly = glycine, Ala = alanine, Pro = proline, Lys = lysine, Tyr = tyrosine, Met = methionine, Val = valine, Ile = isoleucine, Leu = leucine, Phe = phenylalanine) in 217 potato clones.
Weighted standardized multitrait selection indexes
Chipping clones with all other amino acids rich but with low asparagine was found using weighted normalized multitrait selection indexes (Supplementary Table 8). Three clones (NDTX059828-2W, ATTX95490-2W and AOTX95309-2W) were found to have the highest rankings (Z-score > 2) among the chipping potatoes (Supplementary Table 8).
Discussion
This study uncovered the natural variation for various free amino acids in a potato diversity panel from a potato breeding program in the USA54 and linked phenotypic variation for several traits to genetic markers to understand the genetic basis of the traits and obtain genomic-estimated breeding values to improve breeding efficiencies. The panel of potato clones used included reference varieties for various market groups and advanced selections of cultivated tetraploid potatoes from the Texas A&M Potato Breeding Program, developed over 40 years. The outcome of this research should guide breeding efforts in potatoes to improve health, nutritional, and processing qualities.
Significant differences were found between tetraploid potato clones for the relative quantity of all 19 free amino acids evaluated. Variations in the amounts of free amino acids were found in potato tubers from different growing locations and years70. Asparagine and glutamine were the most abundant amino acids, with average relative compositions of 33.0% and 21.0%, respectively in our potato diversity panel. Asparagine levels showed great variation in the panel, from 21.0 to 55.0%. The most prevalent amino acids in potato tubers were asparagine and glutamine, which together made up to 90.0% of the total free amino acid content10. In another study, asparagine accounted for the greatest proportion of free amino acids (up to 30.0–36.0%)71. Asparagine and reducing sugars form acrylamide, a carcinogenic chemical that is a concern, especially in processed potato products28. Both asparagine and reducing sugars should be reduced to decrease acrylamide content in processing potatoes42. According to this study, targeted breeding efforts would be required to reduce the asparagine content of conventionally bred potatoes to the range required for acrylamide mitigation. Since potatoes had never been bred for low tuber asparagine content, the effectiveness of this strategy was questionable42. Our study found that the chipping (processing) market group had significantly higher asparagine content in raw tubers than other market groups (Supplementary Table 4). A previous study showed a negative correlation between tuber sucrose and amino acid content45. It reported that tubers with a reduced sucrose transporter expression displayed significant increases in alanine, arginine, asparagine, glutamine, glycine, serine, threonine, and valine45. In selecting chip potatoes with low-reducing sugars and cold-sweetening resistance, these breeding efforts may have favored indirect selection for higher accumulation of free amino acids including asparagine, but additional research is needed to confirm this relationship. Some research has shown that asparagine content in potatoes is larger and less variable than glucose and fructose, which helps to explain why reducing asparagine could be challenging22. On the other hand, researchers have shown that specific genotypes with low asparagine contents can be exploited in breeding efforts to lower tuber asparagine content41,72.
Ideally, breeders should aim for the identification and development of processing potato cultivars with low free-asparagine and reducing sugar as desirable characteristics for processing purposes. Recently, in a processing cultivar, FL-1533, a distinctive metabolite combination was found to have the least amounts of reducing sugar and asparagine compared to the other cultivars73. The availability of such germplasm would make it possible to introduce the low sugar and low asparagine traits into varieties acceptable for broad use in the food industry. The implications for low asparagine on potato yield, disease suceptibilty, starch properties etc. are limited but it would be worth exploring in the future. However, previous studies indicated that certain amino acids affect the physicochemical properties of potato starch74. Charge-carrying amino acids (lysine, arginine, aspartic acid, and glutamic acid) can potentially alter the physical and chemical characteristics of starch-based products derived from potatoes74. Increased amounts of charge-carrying amino acids during heating can lead to notable improvements in their nutritional qualities, achieved by reducing the swelling power, solubility, light transmittance, and gel strength of the potato starch74. Another study found that amino acids, particularly glutamic acid and aspartic acid, can affect the gelatinization, pasting, and rheological properties of potato starches75, potentially resulting in effects on food texture and mouthfeel, and influencing consumer acceptance.
Another essential amino acid, methionine, was significantly higher in russets and lowest in purples (Supplementary Table 4). The potato industry is interested in boosting the tuber methionine content because of the related nutritional and fragrance benefits32. Even though methionine is an important amino acid, no related QTL were found in the analysis. Increasing the sample size and studying more diverse populations could help capture a broader range of genetic variation and improve the chances of identifying relevant associations. If unearthing natural variation becomes a limitation, the biotechnological interventions to manipulate methionine metabolism is a parallel strategy. Lysine’s average relative composition in the diversity panel was 3.3%, and the clone with the highest proportion of lysine (5.6%) was COTX03134-1Y/W. Potato is a good source of lysine and thus can be used as a supplement to cereals deficient in this amino acid76.
Despite using similar production practices across the Texas potato field locations, the analysis of variance indicated significant environmental effects when comparing locations for histidine and proline content. On average, tubers produced at the Springlake location have lower histidine and higher proline content than those produced in Dalhart. Quantitative differences in the amino acid content between varieties based on location were previously reported77. Proline was the amino acid that varied the greatest between the two regions which differed in rainfall and humidity77. The Springlake and Dalhart locations differ for altitude, latitude, and longitude; day/night temperatures, photoperiod, radiation, type of soil, and biotic and abiotic stresses. The Springlake location regularly has higher night temperatures (often > 20 °C near harvest), resulting in lower marketable yield and more external and internal tuber defects as result of having more heat stress than the Dalhart location. Experiments under controlled greenhouse conditions showed that heat stress-induced chemical changes in potato tubers78. Proline is crucial for plant growth and differentiation throughout the whole life cycle and also serves as a superior osmolyte and also performs three other crucial functions under stress, including metal chelation, antioxidant defense, and signaling79. To protect plants from heat stress, proline functions as a signaling molecule80. Our findings indicated that proline levels in tubers produced at Springlake, a location suffering abiotic stresses (mainly heat stress), were significantly higher (129% higher) than a more favorable location (Dalhart) that, despite having hot days during the day, has more cool nights that favor the accumulation of starch in tubers and less external and internal tuber defects. Proline is a precursor for other downstream metabolites, including polyamines and secondary metabolites and thus may affect tuber quality. Proline can be produced from arginine and glutamine/glutamate in the majority of animals, including humans and pigs, although rates of endogenous synthesis are insufficient for newborns, fish, and birds81, thus consuming proline-rich produce should be beneficial to contribute to the production of proteins in general, and collagen in particular, and thus help in the formation of skin, cartilages, and bones. Likewise, histidine is an essential amino acid for plant growth and development82. Our findings showed lower level of histidine in tubers in the stressful site Springlake. This result conflicts with the amount of free histidine present in canola cultivars, as histidine, serine, and cysteine were found to rise in the xylem sap of various canola cultivars during nickel toxicity83. Further research is needed to fully understand if stress induces low histidine in tubers.
In our study, years did not significantly affect the content of free amino acids in tubers whereas location had a significant effect in histidine and proline. While breeding for high (or low) amino acids, multilocation evaluations would be more important than multiyear evaluations.
In this study, branched-chain amino acids (isoleucine and valine) displayed high correlation values (r = 0.8). Aromatic amino acids (i.e., tyrosine and phenylalanine) also highly correlated (r = 0.7). High correlations were anticipated due to linked pathways. Previous studies in potatoes reported r > 0.7 between branched-chain amino acids46 and r = 0.7 between serine and glycine84. However, in our research, unexpected correlations were also observed between amino acids that are not closely linked biosynthetically (e.g., phenylalanine and isoleucine, alanine and serine, and alanine and glycine). In previous metabolic profile studies of potato tubers, unexpected amino acid associations were also discovered46,84,85. Asparagine and other amino acids were found to have significant negative correlations, suggesting that decreasing asparagine would probably increase the other amino acids involved in the correlations and vice versa.
In this study, proline content was negatively correlated and histidine content was positively correlated with yield components but the correlations were weak. The weaker correlations would likely make them unsuitable to use as markers for yield potential. The relationship between proline and histidine content and potato yield components are limited. However, the relationship may vary depending on several variables, including the type and degree of stress, the potato cultivar, and other environmental factors. In wheat, grain proline content during a drought was linked to greater thousand grain weight loss and does not seem to act positively in terms of productivity86.
Thanks to recent developments in high-throughput metabolic profiling and sequencing technologies, GWAS has been used as a powerful strategy to unveil crop metabolism’s genetic and biochemical underpinnings. GWASpoly is suited for conducting GWAS in polyploids as it investigates different inheritance models58. For example, in this study, nine SNPs were shown to be linked to eight free amino acids with the additive inheritance model. Each additional copy of the allele can boost the trait’s output. Using GWASpoly, we identified 33 QTL associated with 13 free amino acids in this study. In general, only a small portion (< 10%) of the phenotypic variance could be accounted by each QTL, except for arginine (10.5%). Co-localization of QTL for free amino acids was observed in a few cases independently of the model of inheritance investigated. It is not unusual to see colocalized QTL for biochemically related amino acids. For example, in the present study, isoleucine and valine had the strongest, most positive, and most significant correlation (r = 0.80), and shared a QTL in chromosome 2 (SNPs peak position at 27.9 Mb). Previous research on potatoes has shown a high interdependence of biochemically related amino acids46. A shared QTL could come from closely linked independent regulators or a common upstream regulator46 for amino acids that are not biochemically related. QTL linked to various amino acids can potentially improve multiple amino acid content in potatoes simultaneously, and they deserve further investigation.
By assigning potential candidate genes to GWAS outcomes based on metabolites, it is postulated that the candidate loci directly or indirectly encode intermediate products that would biochemically catalyze, modify, translocate, or regulate the associated metabolites- which thus warrants systemic evaluation to link them to end phenotypes. The success of GWAS in mapping genes is limited in its resolution due to the high level of LD in the breeding germplasm, annotation of the reference genome, and the number of high-quality SNPs. This method provides only indirect evidence for the association of the genomic region with individual amino acids. We anticipate continuing additional research to validate the functional role of these genes in using model species or transgenic approaches. Nevertheless, this study has established a foundation to analyze identified loci and their evolutionary significance in amino acid metabolism in potatoes. In our study, the genes within 100 kbp window size were identified according to the positions of the closest flanking significant SNPs. Several independent studies have used genomic space within ± 100 kb of each peak SNP proximity to identify candidate genes associated with varied traits using association mapping in potato87,88. Expanding the search interval to 100 kbp increases the likelihood of capturing genetic variants in LD with the causal variant, improving the chances of identifying relevant candidate genes. In some instances, narrower or broader intervals may be considered, depending on the context and objectives of the study.
Our analysis did uncover a few candidate genes that are directly connected to major metabolic processes. Based on the KEGG pathway (SOT 102,589,057), S-adenosylmethionine decarboxylase proenzyme (SAMDC)—PGSC0003DMG400010051 was mapped to arginine metabolism. S-adenosylmethionine decarboxylase synthesizes polyamines putrescine, spermidine, and spermine across species89. Arginine is the ornithine precursor, the substrate needed for polyamines biosynthesis. Ornithine decarboxylase converts ornithine to putrescine, which is further converted to spermidine and spermine by consuming decarboxylated S-adenosylmethionine90. Transgenic potato plants expressing antisense SAMDC showed stunted phenotypes, reduced levels of SAMDC transcripts and enzyme activity, and polyamine content, suggesting the significance of polyamines in the tuber formation91. Additionally, GWAS of sorghum grain qualities identified S-adenosylmethionine decarboxylase proenzyme mapped to arginine, cysteine, methionine, proline, and starch metabolism92. It will be intriguing to perform additional genetic characterization to understand the role of this gene in polyamine synthesis or, like sorghum, its relevance in starch synthesis in potatoes. Another candidate gene, Proline oxidase/dehydrogenase (PGSC0003DMG400010050), catalyzes proline oxidation to Δ1—Pyrroline—5-Carboxylate Synthetase (P5C) in the mitochondria. Proline and arginine metabolic pathways are interconnected93 as the second and last step in proline catabolism, P5C to glutamate is shared with arginine catabolism94. Arginine can serve as a precursor and product of ornithine during the urea cycle via arginase, converting arginine to ornithine95. Our study identified limited candidate genes directly associated with primary metabolic pathways, which is plausible given the complexity of their biosynthesis and catabolism involving several intermediates, their transport, and feedback regulations. Detailed characterization of identified genes using functional approaches such as cloning and heterologous expression of genes in model species, tissue-specific knocking out genes using RNAi, antisense methods, or CRISPR-Cas would help understand their relevance in amino acid metabolism in potatoes.
GEBV was obtained for all members of the diversity panel (217 clones). GEBVs will allow faster and more effective selection of superior parents and/or the advancement of better clones. The predicted reliabilities in this study were encouraging, except for histidine. Prediction ability is influenced by the genetic make-up of the traits under investigation, the training population, and how similar the training population is to the validation population. Recently, software programs like StageWise96 are being developed for polyploid crops to manage the numerous experimental designs, heritabilities, and spatial models for nongenetic variation. Individual GEBVs will likely not be used for selection since there are many important traits to consider in selection. To use GEBVs for selection, GEBVs for several traits might be merged into an overall index. In this study we utilized the standardized indexes for the chipping clones. Not all traits will have the same importance, and not all traits are measured using the same units or scales; thus, standardized scores and relative weight should be assigned to each trait to facilitate the selection of superior and inferior genotypes97. Market demands, industry needs, and several other considerations drive the relative importance of traits. Using GEBVs to select potatoes with particular concentrations could significantly improve potatoes’ health and nutritional value. GEBVs for traits such as yield, quality, disease/pest resistance, and tolerance to abiotic stresses should be part of the combined selection index to maximize the chances of selecting the best clones and make selection more efficient. For practical applications, GEBVs should be considered in the context of market groups and selection criteria.
Materials and methods
Plant materials and experimentation
A collection of 217 tetraploid potato clones was used for this study. The diversity panel represents varieties and advanced genotypes selected over four decades by the Texas A&M Potato Breeding Program. Details of the potato diversity panel used were described earlier54. The clones included fresh and processing market classes with variations in tuber shape, skin type, skin and flesh color, biotic and abiotic stress tolerance, and quality traits54.
Three field experiments were conducted at two locations in Texas in commercial potato grower fields. In 2019, the diversity panel was planted in Dalhart (35°58′ N, 102°44′ W), and in 2020 the clones were planted in Dalhart and Springlake (34°6′ N, 102°19′ W) in a 12-hill plot with two replications. Dalhart is approx. 268 km North of Springlake and 92 m higher. Both Texas locations experience high-temperature stress during the growing season, however, Dalhart yields often duplicate the yields in Springlake (https://potato.tamu.edu/reports/), likely due to cooler nights at Dalhart. Trials were planted in early May in Dalhart, Texas, and harvested in early September. At Springlake, trials were planted in late March to early April and harvested in early July. Vine desiccation was done 2–3 weeks before harvesting the trials. Standard production practices were used. Details about the field trials, including environmental conditions, spacing, irrigation, fertilization, weed, and insect control, can be obtained online for the corresponding years and locations from the 2019 and 2020 Texas A&M Potato Breeding Program reports (https://potato.tamu.edu/reports/).
DNA extraction and genotyping
Genomic DNA was extracted from 50 to 80 mg of fresh young potato leaves from tissue culture plantlets using the DNeasy Plant Pro Kit (Qiagen, Valencia, CA, USA) as described earlier54. Samples were analyzed using the Infinium 22 K V3 Potato Array on the Illumina iScan (Illumina Inc., San Diego, CA, USA). 10,106 polymorphic SNPs were selected for analysis after filtering the marker dataset for minor allele frequency and polymorphism, as previously described54.
Measurements of free amino acid content
Three tubers per plot were randomly selected and chopped into small cubes (4 mm per side). About 15 g of freshly chopped samples were collected into 50 mL tubes. Samples were stored in −80 C freezer for a few days before freeze-drying them using a Labconco FreeZone 6-L Freeze-dry System (Labconco, Kansas City, MO, USA) at a collector temperature of − 50 °C for 120 h at 22 Pa. Ceramic grinding cylinders (0.95 cm × 2.22 cm) with angle-cut ends (SPEX SamplePrep, Metuchen, NJ, USA) were used for homogenizing the dry tuber sample in 50 mL centrifuge tubes using SPEX Sample Prep 1600 MiniG tissue homogenizer (SPEX SamplePrep, Metuchen, NJ, USA) for 2 min set at a speed of 1500 strokes per minute. Powdered samples were stored at room temperature in tightly closed Minigrip® Red Line zip bags (Minigrip, Alpharetta, GA) until use.
Free amino acids from samples of potato tubers were extracted from two biological replications per environment (location-year combination) and three technical replicates per biological replication using an established protocol98,99. The Kairos™ Amino Acid Kit was used for amino acid calibration (Waters Corp., Milford, MA, USA). Waters Acquity UPLC H-class system with a Waters Xevo TQ mass spectrometer with an electrospray ionization (ESI) probe was used to detect amino acids. Each amino acid’s multiple reaction monitoring (MRM) transition, collision energy values, and cone voltage were optimized using IntelliStart software (Waters Corp., Milford, MA, USA). For instrument monitoring and data capture, Water’s MassLynxTM software was used. The TargetLynxTM Application Manager (Waters Corp., Milford, MA, USA) was employed for data integration, calibration curves, and amino acid quantification. The relative composition of free amino acid levels (each as a percentage of total free amino acids, e.g., alanine /total) was determined.
Statistical analyses
Descriptive statistics (means, minimums, and maximums) were calculated for free amino acids using the software package META-R100. Phenotypic distributions for the traits were generated and normality tests were conducted by the Shapiro–Wilk W-Test69 using JMP Pro 16® statistical software (SAS Institute, Cary, NC). The free amino acids were separated into groups using a hierarchical clustering analysis using Euclidean distance metric and Ward’s minimal variance method. The effect of location, years, and environment (considered as location-year combination: Dalhart 2019, Dalhart 2020, and Springlake 2020), clone (217 clones), and the corresponding individual amino acid content of raw potato tubers were examined using analysis of variance using a mixed model in JMP Pro 16. Clones were considered fixed effects, while environment (location-year combination), replication within environments, and interactions were considered random.
Similarly, an analysis of variance was used to determine the effect of location (two field sites) on the traits (Dalhart and Springlake field trials conducted in 2020 were used). In this case, clones and locations were treated as fixed effects. Data from 2 years (2019, 2020) from one location (Dalhart) were used to ascertain the impact of years on the traits evaluated. Pearson’s correlation coefficients (r) were obtained, and a correlation network was built to examine and illustrate the relationships among free amino acids. A correlation coefficient threshold was set at 0.5 with a p-value of less than 0.05 to visualize the stronger correlations between free amino acids. A significant correlation matrix was generated using the R program (version 4.1.2), and the network visualization was performed with Cytoscape 3.9.1101, using a previously reported method102. Also, principal component analysis (PCA) was completed using the function prcomp in the stats package and plotted using the function ggbiplot103 and factoextra package (https://github.com/kassambara/factoextra). Confidence ellipses were included, representing 95% confidence intervals around the centroid value of each data cluster. The function fviz_cos2 was used to visualise the quality of representation (cos2) of the variables in the principal components. Pearson correlations (r) between selected amino acid and yield-related traits104 evaluated in Texas (Dalhart in 2019 and 2020, and Springlake in 2020) were also calculated.
Genome-wide association analysis (GWAS)
Association analysis was performed for free amino acids with 10,116 SNPs using the GWASpoly Version 2 package in R58. The leave-one-chromosome-out (LOCO) method105 was used to account for population structure. For each trait, additive and dominant genetic models were tested. A LOD threshold for each trait, corresponding to a genome-wide false-positive rate of 5%, was established based on a Bonferroni test. Manhattan plots were generated using the GWASpoly package. GWASpoly estimated the proportion of phenotypic variance explained by significant SNPs at the peak of each QTL. Contextual sequences of SNPs at the peak of QTL were used in BLAST searches of DM1-3 pseudomolecules (Version 4.03) in the SpudDB database (http://solanaceae.plantbiology.msu.edu/) to identify putative candidate genes located in the 100 kbp (kilobase pair) flanking genomic region.
Genomics selection (GS)—Genomic-estimated breeding values
Genetic variance partitioning and genome-wide prediction were performed using allele dosage information96,97,104,106. Standard errors and the best linear unbiased predictors (BLUP) were calculated using standard methods. For a generic random vector u, BLUP \(\left[\mathbf{u}\right]=\widehat{\mathbf{u}}=\mathbf{C}\mathbf{P}\mathbf{y}\), where \(\mathbf{C}=\mathrm{cov}[\mathbf{u},\mathbf{y}]\), \(\mathbf{P}={\mathbf{V}}^{-1}-{\mathbf{V}}^{-1}\mathbf{X}{\left({\mathbf{X}}^{\mathrm{^{\prime}}}{\mathbf{V}}^{-1}\mathbf{X}\right)}^{-1}{\mathbf{X}}^{\boldsymbol{^{\prime}}}{\mathbf{V}}^{-1}\), X is the incidence matrix for fixed effects, and V is the variance–covariance matrix of the response variable107. The reliability of \({\widehat{u}}_{i}\) is represented by the squared correlation with the true value which equals \({\mathrm{r}}_{\mathrm{i}}^{2}=\frac{\mathrm{var}\left[{\widehat{\mathrm{u}}}_{\mathrm{i}}\right]}{\mathrm{var}\left[{\mathrm{u}}_{\mathrm{i}}\right]},\) and \(\mathrm{var}\left[\widehat{\mathbf{u}}\right]=\mathbf{C}\mathbf{P}{\mathbf{C}}^{\mathbf{^{\prime}}}\).
Weighted standardized multitrait selection indexes
Standardized multi-trait selection indexes were calculated for all amino acids in the chipping class based on the Z values of GEBVs97. The weighted multi-trait selection index for each clone was calculated assigning positive weight for all amino acids except asparagine. Asparagine was assigned a negative weight because lower values of asparagine are preferred by the processing industry.
Data availability
The genotype datasets used during the current study are available in our previous article in Scientific Reports, https://www.nature.com/articles/s41598-021-87284-x#additional-information. All other data generated or analyzed during this study are included in this published article.
References
Beals, K. A. Potatoes, nutrition and health. Am. J. Potato Res. 96, 102–110 (2019).
Camire, M. E., Kubow, S. & Donnelly, D. J. Potatoes and human health. Crit. Rev. Food Sci. Nutr. 49, 823–840 (2009).
Woolfe, J. A. The Potato in the Human Diet (Cambridge University Press, 1987).
Muller, C., Pretorius, B. & Schönfeldt, H. C. Protein quality of South African potatoes to inform dietary choices. Potato Res. https://doi.org/10.1007/s11540-021-09538-5 (2022).
Hussain, M. et al. Potato protein: An emerging source of high quality and allergy free protein, and its possible future based products. Food Res. Int. 148, 110583 (2021).
McGill, C. R., Kurilich, A. C. & Davignon, J. The role of potatoes and potato components in cardiometabolic health: A review. Ann. Med. 45, 467–473 (2013).
Gorissen, S. H. M. et al. Protein content and amino acid composition of commercially available plant-based protein isolates. Amino Acids 50, 1685–1695 (2018).
Wegener, C. B., Jansen, G. & Jürgens, H.-U. Bioactive compounds in potatoes: Accumulation under drought stress conditions. Funct. Foods Health Dis. 5, 108–116 (2015).
Koch, W. et al. Reduced amino acid content in transgenic potato tubers due to antisense inhibition of the leaf H+/amino acid symporter StAAP1. Plant J. Cell Mol. Biol. 33, 211–220 (2003).
Brierley, E. R., Bonner, P. L. R. & Cobb, A. H. Aspects of amino acid metabolism in stored potato tubers (cv. Pentland Dell). Plant Sci. 127, 17–24 (1997).
Brierley, E. R., Bonner, P. L. R. & Cobb, A. H. Factors influencing the free amino acid content of potato (Solanum tuberosum L.) tubers during prolonged storage. J. Sci. Food Agric. 70, 515–525 (1996).
Jaynes, J. M., Yang, M. S., Espinoza, N. & Dodds, J. H. Plant protein improvement by genetic engineering: Use of synthetic genes. Trends Biotechnol. 4, 314–320 (1986).
Waglay, A., Karboune, S. & Alli, I. Potato protein isolates: Recovery and characterization of their properties. Food Chem. 142, 373–382 (2014).
Mottram, D. S. The Maillard reaction: Source of flavour in thermally processed foods. In Flavours and Fragrances (ed. Berger, R. G.) 269–283 (Springer Berlin Heidelberg, 2007). https://doi.org/10.1007/978-3-540-49339-6_12.
Nursten, H. E. Maillard reaction (The Royal Society of Chemistry, 2005). https://doi.org/10.1039/9781847552570.
Mottram, D. S., Wedzicha, B. L. & Dodson, A. T. Acrylamide is formed in the Maillard reaction. Nature 419, 448–449 (2002).
Stadler, R. H. et al. Acrylamide from Maillard reaction products. Nature 419, 449–450 (2002).
Muttucumaru, N. et al. Evidence for the complex relationship between free amino acid and sugar concentrations and acrylamide-forming potential in potato. Ann. Appl. Biol. 164, 286–300 (2014).
Parker, J. K. et al. Kinetic model for the formation of acrylamide during the finish-frying of commercial french fries. J. Agric. Food Chem. 60, 9321–9331 (2012).
Elmore, J. S. et al. Changes in free amino acids and sugars in potatoes due to sulfate fertilization and the effect on acrylamide formation. J. Agric. Food Chem. 55, 5363–5366 (2007).
Becalski, A. et al. Acrylamide in french fries: influence of free amino acids and sugars. J. Agric. Food Chem. 52, 3801–3806 (2004).
Amrein, T. M. et al. Potential of acrylamide formation, sugars, and free asparagine in potatoes: A comparison of cultivars and farming systems. J. Agric. Food Chem. 51, 5556–5560 (2003).
Chawla, R., Shakya, R. & Rommens, C. M. Tuber-specific silencing of asparagine synthetase-1 reduces the acrylamide-forming potential of potatoes grown in the field without affecting tuber shape and yield. Plant Biotechnol. J. 10, 913–924 (2012).
Rommens, C. M., Yan, H., Swords, K., Richael, C. & Ye, J. Low-acrylamide french fries and potato chips. Plant Biotechnol. J. 6, 843–853 (2008).
Zyzak, D. V. et al. Acrylamide formation mechanism in heated foods. J. Agric. Food Chem. 51, 4782–4787 (2003).
Elmore, J. S., Dodson, A. T., Briddon, A., Halford, N. G. & Mottram, D. S. The effects of storage on the formation of aroma and acrylamide in heated potato. in Controlling Maillard Pathways To Generate Flavors vol. 1042 95–109 (American Chemical Society, 2010).
De Wilde, T. et al. Influence of storage practices on acrylamide formation during potato frying. J. Agric. Food Chem. 53, 6550–6557 (2005).
Halford, N. G. et al. Concentrations of free amino acids and sugars in nine potato varieties: Effects of storage and relationship with acrylamide formation. J. Agric. Food Chem. 60, 12044–12055 (2012).
Ly, D. N. P., Iqbal, S., Fosu-Nyarko, J., Milroy, S. & Jones, M. G. K. Multiplex CRISPR-Cas9 gene-editing can deliver potato cultivars with reduced browning and acrylamide. Plants 12, 379 (2023).
Fennema, O. R. Food Chemistry 3rd edn. (CRC Press, 1996).
Kumar, P. & Jander, G. Concurrent overexpression of Arabidopsis thaliana cystathionine γ-synthase and silencing of endogenous methionine γ-lyase enhance tuber methionine content in Solanum tuberosum. J. Agric. Food Chem. 65, 2737–2742 (2017).
Huang, T., Joshi, V. & Jander, G. The catabolic enzyme methionine gamma-lyase limits methionine accumulation in potato tubers. Plant Biotechnol. J. 12, 883–893 (2014).
Rinder, J., Casazza, A. P., Hoefgen, R. & Hesse, H. Regulation of aspartate-derived amino acid homeostasis in potato plants (Solanum tuberosum L.) by expression of E. coli homoserine kinase. Amino Acids 34, 213–222 (2008).
Dancs, G., Kondrák, M. & Bánfalvi, Z. The effects of enhanced methionine synthesis on amino acid and anthocyanin content of potato tubers. BMC Plant Biol. 8, 65 (2008).
Kreft, O., Hoefgen, R. & Hesse, H. Functional analysis of cystathionine γ-synthase in genetically engineered potato plants. Plant Physiol. 131, 1843–1854 (2003).
Di, R. et al. Enhancement of the primary flavor compound methional in potato by increasing the level of soluble methionine. J. Agric. Food Chem. 51, 5695–5702 (2003).
Zeh, M. et al. Antisense inhibition of threonine synthase leads to high methionine content in transgenic potato plants. Plant Physiol. 127, 792–802 (2001).
Sverrisdóttir, E. et al. Genomic prediction of starch content and chipping quality in tetraploid potato using genotyping-by-sequencing. Theor. Appl. Genet. 130, 2091–2108 (2017).
Ahmed, S., Zhou, X., Pang, Y., Jin, L. & Bao, J. Improving starch-related traits in potato crops: Achievements and future challenges. Starch - Stärke 70, 1700113 (2018).
Carrillo, L. et al. Ectopic expression of the AtCDF1 transcription factor in potato enhances tuber starch and amino acid contents and yield under open field conditions. Front. Plant Sci. 14, 1010669 (2023).
Shepherd, L. V. T. et al. Variation in acrylamide producing potential in potato: Segregation of the trait in a breeding population. Food Chem. 123, 568–573 (2010).
Bethke, P. C. Progress and successes of the specialty crop research initiative on acrylamide reduction in processed potato products. Am. J. Potato Res. 95, 328–337 (2018).
Jansky, S., Navarre, R. & Bamberg, J. Introduction to the special issue on the nutritional value of potato. Am. J. Potato Res. 96, 95–97 (2019).
Friedman, M. Potato glycoalkaloids and metabolites: Roles in the plant and in the diet. J. Agric. Food Chem. 54, 8655–8681 (2006).
Roessner-Tunali, U. et al. De novo amino acid biosynthesis in potato tubers is regulated by sucrose levels. Plant Physiol. 133, 683–692 (2003).
Carreno-Quintero, N. et al. Cross-platform comparative analyses of genetic variation in amino acid content in potato tubers. Metabolomics 10, 1239–1257 (2014).
Galdón, B. R., Mesa, D. R., Rodríguez, E. M. R. & Romero, C. D. Amino acid content in traditional potato cultivars from the Canary Islands. J. Food Compos. Anal. 23, 148–153 (2010).
Synge, R. L. M. Free amino acids of potato tubers: A survey of published results set out according to potato variety. Potato Res. 20, 1–7 (1977).
Kloosterman, B. et al. From QTL to candidate gene: Genetical genomics of simple and complex traits in potato using a pooling strategy. BMC Genom. 11, 158 (2010).
Zhang, F., Qu, L., Gu, Y., Xu, Z.-H. & Xue, H.-W. Resequencing and genome-wide association studies of autotetraploid potato. Mol. Hortic. 2, 6 (2022).
Wilson, S. et al. Understanding the effectiveness of genomic prediction in tetraploid potato. Front. Plant Sci. 12, 672417 (2021).
Wang, F., Zou, M., Zhao, L., Xia, Z. & Wang, J. Genome-wide association mapping of late blight tolerance trait in potato (Solanum tuberosum L.). Front. Genet. 12, 714575 (2021).
Díaz, P., Sarmiento, F., Mathew, B., Ballvora, A. & Vásquez, T. M. Genomic regions associated with physiological, biochemical and yield-related responses under water deficit in diploid potato at the tuber initiation stage revealed by GWAS. PLoS ONE 16, e0259690 (2021).
Pandey, J. et al. Genetic diversity and population structure of advanced clones selected over 40 years by a potato breeding program in the USA. Sci. Rep. 11, 8344 (2021).
Khlestkin, V. K., Erst, T. V., Rozanova, I. V., Efimov, V. M. & Khlestkina, E. K. Genetic loci determining potato starch yield and granule morphology revealed by genome-wide association study (GWAS). PeerJ 8, e10286 (2020).
Klaassen, M. T. et al. Genome-wide association analysis in tetraploid potato reveals four QTLs for protein content. Mol. Breed. 39, 151 (2019).
Sharma, S. K. et al. Linkage disequilibrium and evaluation of genome-wide association mapping models in tetraploid potato. G3 Genes Genomes Genetics 8, 3185–3202 (2018).
Rosyara, U. R., De Jong, W. S., Douches, D. S. & Endelman, J. B. Software for genome-wide association studies in autopolyploids and its application to potato. Plant Genome 9, 1–10 (2016).
Hirsch, C. N. et al. Retrospective view of north american potato (Solanum tuberosum L.) breeding in the 20th and 21st centuries. G3 Genes Genomes Genetics 3, 1003–1013 (2013).
Levina, A. V., Hoekenga, O., Gordin, M., Broeckling, C. & De Jong, W. S. Genetic analysis of potato tuber metabolite composition: Genome-wide association studies applied to a nontargeted metabolome. Crop Sci. 61, 591–603 (2021).
Jayanty, S. S., Diganta, K. & Raven, B. Effects of cooking methods on nutritional content in potato tubers. Am. J. Potato Res. 96, 183–194 (2019).
Talley, E. A., Toma, R. B. & Orr, P. H. Composition of raw and cooked potato peel and flesh: Amino acid content. J. Food Sci. 48, 1360–1361 (1983).
Golan-Goldhirsh, A. Effect of the add-back process on the free amino acid pool of potatoes. Z. Für Lebensm.-Unters Forsch. 182, 29–32 (1986).
Jaswal, A. S. Effects of various processing methods on free and bound amino acid contents of potatoes. Am. Potato J. 50, 86–95 (1973).
Drapal, M. et al. Identification of metabolites associated with water stress responses in Solanum tuberosum L. clones. Phytochemistry 135, 24–33 (2017).
Choi, S.-H., Kozukue, N., Kim, H.-J. & Friedman, M. Analysis of protein amino acids, non-protein amino acids and metabolites, dietary protein, glucose, fructose, sucrose, phenolic, and flavonoid content and antioxidative properties of potato tubers, peels, and cortexes (pulps). J. Food Compos. Anal. 50, 77–87 (2016).
Muttucumaru, N., Powers, S. J., Elmore, J. S., Mottram, D. S. & Halford, N. G. Effects of water availability on free amino acids, sugars, and acrylamide-forming potential in potato. J. Agric. Food Chem. 63, 2566–2575 (2015).
Eppendorfer, W. H. & Eggum, B. O. Effects of sulphur, nitrogen, phosphorus, potassium, and water stress on dietary fibre fractions, starch, amino acids and on the biological value of potato protein. Plant Foods Hum. Nutr. 45, 299–313 (1994).
Shapiro, S. S. & Wilk, M. B. An analysis of variance test for normality (complete samples). Biometrika 52, 591–611 (1965).
Talley, E. A. & Porter, W. L. Chemical composition of potatoes. VII Relationship of the free amino acid concentrations to specific gravity and storage time. Am. Potato J. 47, 214–224 (1970).
Pęksa, A., Miedzianka, J., Nemś, A. & Rytel, E. The free-amino-acid content in six potatoes cultivars through storage. Molecules 26, 1322 (2021).
Bethke, P. C. & Bussan, A. J. Acrylamide in processed potato products. Am. J. Potato Res. 90, 403–424 (2013).
Datir, S. S. et al. Cold storage reveals distinct metabolic perturbations in processing and non-processing cultivars of potato (Solanum tuberosum L.). Sci. Rep. 10, 6268 (2020).
Cui, M., Fang, L., Zhou, H. & Yang, H. Effects of amino acids on the physiochemical properties of potato starch. Food Chem. 151, 162–167 (2014).
Wang, W., Chen, W., Yang, H. & Cui, M. Textural and rheological properties of potato starch as affected by amino acids. Int. J. Food Prop. 20, S3123–S3134 (2017).
Kowalczewski, P. Ł et al. The nutritional value and biological activity of concentrated protein fraction of potato juice. Nutrients 11, 1523 (2019).
Davies, A. M. C. The free amino acids of tubers of potato varieties grown in England and Ireland. Potato Res. 20, 9–21 (1977).
Gautam, S. et al. Raman spectroscopy detects chemical differences between potato tubers produced under normal and heat stress growing conditions. Front. Plant Sci. 14, 1105603 (2023).
Hayat, S. et al. Role of proline under changing environments. Plant Signal. Behav. 7, 1456–1466 (2012).
Iqbal, N., Fatma, M., Khan, N. A. & Umar, S. Chapter 28 - Regulatory role of proline in heat stress tolerance: modulation by salicylic acid. In Plant Signaling Molecules (eds Khan, M. I. R. et al.) 437–448 (Woodhead Publishing, 2019).
Wu, G. et al. Proline and hydroxyproline metabolism: implications for animal and human nutrition. Amino Acids 40, 1053–1063 (2011).
Ingle, R. A. Histidine biosynthesis. Arab. Book Am. Soc. Plant Biol. 9, (2011).
Ali, M. A., Ashraf, M. & Athar, H. R. Influence of nickel stress on growth and some important physiological/biochemical attributes in some diverse canola (Brassica napus L.) cultivars. J. Hazard. Mater. 172, 964–969 (2009).
Dobson, G. et al. Phytochemical diversity in tubers of potato cultivars and landraces using a GC–MS metabolomics approach. J. Agric. Food Chem. 56, 10280–10291 (2008).
Roessner, U. et al. Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13, 11–29 (2001).
Griffiths, C. A., Reynolds, M. P. & Paul, M. J. Combining yield potential and drought resilience in a spring wheat diversity panel. Food Energy Secur. 9, e241 (2020).
Parra-Galindo, M.-A., Piñeros-Niño, C., Soto-Sedano, J. C. & Mosquera-Vasquez, T. Chromosomes I and X harbor consistent genetic factors associated with the anthocyanin variation in potato. Agronomy 9, 366 (2019).
Jo, K. R., Choi, J.-G., Kwon, D.-H., Park, Y.-E. & Kim, S.-J. Revealing genetic variations associated with chip-processing properties in potato (Solanum tuberosum L.). Agronomy 13, 642 (2023).
Pegg, A. E., Xiong, H., Feith, D. J. & Shantz, L. M. S-Adenosylmethionine decarboxylase: Structure, function and regulation by polyamines. Biochem. Soc. Trans. 26, 580–586 (1998).
Pegg, A. E. Mammalian polyamine metabolism and function. IUBMB Life 61, 880–894 (2009).
Kumar, A., Taylor, M. A., Arif, S. A. M. & Davies, H. V. Potato plants expressing antisense and sense S-adenosylmethionine decarboxylase (SAMDC) transgenes show altered levels of polyamines and ethylene: antisense plants display abnormal phenotypes. Plant J. 9, 147–158 (1996).
Kimani, W., Zhang, L.-M., Wu, X.-Y., Hao, H.-Q. & Jing, H.-C. Genome-wide association study reveals that different pathways contribute to grain quality variation in sorghum (Sorghum bicolor). BMC Genom. 21, 112 (2020).
Anwar, A., She, M., Wang, K., Riaz, B. & Ye, X. Biological roles of ornithine aminotransferase (OAT) in plant stress tolerance: Present progress and future perspectives. Int. J. Mol. Sci. 19, 3681 (2018).
Forlani, G., Bertazzini, M., Zarattini, M. & Funck, D. Functional characterization and expression analysis of rice δ1-pyrroline-5-carboxylate dehydrogenase provide new insight into the regulation of proline and arginine catabolism. Front. Plant Sci. 6, 591 (2015).
Verslues, P. E. & Sharma, S. Proline metabolism and its implications for plant-environment interaction. Arab. Book Am. Soc. Plant Biol. 8, e0140 (2010).
Endelman, J. B. Fully efficient, two-stage analysis of multi-environment trials with directional dominance and multi-trait genomic selection. Theor. Appl. Genet. 136, 65 (2023).
Pandey, J., Scheuring, D. C., Koym, J. W., Endelman, J. B. & Vales, M. I. Genomic selection and genome-wide association studies in tetraploid chipping potatoes. Plant Genome 16, e20297 (2023).
Joshi, V. et al. Genome-wide association study and population structure analysis of seed-bound amino acids and total protein in watermelon. PeerJ 9, e12343 (2021).
Song, Q., Joshi, M., DiPiazza, J. & Joshi, V. Functional relevance of citrulline in the vegetative tissues of watermelon during abiotic stresses. Front. Plant Sci. 11, 512 (2020).
Alvarado, G. et al. META-R: A software to analyze data from multi-environment plant breeding trials. Crop J. 8, 745–756 (2020).
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Batushansky, A., Toubiana, D. & Fait, A. Correlation-based network generation, visualization, and analysis as a powerful tool in biological studies: A case study in cancer cell metabolism. BioMed. Res. Int. 2016, e8313272 (2016).
Wickham, H. Data analysis. In ggplot2: Elegant Graphics for Data Analysis (ed. Wickham, H.) 189–201 (Springer International Publishing, 2016).
Pandey, J., Scheuring, D. C., Koym, J. W. & Vales, M. I. Genomic regions associated with tuber traits in tetraploid potatoes and identification of superior clones for breeding purposes. Front. Plant Sci. 13, 952263 (2022).
Yang, J., Zaitlen, N. A., Goddard, M. E., Visscher, P. M. & Price, A. L. Advantages and pitfalls in the application of mixed model association methods. Nat. Genet. 46, 100–106 (2014).
Endelman, J. B. et al. Genetic variance partitioning and genome-wide prediction with allele dosage information in autotetraploid potato. Genetics 209, 77–87 (2018).
Searle, S. R., Casella, G. & McCulloch, C. E. Variance Components (Wiley, 2009).
Acknowledgements
The USDA-NIFA (Grant No. 2019-03814), Hatch Project TEX09647, and Texas A&M AgriLife Vegetable Strategic Initiative provided funding for this research project. We acknowledge potato breeding programs at USDA-ARS Centers (Beltsville, Maryland; Aberdeen, Idaho; and Prosser, Washington) and State Universities (University of Wisconsin, Colorado State University, University of Minnesota, and North Dakota State University) for contributing unselected potato families to the Texas A&M University Potato Breeding Program. Additionally, we thank Sanjeev Gautam, Nina Rau, Taylor Swoboda, Mike Jensen, Angel Chappell, and Mythreyi Jamadagni for technical assistance.
Author information
Authors and Affiliations
Contributions
J.P.: conceptualization, original draft, data collection, analysis, review, and editing; D.T. and M.J.: extraction and method development, analysis of amino acids; D.C.S. and J.W.K.: data collection, review, and editing; V.J.: conceptualization, funding acquisition, quantification of amino acid, analysis, review, and editing; M.I.V.: conceptualization, funding acquisition, data collection, analysis, review, and editing; All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pandey, J., Thompson, D., Joshi, M. et al. Genetic architecture of tuber-bound free amino acids in potato and effect of growing environment on the amino acid content. Sci Rep 13, 13940 (2023). https://doi.org/10.1038/s41598-023-40880-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-40880-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.