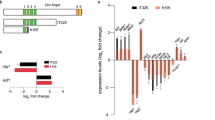
Abstract
Krüppel-like transcription factors (Klfs), which are characterized by the three conserved C-terminal zinc fingers, are involved in various biological processes, such as haematopoiesis and angiogenesis. However, how the Klf family of transcription factors cooperate in organogenesis remains elusive. During zebrafish embryogenesis, both klf1 and klf17 are expressed in the intermediate cell mass (ICM), where primitive erythroid cells are produced. Using CRISPR–Cas9 genome editing technology, we established klf1-klf17 double mutant zebrafish to investigate the functionally interactive roles of the klf1 and klf17 genes. The klf1-klf17 mutant exhibited a diminished number of circulating primitive erythroid cells at 2 days postfertilization (dpf), while klf1 or klf17 single mutants and wild-type embryos produced comparable numbers of primitive erythroid cells. Circulating erythroid cells from the klf1-klf17 mutant possessed larger nuclei at 2 dpf than wild-type cells, suggesting the impairment of primitive erythroid cell maturation. The expression of the erythroid cell maturation markers band3 and mitoferrin, but not the haematopoietic progenitor markers c-myb and scl, was decreased in the klf1-klf17 mutant at 1 dpf. Thus, these results illustrate the cooperative function of klf1 and klf17 in the maturation processes of zebrafish primitive erythroid cells.
Similar content being viewed by others
Introduction
The molecular mechanism underlying haematopoietic development is well conserved between zebrafish and mammals1. In fact, haematopoietic cells are produced from two consecutive waves of primitive and definitive haematopoiesis during vertebrate embryogenesis2. Although myeloid precursors originate from the anterior lateral mesoderm (ALM) during zebrafish primitive haematopoiesis, erythroid precursors originate from the posterior lateral mesoderm (PLM)1,3, which later becomes the blood island/intermediate cell mass (ICM). Erythroid precursors differentiate into primitive erythroid cells in the ICM and migrate into vascular vessels to circulate at approximately 25 h postfertilization (hpf), when the heartbeat begins. Haematopoietic stem cells (HSCs) in definitive haematopoiesis are derived from the aorta-gonad-mesonephros (AGM) and start to produce various differentiated blood cells, including definitive erythroid, myeloid and lymphoid cells, at approximately 3–5 days postfertilization (dpf) in the caudal haematopoietic tissue (CHT) and later in the kidney4.
Krüppel-like transcription factors (Klfs), which possess Cys2/His2 zinc fingers at the C-terminus that interact with a CACCC box in the promoter region of their target genes, function as key transcriptional regulators in various organogenesis processes, including haematopoiesis5. The Klf1 gene known as EKLF in mice is specifically expressed in the yolk sac blood island and foetal liver; both sites generate primitive erythroid cells6,7. Klf1-deficient mice die from β-thalassemia at embryonic Day 14 (E14.5); however, the developmental function of the klf1 gene in other vertebrates is not fully understood. During mouse embryogenesis, Klf6 is widely expressed in several tissues, including the kidney, heart and lung. Klf6-deficient mice die at E12.5 from defects in erythropoiesis and yolk sac vascularization8. Thus, multiple Klf genes contribute to the control of mammalian haematopoietic development.
We have previously isolated zebrafish klf17/biklf (blood island-enriched Krüppel-like factor), which is expressed in the blood island/ICM9. Although transient knockdown analysis of the klf17 gene using antisense morpholino injection caused defects in primitive erythropoiesis10, zebrafish with CRISPR–Cas9-mediated klf17 deficiency exhibited blood circulation without obvious haematopoietic defects11. In contrast, both hatching defects and abnormal lateral neuromast depositions are observed in the mutant11. Other klf genes, such as klf1, klf2a, klf3, klf6a and klf8, are expressed in the ICM12, raising the possibility that these genes serve redundant functions in zebrafish haematopoiesis. A recent study has shown that knockdown of klf3 or klf6a using antisense morpholinos causes fewer mature erythroid cells12. However, genetic ablations of ICM-expressed klf genes, except for klf17, have not been analysed during zebrafish haematopoietic development.
At present, the function of ICM-expressed klf genes in haematopoietic development in zebrafish is not clear. Therefore, we established the klf1 single mutant and klf1-klf17 double mutant using CRISPR–Cas9 and investigated the morphological phenotypes of the mutants during zebrafish haematopoiesis. The klf1 mutant and klf17 mutant exhibited normal blood circulation at 2 dpf, whereas the number of circulating blood cells was reduced in the klf1-klf17 double mutant. Primitive erythroid cells from the klf1-klf17 mutant contained a large nucleus at 2 dpf, suggesting maturation defects in primitive erythroid cells. These results suggest that the function of the klf1 and klf17 genes is required for zebrafish primitive erythropoiesis.
Results
Establishment of the klf1 mutant and klf1-klf17 double mutant and phenotypic analysis of these mutants in zebrafish haematopoietic development
Both the klf1 and klf17 genes are specifically expressed in the blood island/ICM during zebrafish embryogenesis9,13. The expression of klf1 was maintained in the CHT, whereas the expression of klf17 was diminished in the CHT. The production of primitive erythroid cells is observed in klf17-deficient fish as well as wild-type fish11. Because there has been no report on the phenotype of klf1-deficient zebrafish in haematopoietic development, we established klf1 single mutants and klf1-klf17 double mutants using CRISPR–Cas9 genome editing technology. The klf1 mutant allele uy19 possesses a deletion of 35 base pairs (bp). The Klf1 mutant protein was thus functionally disrupted because it lacked most of the C-terminal domain, including the three zinc fingers (Fig. S1 and S2).
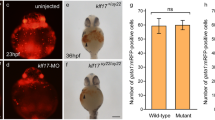
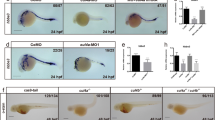
We first examined the production of primitive haematopoietic cells in the klf1 single mutant, klf17 single mutant and klf1-klf17 double mutant at 48 hpf. Primitive haematopoiesis in vertebrates mainly produces primitive erythroid and myeloid cells, and most blood-circulating haematopoietic cells in zebrafish are primitive erythroid cells. The presence of primitive erythroid cells around the heart at 48 hpf was observed by the presence of a red colour in the klf1 mutant (n = 3) as well as wild-type (n = 6) or the klf17 mutant (n = 3) (Fig. 1), whereas the red colour of primitive erythroid cells in the klf1-klf17 mutant (n = 5) was faint. Consistently, the number of blood-circulating erythroid cells in the trunk of the klf1-klf17 mutant was reduced compared to those of the wild-type, klf1 mutant and klf17 mutant (Supplemental Videos S1–S4). We counted the number of blood-circulating erythroid cells in the intersegmental vessels (ISVs) (Supplemental Videos S5, S6). We found that the number of blood-circulating erythroid cells in the ISV of klf1-klf17 mutant (n = 5) was reduced compared to that of wild-type (n = 6) (Supplemental Fig. S3). Next, we examined haemoglobin production in primitive erythroid cells by o-dianisidine staining. Haemoglobin production at 48 hpf was decreased in the klf1-klf17 mutant, while haemoglobin production was comparable among the wild-type, klf1 mutant and klf17 mutant. It has been shown that both mitoferrin-deficient zebrafish and alas2-deficient zebrafish exhibit anaemia, and the haemoglobin production defect in mitoferrin-deficient zebrafish, but not alas2-deficient zebrafish, is restored by the treatment with hinokitiol (1 μM) and ferric citrate (10 μM)14. We confirmed that haemoglobin production was decreased in the klf1-klf17 mutant (Supplemental Fig. S4E, wild-type vs klf1-klf17 mutant). Haemoglobin production of the klf1-klf17 mutant treated with dimethyl sulfoxide (DMSO) (n = 10) or the klf1-klf17 mutant treated with hinokitiol (1 μM) and ferric citrate (10 μM) (n = 8) equivalently decreased compared to that of the wild-type treated with DMSO (n = 12) or the wild-type treated with hinokitiol (1 μM) and ferric citrate (10 μM) (n = 11) (Supplemental Fig. S4). The status of primitive myelopoiesis was evaluated by the expression of the lyz:EGFP transgene15. The number of lyz:EGFP-positive cells was comparable among the wild-type (n = 7), klf1 mutant (n = 7), klf17 mutant (n = 5) and klf1-klf17 mutant (n = 4) (Fig. 2). Consistently, the number of lyz-positive cells visualized by whole-mount in situ hybridization (WISH) was comparable between the wild-type (n = 7) and klf1-klf17 mutant (n = 3) (Supplemental Fig. S5. Because the klf1-klf17 mutant, but not the klf1 mutant or klf17 mutant showed primitive erythropoiesis defects, we focused on the cooperative function of klf1 and klf17 in primitive erythropoiesis.
The klf1-klf17 double mutant exhibited impairment of primitive erythropoiesis. (A–D) Live embryos at 48 hpf. The red colour of blood-circulating erythroid cells (arrowheads) was faint in the klf1-klf17 mutant (klf1-/-klf17-/-) (D) compared to those in the wild-type (klf1+/-klf17+/+) (A), klf1 mutant (klf1-/-klf17+/-) (B) and klf17 mutant (klf1+/+klf17-/-) (C). (E–H) Detection of haemoglobin (arrowheads) at 48 hpf. Haemoglobin production was examined by o-dianisidine staining. Haemoglobin production in the yolk was reduced in the klf1-klf17 mutant (H) compared to the wild-type (E), klf1 mutant (F) and klf17 mutant (G). Genotyping was performed by genomic PCR using locus-specific primers. Scale bar, 200 μm (A, E).
Primitive myelopoiesis in the klf1-klf17 mutant. (A) Wild-type (klf1+/-klf17+/+), 22 hpf. (B) klf1 (klf1-/-klf17+/+) mutant, 22 hpf. (C) klf17 (klf1+/-klf17-/-) mutant, 22 hpf. (D) klf1-klf17 (klf1-/-klf17-/-) mutant, 22 hpf. Primitive myeloid cells were visualized as lyz:EGFP-positive cells on the yolk and were comparable in the wild-type, klf1 mutant, klf17 mutant and klf1-klf17 mutant (arrowheads). Genotyping of individual embryos was performed by genomic PCR. Scale bar, 200 μm (A).
Blood circulation and maturation of primitive erythroid cells in the klf1-klf17 mutant
Because erythroid cells can be visualized by the erythroid cell-specific expression of gata1:mRFP15, we used this strategy to examine the blood circulation of primitive erythroid cells in wild-type and klf1-klf17 mutant zebrafish. The area of gata1:mRFP-positive erythroid cells in the ICM was comparable between the wild-type (n = 3) and klf1-klf17 mutant (n = 7) at 24 hpf (Fig. 3A,B,C). The gata1:mRFP-positive erythroid cells in wild-type (n = 3) and klf1-klf17 mutant (n = 7) migrated into the dorsal aorta and started circulation on the yolk at 28 hpf (Fig. 3D,E, Supplemental Videos S7, S8). The number of gata1:mRFP-positive neurons in the neural tube was comparable between wild-type and klf1-klf17 mutant. On the other hand, the number of gata1:mRFP-positive erythroid cells on the yolk of the klf1-klf17 mutant (n = 7) was reduced compared to that of wild-type (n = 9) (Fig. 3F,G,H).
Blood circulation in wild-type and klf1-klf17 mutant embryos. Fluorescence microscopy images of the wild-type (A, D, F) and klf1-klf17 mutant (B, E, G). Lateral views, with the anterior to the right (A, B, D, E). Ventral views, with the anterior at the top (F, G). (A, B) gata1:mRFP-positive erythroid cells (white arrowheads) in the ICM at 24 hpf. Genotyping of individual embryos was performed by genomic PCR. Scale bar, 200 μm (A). (C) Area existing gata1:mRFP-positive erythroid cells in the ICM at 24 hpf. The area of gata1:mRFP-positive erythroid cells in the ICM was comparable between wild-type (n = 3) and klf1-klf17 mutant (n = 7). The data shown are the mean ± standard deviation (SD). ns, not significant. (D, E) gata1:mRFP-positive erythroid cells (white arrowheads) in the dorsal aorta and gata1:mRFP-positive neurons (asterisks) in the neural tube at 28 hpf. Scale bar, 100 μm (D). (F, G) gata1:mRFP-positive erythroid cells (white arrowheads) on the yolk at 28 hpf. Scale bar, 100 μm (F). (H) Number of gata1:mRFP-positive erythroid cells on the yolk. The number of gata1:mRFP-positive erythroid cells was reduced in the klf1-klf17 mutant (n = 7) compared to that of wild-type (n = 9). The data shown are the mean ± standard deviation (SD). *** P < 0.001 was considered significant.
The morphological phenotype of erythroid cells in the klf1-klf17 mutant was determined by Wright–Giemsa staining, which is useful for the characterization of the developmental stages of the erythroid lineage. The nucleus-to-cytoplasm ratio of erythroid cells is decreased during erythroid cell maturation16. We quantified the nucleus-to-cytoplasm (N:C) ratio in the wild-type (n = 6) and the klf1-klf17 mutant (n = 4) at 52 hpf (Fig. 4). We found that the size of erythrocyte nuclei from the klf1-klf17 mutant was larger than that of wild-type at 52 hpf (Fig. 4C), suggesting maturation defects of erythroid cells during zebrafish primitive erythropoiesis.
Wright–Giemsa staining of erythroid cells from wild-type and klf1-klf17 mutant embryos. (A, B) Wright–Giemsa staining at 52 hpf. Erythroid cells in the klf1-klf17 mutant (B) had larger nuclei and more basophilic cytoplasm than wild-type cells (A) at 52 hpf. Scale bar, 10 μm (A). Genotyping was performed by genomic PCR using locus-specific primers. (C) Scatter plots of the nucleus-to-cytoplasm (N:C) ratio in the wild-type (n = 6) and the klf1-klf17 mutant (n = 4) at 52 hpf. The N:C ratio in the klf1-klf17 mutant was larger than that in the wild-type. The central horizontal lines indicate the mean values. The data shown are the mean ± standard deviation (SD). Each wild-type and klf1-klf17 mutant sample contained 10 cells. P < 0.001 was considered significant.
Expression of haematopoietic genes in the klf1-klf17 mutant
To examine what kind of genes are affected in the klf1-klf17 mutant, we performed WISH analysis for haematopoietic genes. The myeloid cell-specific genes cebp1 (wild-type: n = 9, mutant: n = 8) and l-plastin (wild-type: n = 9, mutant: n = 11) were expressed at comparable levels in the wild-type and klf1-klf17 mutant at 20 hpf (Fig. S6). The expression of the haematopoietic progenitor genes c-myb (wild-type: n = 11, mutant: n = 11), draculin (wild-type: n = 11, mutant: n = 12) and scl (wild-type: n = 11, mutant: n = 12) and the erythroid cell-specific genes βe1-globin (wild-type: n = 7, mutant: n = 7) and gata1 (wild-type: n = 7, mutant: n = 9) at 24 hpf was comparable between wild-type and klf1-klf17 mutant embryos (Fig. 5 and Fig. S6). During erythroid cell maturation, gata1 expression is downregulated at 48 hpf17. We found that the expression of gata1 in circulating erythroid cells of the klf1-klf17 mutant (n = 9) was maintained at a high level compared to that in wild-type cells (n = 11) (Fig. S7). The expression of the haem synthesis enzyme genes 5′-aminolevulinate synthase 2 (alas2) (wild-type: n = 20, mutant: n = 19), coproporphyrinogen oxidase (cpo) (wild-type: n = 9, mutant: n = 10) and porphobilinogen deaminase (pbgd) (wild-type: n = 11, mutant: n = 9) was decreased in the klf1-klf17 mutant, whereas the expression of ferrochelatase (fech) (wild-type: n = 7, mutant: n = 8) and uroporphyrinogen decarboxylase (urod) (wild-type: n = 7, mutant: n = 8) was not affected. The expression of erythroid anion exchanger band3 (wild-type: n = 19, mutant: n = 21) was reduced in the klf1-klf17 mutant, while the expression of the cytoskeletal protein gene β-spectrin (sptb) (wild-type: n = 11, mutant: n = 13) was not affected. The expression of the mitochondrial iron transporter mitoferrin (wild-type: n = 7, mutant: n = 8), but not divalent metal transporter 1 (dmt1) (wild-type: n = 12, mutant: n = 12), was diminished in the klf1-klf17 mutant. Quantitative real-time PCR (qPCR) analysis confirmed that the expression of the erythrocyte genes alas2, band3 and mitoferrin, but not fech, was diminished in the klf1-klf17 mutant (Fig. 6). Thus, the levels of the erythroid cell maturation markers band3 and mitoferrin, but not the haematopoietic progenitor genes c-myb and scl, were decreased in the klf1-klf17 mutant.
Differential expression of haematopoietic genes in the klf1-klf17 mutant. (A,C,E,G,I,K,M,O) Wild-type embryos with wild-type alleles of klf1 and klf17 at 24 hpf. (B,D,F,H,J,L,N,P) klf1-klf17 mutant at 24 hpf. Whole-mount in situ hybridization (WISH) with scl (A,B), c-myb (C,D), fech (E,F), alas2 (G,H), sptb (I,J), band3 (K,L), dmt1 (M,N) and mitoferrin (O,P). All pictures show lateral views, with the anterior to the left. Scale bar, 200 μm (A). Arrowheads indicate the position of the ICM. The expression of scl, c-myb, fech, sptb and dmt1 at 24 hpf was comparable between the wild-type and klf1-klf17 mutant. In contrast, the expression of alas2, band3 and mitoferrin was decreased in the klf1-klf17 mutant. Genotyping of individual embryos was performed by genomic PCR.
Expression levels of haematopoietic genes in the klf1-klf17 mutant. Quantitative real-time PCR (qPCR) represents the indicated gene expression relative to that of the tubulin α1 (tuba1) gene. alas2, *** p < 0.001. band3, *** p < 0.001. mitoferrin, * p < 0.05. fech, ns, not significant. The results are expressed as the mean ± standard deviation (SD). The fech expression level was comparable between the wild-type and the klf1-klf17 mutant, whereas the expression levels of alas2, band3 and mitoferrin were diminished in the klf1-klf17 mutant.
Discussion
In this study, we established klf1-klf17 double-deficient zebrafish with erythroid cell maturation defects and presented the cooperative function of klf1 and klf17 in zebrafish primitive erythropoiesis. Both zebrafish klf1 and klf17 genes are predominantly expressed in the blood island/ICM where primitive erythroid cells are produced9,13. In mice, Klf1 is expressed in yolk sac blood islands, while the expression of Klf17 during mouse embryogenesis is not well characterized. Klf1-deficient mice exhibit a lethal anaemic phenotype6,7. In contrast, klf1-deficient zebrafish and klf17-deficient zebrafish did not show obvious primitive haematopoietic defects (Fig. 1). We found that the number of primitive erythroid cells was reduced in the klf1-klf17 double mutant (Fig. 1, 3 and Fig. S3), whereas primitive myeloid cells derived from ALM were produced at comparable levels in the wild-type and klf1-klf17 mutant (Fig. 2 and Fig. S5). These results suggest the cooperative function of klf1 and klf17 in zebrafish primitive erythropoiesis. Multiple zebrafish klf genes, klf1, klf2a, klf3, klf6a, klf8 and klf17, are expressed in the ICM13. Although klf1, klf2a and klf6a are substantially expressed in the CHT, the expression of klf3, klf8 and klf17 in the CHT is weak, suggesting that the cooperative function of klf1, klf2a and klf6a genes is required for definitive erythropoiesis. Because loss-of-function analyses of klf genes remain restricted at present, further analysis is required to investigate the relationship among the functional roles of klf genes in haematopoietic development.
During primitive erythroid cell maturation, haemoglobin production visualized by o-dianisidine staining was decreased in the klf1-klf17 double mutant (Fig. 1 and Fig. S4). Consistent with such haemoglobin production defects, the expression of the haem synthesis genes alas2, cpo and pbgd was decreased in the klf1-klf17 mutant (Fig. 5 and Fig. S6). Human ALAS2 is well known as the rate-limiting enzyme in haem biosynthesis18. The first 300 base pairs of the promoter sequence in the 5'-flanking region of the human ALAS2 gene are required for maximal expression in erythroid cells and include GATA motifs and CACCC boxes19. Gel shift analysis and transactivation analysis revealed that human GATA1 and EKLF/KLF1 can bind to and activate the GATA motif and CACCC box, respectively, in the promoter. Because alas2 expression was decreased in the klf1-klf17 mutant, both klf1 and klf17 genes may be involved in the initial erythroid-specific induction of the alas2 gene in zebrafish. The expression of gata1 is downregulated during zebrafish primitive erythroid cell maturation17. Initial gata1 induction at the ICM was comparable between the wild-type and klf1-klf17 mutant, whereas gata1 expression was maintained at a high level in the blood-circulating erythroid cells of the klf1-klf17 mutant (Fig. S7). During zebrafish erythroid cell maturation, the size of the nucleus of erythroid cells is reduced16. The nucleus to cytoplasm ratio of erythroid cells, judged by Wright–Giemsa staining, was increased in the klf1-klf17 mutant compared to the wild-type (Fig. 4). These results suggest that the maturation of primitive erythroid cells is impaired in the klf1-klf17 mutant.
WISH analysis revealed that the haematopoietic progenitor genes c-myb and scl were expressed at comparable levels in the ICM of the klf1-klf17 mutant. In contrast, the expression of the erythroid anion exchanger band3 and a mitochondrial iron transporter mitoferrin, known erythroid cell maturation markers, was selectively decreased in the klf1-klf17 mutant. As previously reported, band3-deficient zebrafish exhibit anaemia with dyserythropoiesis20, while mitoferrin-deficient zebrafish develop hypochromic anaemia21. The transcription of Band3 mRNA, but not β-Spectrin mRNA, was diminished in EKLF/KLF1-deficient mice22. Mitoferrin expression is markedly downregulated in the Klf1-deficient mouse foetal liver23. Therefore, the impairment of the initial induction of band3 and mitoferrin in primitive erythroid cells of the klf1-klf17 mutant may be associated with erythroid cell circulation and/or erythroid cell maturation defects. Both mitoferrin-deficient zebrafish and alas2-deficient zebrafish exhibit anaemia14,24. The haemoglobin production defects in mitoferrin-deficient zebrafish, but not the alas2-deficient zebrafish, were restored by treatment with hinokitiol and ferric citrate14, suggesting that hinokitiol selectively affects iron transport processes in erythroid cell maturation. On the other hand, treatment with hinokitiol and ferric citrate did not restore the defects in haemoglobin production in the klf1-klf17 mutant (Fig. S4). Various erythroid cell maturation defects, including the diminished expression of alas2 and band3, are impaired in the klf1-klf17 mutant, suggesting that hinokitiol and ferric citrate treatment is not sufficient to restore the haemoglobin production defects in the mutant. From these functional analyses of the klf1-klf17 mutant, we propose that klf1 and klf17 cooperatively play important roles in the processes of erythroid cell maturation during zebrafish primitive erythropoiesis.
Methods
Ethics statement
All animal experiments were performed in accordance with institutional and national guidelines and regulations. The study was carried out in compliance with the ARRIVE guidelines25. The study was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Yamanashi and the Use Committee of the University of Yamanashi (Approval Identification Number: A30-25).
Synthetic crRNA and tracrRNA, recombinant Cas9 protein, and microinjection
To disrupt the klf1 genomic locus, we used the ready-to-use CRISPR–Cas9 system composed of CRISPR RNA (crRNA), trans-activating crRNA (tracrRNA) and recombinant Cas9 protein26. Synthetic crRNAs and tracrRNA (Supplementary Table S1) and recombinant Cas9 protein were obtained from Integrated Device Technology, Inc. (IDT). Synthetic klf17-crRNA1 (25 pg), klf17-crRNA2 (25 pg) and tracrRNA (100 pg) were coinjected together with recombinant Cas9 protein (1 ng) into 1-cell stage zebrafish embryos.
Genotyping for the klf1 and klf17 loci and genomic sequencing
The klf17uy21 allele bears a deletion of bp 278–332 in the klf17 coding region, and an unrelated 35 bp was inserted into the site11. The klf1uy19 allele bears a deletion of bp 220–254 in the klf1 coding region. To prepare the genomic DNA, the embryos at the indicated stages were incubated in 108 μl of 50 mM NaOH at 98 °C for 10 min. Subsequently, 12 μl of 1 M Tris–HCl (pH 8.0) was added to the solution26. Genomic fragments at the targeted sites were amplified by PCR with PrimeTaq (Primetech) with the locus-specific primers listed in Supplementary Table S2. The PCR conditions were as follows: 95 °C for 3 min, and 98 °C for 10 s, 55 °C for 30 s and 72 °C for 30 s (40 cycles). To perform a heteroduplex mobility assay (HMA), the resultant PCR amplicons were electrophoresed on a 12.5% polyacrylamide gel27. To confirm the presence of the mutations in the klf1 genomic locus, genomic fragments of the targeted genomic locus were amplified from the solution (1 μl) using PCR (Supplementary Table S2). The resultant PCR fragments were subcloned into the pGEM-T Easy vector (Promega), and the genomic sequences were determined by sequence analysis.
Imaging
We counted the number of blood-circulating erythroid cells in the ISV using a high-speed camera (HAS-U1, DITECT Co. Ltd.). We established klf1uy19, klf17uy21 and klf1 uy19-klf17 uy21 mutant lines containing the transgene from Tg(gata1:mRFP)ko05 or containing the transgene from Tg(lyz:EGFP)ko0215. Zebrafish larvae were anaesthetized with tricaine (MS-222) and mounted in 3% methylcellulose. Zebrafish were imaged by a ZEISS Axio Zoom V16 fluorescence stereomicroscope.
Wright–Giemsa staining and o-dianisidine staining
Zebrafish erythroid cells were prepared from the heart or vessels at 52 hpf and attached to slide glass as previously described28. Wright–Giemsa staining was performed to label erythroid cells.
Dechorionated zebrafish embryos at 48 hpf were incubated for 15 min with staining buffer (0.6 mg/ml o-dianisidine, 10 mM sodium acetate [pH 5.2], 0.65% hydrogen peroxide and 40% ethanol) to detect haemoglobin10.
Whole-mount in situ hybridization (WISH)
We examined the expression of scl, gata1, c-myb, sptb, fech, cebp1, l-plastin, dmt1, band3, alas2, mitoferrin, draculin, lmo2, βe1-globin, urod, pbgd and cpo. WISH was performed as previously described29. Zebrafish embryos hybridized with the digoxygenin (DIG)-labelled RNA probe were incubated with alkaline phosphatase-conjugated anti-DIG antibody. To visualize the RNA probe recognized by the anti-DIG antibody, the samples were subsequently incubated with BM Purple (Roche) as the substrate. After three washes with PBST, the samples were fixed in 4% paraformaldehyde.
RNA extraction and quantitative real-time PCR
Total RNA was isolated from wild-type and klf1-klf17 mutant embryos at 24 hpf using TRIzol reagent (Thermo Fisher Scientific) and cDNA was synthesized using ReverTra Ace (Toyobo). Quantitative real-time PCR (qPCR) was performed with KAPA SYBR FAST Master Mix (Kapa Biosystems). Reaction conditions were as follows: 95 °C for 3 min, and 95 °C for 3 s and 62 °C for 30 s (40 cycles), and then a melting curve analysis at 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s. qPCR was performed in duplicate and in at least three independent experiments. Quantification of mRNA was normalized with tubulin α1 (tuba1) as the housekeeping gene and calculated according to the 2 − ΔΔCT method.30. The primers used in this study are listed in Table S3.
Drug treatment
Wild-type or klf1-klf17 mutant embryos were dechorionated at 24 hpf and incubated in (i) 1% DMSO as a vehicle control, or (ii) hinokitiol (1 μM) and ferric citrate (10 μM) until 48 hpf14. Haemoglobin production was determined by o-dianisidine staining as described above.
Data availability
All data generated or analysed during this study are included in this published article and its Supplemental Information files. Additional raw data files can be available from the corresponding author upon request.
References
Orkin, S. H. & Zon, L. I. Hematopoiesis: An evolving paradigm for stem cell biology. Cell 132, 631–644. https://doi.org/10.1016/j.cell.2008.01.025 (2008).
Clements, W. K. & Traver, D. Signalling pathways that control vertebrate haematopoietic stem cell specification. Nat. Rev. Immunol. 13, 336–348. https://doi.org/10.1038/nri3443 (2013).
Berman, J. N., Kanki, J. P. & Look, A. T. Zebrafish as a model for myelopoiesis during embryogenesis. Exp. Hematol. 33, 997–1006. https://doi.org/10.1016/j.exphem.2005.06.010 (2005).
Chen, A. T. & Zon, L. I. Zebrafish blood stem cells. J. Cell Biochem. 108, 35–42. https://doi.org/10.1002/jcb.22251 (2009).
McConnell, B. B. & Yang, V. W. Mammalian Krüppel-like factors in health and diseases. Physiol. Rev. 90, 1337–1381. https://doi.org/10.1152/physrev.00058.2009 (2010).
Nuez, B., Michalovich, D., Bygrave, A., Ploemacher, R. & Grosveld, F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375, 316–318. https://doi.org/10.1038/375316a0 (1995).
Perkins, A. C., Sharpe, A. H. & Orkin, S. H. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375, 318–322. https://doi.org/10.1038/375318a0 (1995).
Matsumoto, N. et al. Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood 107, 1357–1365. https://doi.org/10.1182/blood-2005-05-1916 (2006).
Kawahara, A. & Dawid, I. B. Expression of the Krüppel-like zinc finger gene biklf during zebrafish development. Mech. Dev. 97, 173–176. https://doi.org/10.1016/s0925-4773(00)00404-4 (2000).
Kawahara, A. & Dawid, I. B. Critical role of biklf in erythroid cell differentiation in zebrafish. Curr Biol. 11, 1353–1357. https://doi.org/10.1016/s0960-9822(01)00398-0 (2001).
Suzuki, H. et al. Characterization of biklf/klf17-deficient zebrafish in posterior lateral line neuromast and hatching gland development. Sci. Rep. 9, 13680. https://doi.org/10.1038/s41598-019-50149-5 (2019).
Xue, Y., Gao, S. & Liu, F. Genome-wide analysis of the zebrafish Klf family identifies two genes important for erythroid maturation. Dev. Biol. 403, 115–127. https://doi.org/10.1016/j.ydbio.2015.05.015 (2015).
Oates, A. C. et al. The zebrafish klf gene family. Blood 98, 1792–1801. https://doi.org/10.1182/blood.v98.6.1792 (2001).
Grillo, A. S. et al. Restored iron transport by a small molecule promotes absorption and hemoglobinization in animals. Science 356, 608–616. https://doi.org/10.1126/science.aah3862 (2017).
Kitaguchi, T., Kawakami, K. & Kawahara, A. Transcriptional regulation of a myeloid-lineage specific gene lysozyme C during zebrafish myelopoiesis. Mech. Dev. 126, 314–323. https://doi.org/10.1016/j.mod.2009.02.007 (2009).
Qian, F. et al. Distinct functions for different scl isoforms in zebrafish primitive and definitive hematopoiesis. PLoS Biol. 5, e132. https://doi.org/10.1371/journal.pbio.0050132 (2007).
Amatruda, J. F. & Zon, L. I. Dissecting hematopoiesis and disease using the zebrafish. Dev. Biol. 216, 1–15. https://doi.org/10.1006/dbio.1999.9462 (1999).
Chiabrando, D., Mercurio, S. & Tolosano, E. Heme and erythropoieis: More than a structural role. Haematologica 99, 973–983. https://doi.org/10.3324/haematol.2013.091991 (2014).
Surinya, K. H., Cox, T. C. & May, B. K. Transcriptional regulation of the human erythroid 5-aminolevulinate synthase gene. Identification of promoter elements and role of regulatory proteins. J. Biol. Chem. 272, 26585–26594. https://doi.org/10.1074/jbc.272.42.26585 (1997).
Paw, B. H. et al. Cell-specific mitotic defect and dyserythropoiesis associated with erythroid band 3 deficiency. Nat. Genet. 34, 59–64. https://doi.org/10.1038/ng1137 (2003).
Shaw, G. C. et al. Mitoferrin is essential for erythroid iron assimilation. Nature 440, 96–100. https://doi.org/10.1038/nature04512 (2006).
Nilson, D. G., Sabatino, D. E., Bodine, D. M. & Gallagher, P. G. Major erythrocyte membrane protein genes in EKLF-deficient mice. Exp. Hematol. 34, 705–712. https://doi.org/10.1016/j.exphem.2006.02.018 (2006).
Tallack, M. R. et al. A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome. Res. 20, 1052–1063. https://doi.org/10.1101/gr.106575.110 (2010).
Brownlie, A. et al. Positional cloning of the zebrafish sauternes gene: A model for congenital sideroblastic anaemia. Nat. Genet. 20, 244–250. https://doi.org/10.1038/35001596 (1998).
Kilkenny, C., Browne, W. J., Cuthill, I. C., Emerson, M. & Altman, D. G. Improving bioscience research reporting: The ARRIVE guidelines for reporting animal research. Osteoarthr. Cartil. 20, 256–260. https://doi.org/10.1016/j.joca.2012.02.010 (2012).
Kotani, H., Taimatsu, K., Ohga, R., Ota, S. & Kawahara, A. Efficient multiple genome modifications induced by the crRNAs, tracrRNA and Cas9 protein complex in zebrafish. PLoS One 10, e0128319. https://doi.org/10.1371/journal.pone.0128319 (2015).
Ota, S., Hisano, Y., Ikawa, Y. & Kawahara, A. Multiple genome modifications by the CRISPR/Cas9 system in zebrafish. Genes. Cells 19, 555–564. https://doi.org/10.1111/gtc.12154 (2014).
Weinstein, B. M. et al. Hematopoietic mutations in the zebrafish. Development 123, 303–309. https://doi.org/10.1242/dev.123.1.303 (1996).
Hanaoka, R., Katayama, S., Dawid, I. B. & Kawahara, A. Characterization of the heme synthesis enzyme coproporphyrinogen oxidase (CPO) in zebrafish erythrogenesis. Genes. Cells. 11, 293–303. https://doi.org/10.1111/j.1365-2443.2006.00939.x (2006).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25(4), 402–408. https://doi.org/10.1006/meth.2001.1262 (2001).
Acknowledgements
We would like to thank R. Ohga, S. Fujimaki and A. Nagase for zebrafish maintenance. We would like to thank NBRP for sperm freezing and storage of klf1 mutant and klf17 mutant zebrafish. This work was supported by the Japan Society for the Promotion of Science (KAKENHI: 22K06235) and Takeda Science Foundation (A.K.).
Author information
Authors and Affiliations
Contributions
A.K., H.S. and S.R. conceived and designed the work. A.K., H.S. and S.R. wrote the manuscript. H.S., O.T., S.F., S.R. and A.K. performed the experiments. All authors performed the data analysis and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Video 1.
Supplementary Video 2.
Supplementary Video 3.
Supplementary Video 4.
Supplementary Video 5.
Supplementary Video 6.
Supplementary Video 7.
Supplementary Video 8.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Suzuki, H., Ogawa, T., Fujita, S. et al. Cooperative contributions of the klf1 and klf17 genes in zebrafish primitive erythropoiesis. Sci Rep 13, 12279 (2023). https://doi.org/10.1038/s41598-023-39196-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-39196-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.