Abstract
Mammalian sex chromosomes are enriched for large, nearly-identical, palindromic sequences harboring genes expressed predominately in testicular germ cells. Discerning if individual palindrome-associated gene families are essential for male reproduction is difficult due to challenges in disrupting all copies of a gene family. Here we generate precise, independent, deletions to assess the reproductive roles of two X-linked palindromic gene families with spermatid-predominant expression, 4930567H17Rik and Mageb5. Sequence analyses reveals mouse 4930567H17Rik and Mageb5 are orthologs of human HSFX3 and MAGEB5, respectively, where 4930567H17Rik/HSFX3 is harbored in a palindrome in humans and mice, while Mageb5 is not. Additional sequence analyses show 4930567H17Rik and HSFX3 are rapidly diverging in rodents and primates, respectively. Mice lacking either 4930567H17Rik or Mageb5 gene families do not have detectable defects in male fertility, fecundity, spermatogenesis, or in gene regulation, but do show differences in sperm head morphology, suggesting a potential role in sperm function. We conclude that while all palindrome-associated gene families are not essential for male fertility, large palindromes influence the evolution of their associated gene families.
Similar content being viewed by others
Introduction
Large (> 8 kb) palindromes are inverted segmental duplications that contain nearly-identical (> 99%) DNA sequences. Large palindromes are enriched on the X and Y chromosomes in mammals1,2 and harbor gene families (≥ 2 copies of nearly-identical gene sequence) expressed predominantly during spermatogenesis3,4. Males are more susceptible than females to deleterious mutations in single-copy X-linked genes, because males are hemizygous (carry only one X compared to females two X chromosomes)5. Having a second copy of a sex-linked gene could potentially provide protection against hemizygotic susceptibility to new deleterious mutations. The presence of genes in two copies on either the X or Y in large palindromes may have evolved to provide protective roles for genes that are important for male fertility6. For example, in mice, three of four independent deletions of large palindrome arrays result in male infertility7,8,9. Additionally, large deletions of Y chromosome palindromes in men can result in male infertility10,11. These previous findings suggest genes harbored within large palindromes may be essential for male fertility.
Deletions of large palindrome arrays also remove both associated gene families and the palindrome structures, making it difficult to separate whether the loss of the gene families or palindrome structures contribute to male infertility. Unlike palindrome arrays, which contain multiple segmental duplications in each arm, singleton palindromes contain a single segmental duplication in each arm. To investigate the importance of palindrome structure, previous studies have disrupted arms of two independent singleton palindromes, carrying the X-linked gene families 4930567H17Rik and Mageb512. Deletion or inversion of arms within singleton palindromes did not alter overall fertility12. These studies did not, however, investigate whether gene families within these palindromes are necessary for male fertility since one copy of the gene family was left intact within the remaining palindrome arm. If large palindromes provide protective functions for genes that are essential for male fertility, then complete loss of singleton palindrome-associated gene families could result in defects in spermatogenesis and male fertility.
To test if singleton palindrome-associated gene families 4930567H17Rik or Mageb5 are important for male fertility, we independently deleted both gene copies. In addition to building on our prior studies12 on the palindrome structure of the 4930567H17Rik and Mageb5 gene families, we chose to independently delete both gene families because they exhibit canonical features found in other X-palindrome gene families (e.g. predominant expression in spermatids, high level of nucleotide identity between palindrome arms)3. Moreover, the presence of a single protein-coding gene in each palindrome enables us to more confidently ascribe associated reproductive phenotypes to loss of the deleted gene family12. We find both 4930567H17Rik or Mageb5 gene families in mice have orthologs in humans, but despite this conservation, mice lacking either gene family do not exhibit detectable defects in male fertility and post-meiotic spermatogenesis. We observe several abnormalities in sperm head morphology, indicating that while 4930567H17Rik or Mageb5 are not necessary for male fertility, both genes likely play a role in post-meiotic sperm development. Overall, our study supports that the 4930567H17Rik and Mageb5 gene families may play roles in spermatogenesis, but are not necessary for overall male fertility in C57BL/6 J mice. Our findings that 4930567H17Rik or Mageb5 are dispensable for male fertility is consistent with previous efforts demonstrating many single-copy testes-specific genes are also dispensable for male fertility in mice13,14,15. Our studies add to previous findings suggesting 4930567H17Rik and Mageb5 palindrome structures are not essential for male fertility or spermatogenesis12.
Results
Mouse 4930567H17Rik is a highly diverged ortholog of human HSFX3, while mouse Mageb5 is a conserved ortholog of human MAGEB5
X-linked gene families associated with large palindromes can have orthologs in other species or be independently acquired4. To assess possible orthologs of mouse 4930567H17Rik and Mageb5 in humans, we compared their protein sequence via BLASTP and found mouse 4930567H17Rik is orthologous to human Heat Shock Transcription Factor X linked Member 3 (HSFX3) and mouse MAGEB5 is orthologous to human MAGEB5 (Fig. 1A). We further examined whether the genomic regions between mouse and human are syntenic (i.e. if they share flanking orthologous genes). In mouse, 4930567H17Rik is flanked by the genes Iduronate 2-sulfatase (Ids) and Transmembrane Protein 185a (Tmem185a) which also flank HSFX3 in humans. Interleukin 1 receptor accessory protein-like 1 (Il1RAPl1) and Aristaless related homeobox (Arx) flank Mageb5 in both human and mouse (Fig. 1A). This data supports that 4930567H17Rik and HSFX3 and Mageb5 and MAGEB5 are orthologous between human and mice.
4930567H17RIK and MAGEB5 share orthologs between mouse and humans. (A) Syntenic region of 4930567H17RIK and MAGEB5 between mouse and human with percent amino acid identity in the shaded region. 4930567H17Rik shares 35% amino acid identity (across 18% of the protein) with HSFX3. Mageb5 shares 53% amino acid identity (across 81% of the protein) with MAGEB5 (B) Palindrome structure of the regions containing 4930567H17Rik, HSFX3, Mageb5, and MAGEB5. Palindrome arms are represented as blue arrows. HSFX3 is amplified compared to 4930567H17Rik. Mageb5 does not share palindrome structure between mouse and human. (C) Intron–exon structure for 4930567H17Rik/HSFX3, and MAGEB5/MAGEB5 showing protein domains (HSF DNA binding domain (black), glutamic acid repeats (dark grey), MAGE domain (light grey)) and amino acid sequence similarity (grey shading between species). (D) Reanalyzed ribosome profiling and natural log values of UMI counts from single-cell RNA-seq data for 4930567H17Rik and Mageb5 (data sets taken from Wang, et. al and Green, et. al.18,19). Ribosome association and mRNA expression is most strongly seen in post meiotic cells for both 4930567H17Rik and Mageb5. SG spermatogonia, SC spermatocyte, RS round spermatid, ES elongating spermatid, SZ spermatozoa.
In mouse, two gene copies of 4930567H17Rik and Mageb5 exist within palindromic sequences (Fig. 1B), however, the copy number of both genes is different on the human X chromosome. In humans, HSFX3 is present in four copies (annotated as HSFX1-4) and MAGEB5 is inverted in a unique non-palindromic sequence. Human MAGEB5 has additional neighboring MAGEB gene family copies, but none of the gene family members are within palindromes (Fig. 1B). BLASTP alignments and synteny of 4930567H17Rik/HSFX3 and Mageb5/MAGEB5 suggest both X-linked gene families were present in the common ancestor of mouse and humans ~ 80 million years ago (MYA), but diverged at the sequence level, as in the case of 4930567H17Rik, or at the level of palindrome structure, as in the case of Mageb5.
Mouse 4930567H17Rik and human HSFX3 are rapidly diverging protein-coding genes
To further understand how 4930567H17Rik and HSFX3 diverged, we compared the evolutionary dynamics and intron–exon structures of 4930567H17Rik in rodents and HSFX3 in primates. Previous studies have shown both copies of 4930567H17Rik (gene accession #’s NM_001033807, NM_001081476.1) exhibit rapid sequence divergence in rodents; having a Ka/Ks value of 1.81 when compared across three Mus lineages and R. norvegicus as an outgroup16. We find primate HSFX3 has a Ka/Ks value of 1.20, indicating 4930567H17Rik and HSFX3 sequence is rapidly diverging in both rodents and primates. The rapid sequence divergence of 4930567H17Rik in rodents may have been facilitated by loss of an exon. Other mammalian HSFX3 orthologs, including human HSFX3, encode two exons, while 4930567H17Rik encodes a single exon (Fig. 1C). HSFX3 has a DNA binding domain spanning the splice junction between exon 1 and 2. 4930567H17Rik produces a predicted protein that shares amino acid identity only with HSFX3. While 4930567H17RIK lacks the DNA-binding domain, it possesses an expanded glutamic acid repeat at the C-terminus end (Fig. 1C). Interestingly, the first exon of mouse 4930567H17Rik is pseudogenized and remnants of the first exon are found outside of the palindrome, via sequence comparisons (Fig S1). RNA-seq data supports the ancestral first exon of HSFX is not transcribed in mice and thus the second exon is the only remaining functional exon in Mus musculus (Fig S1).
The predicted annotation of 4930567H17Rik is a long non-coding RNA17. However, we find evidence to support that 4930567H17Rik is a protein-coding gene despite loss of an exon and rapid sequence divergence. First, 4930567H17Rik encodes a large open reading frame (233 amino acids), conserved across rodents (Fig. 1C). Despite the rapid sequence divergence, 4930567H17Rik has not acquired new nonsense mutations typical of pseudogenes or long non-coding RNAs. Second, reanalysis of ribosome profiling data18 from sorted spermatogenic cells demonstrates 4930567H17Rik RNA is most highly associated with ribosomes in round spermatids and elongating spermatids, consistent with single cell RNA-seq studies, altogether indicating 4930567H17Rik mRNA is likely translated (Fig. 1D). Overall, we conclude that the protein-coding sequence of 4930567H17Rik/HSFX3 is rapidly diverging across mammals, and the exon containing the HSF DNA-binding domain has been lost along the Mus lineage, after divergence with rat.
Generation of mice lacking both copies of the palindrome associated gene families 4930567H17Rik or Mageb5
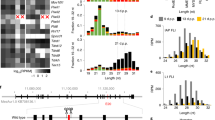
To determine whether the 4930567H17Rik and Mageb5 gene families are necessary for male fertility, we generated complete null mutant mice for both 4930567H17Rik and Mageb5 by using CRISPR/Cas9 (Fig. 2A). We deleted both 4930567H17Rik and Gm14725 protein-coding gene copies (mm10 chrX:70,394,006:70,394,659 and chrX:70,545,638:70,546,279), resulting in a null mutant mouse line (4930567H17Rik∆CDS/Y), as evidenced by RT-PCR and RNA-seq (Fig. 2C,D). For Mageb5, we utilized Mageb5∆Arm/+ mice, which already had one Mageb5 palindrome arm deleted12. We deleted the second copy of Mageb5, annotated as Gm14781, by specifically targeting the remaining Gm14781 protein-coding gene copy in Mageb5∆Arm/+ mice (Fig. 2A), to generate two null mutant Mageb5∆Arm∆CDS/Y lines (L1 and L2). In L1 mice, ~ 860 bp were removed (mm10 chrX:91,634,446–91,635,305), while in L2 ~ 430 bp were removed (mm10: chrX:91,634,446–91,634,878) (Fig. 2B, right), yielding PCR products of 166 bp (L1) and 596 bp (L2). The translational start site was removed in both lines. To assess if Mageb5∆Arm∆CDS/Y L1 and L2 produced RNA, we performed RT-PCR, followed by Sanger sequencing. We find that Mageb5∆Arm∆CDS/Y L1 yields a highly truncated RNA product (322 bp) and L2 does not yield any detectable RNA product (Fig. 2C). Sequencing of the 322 bp cDNA from L1 reveals a small, predicted peptide (< 80 amino acids) (Fig S3). RNA-seq analyses of L1 support a lack of mRNA expression for both copies of the Mageb5 gene in Mageb5∆Arm∆CDS/Y L1 mice (Fig. 2D). Our results support the successful removal of ~ 650 bp of both copies of 4930567H17Rik and ~ 860 bp and ~ 430 bp of Mageb5, and 30 Kb of a palindromic arm containing Mageb5, to yield 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y null mutant mice, respectively.
Creation of 4930567H17Rik∆CDS/Y, and Mageb5∆Arm∆CDS/Y mice. (A) Top: Schematic of the mouse X chromosome showing singleton palindromes (blue) the pseudoautosomal region (green, right end), and centromere (orange, left end). Middle: Diagrams of the 4930567H17Rik and Mageb5 palindromes showing palindrome arms as blue arrows. Bottom: Coding sequence of 4930567H17Rik and one copy of Mageb5 with CRISPR cut sites (red lightning bolts) and genotyping primers (black arrows with numbers). (B) PCR genotyping of DNA from mutant and wild type 4930567H17Rik and Mageb5 mice. Numbered primers correspond to panel A. (C) RT-PCR of 4930567H17Rik and Mageb5 cDNA from one 4930567H17Rik line and Mageb5 L1 and L2. L1 shows a product due to a small portion of the transcript still being produced. (D) RNA-seq data for 4930567H17Rik∆CDS/Y, and Mageb5∆Arm∆CDS/Y L1 mice. GM14725 and Gm14781 are the palindrome arm gene copies of 4930567H17Rik and Mageb5, respectively. See Fig S2 for full-size images of (B,C).
4930567H17Rik and Mageb5 do not play major transcriptional regulatory roles during mouse spermatogenesis
We tested whether mouse 4930567H17RIK or MAGEB5 protein regulates transcription, since 4930567H17RIK is orthologous to a heat shock transcription factor, HSFX3, and MAGE proteins are known to regulate transcription20,21. We assessed whether mice lacking 4930567H17Rik or Mageb5 exhibit differences in gene expression. We performed whole testis RNA-seq to identify differentially expressed genes between 4930567H17Rik∆CDS/Y or Mageb5∆Arm∆CDS/Y mice and their wild-type littermate controls. We identified 21 and 27 differentially expressed (p-value < 0.0001) genes for 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice, respectively (Fig S4, Table S1 + S2). Consistent with our above analyses supporting a lack of 4930567H17Rik and Mageb5 expression, both copies of each gene family (4930567H17Rik and Gm14725 and Mageb5 and Gm14781) are the top two differentially expressed genes and are significantly downregulated in 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y L1 mice (Fig S4, Table S1 + S2). 4930567H17Rik or Mageb5 thus influence the transcription of a limited number of genes, suggesting that they have a minimal, if any role, in regulating gene transcription during spermatogenesis.
4930567H17Rik ∆CDS/Y and Mageb5 ∆Arm∆CDS/Y mice display testis histology, sperm counts, and sperm motility similar to wild-type mice
To assess if 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice exhibit defects in spermatogenesis or sperm biology, we examined if 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice display defects in post-meiotic sperm development via testis histological sections. Periodic acid-shift and hematoxylin (PAS + H) stained slides of testis tubules revealed that all stages of spermatogenesis were present and apparently similar to that of wild-type controls (Fig. 3A) (Fig S5), including a normal appearance of round spermatids, the cell type in which 4930567H17Rik and Mageb5 are predominantly expressed3,12. In agreement with this finding, no significant differences in testis size (assayed via testis/body mass ratios) were detected (4930567H17Rik∆CDS/Y = 0.006428 ± 0.00086, 4930567H17Rik+/Y = 0.006167 ± 0.00067, p = 0.47; Mageb5∆Arm∆CDS/Y = 0.007342 ± 0.00050, Mageb5+/Y = 0.007163 ± 0.00069, p = 0.54) (Fig. 3B). Thus, in the absence of 4930567H17Rik and Mageb5 gene families, sperm development proceeds similar to wild-type testes.
4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice display testis histology, testis size, sperm counts, and motility similar to 4930567H17Rik+/Y and Mageb5+/Y mice. (A) Periodic Acid-Schiff and Hemoxylin stained sections showing normal progression of spermatogenesis in 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice. Arrows indicate presence of round spermatids. (B) Total testis weight normalized to total body weight (g). 4930567H17Rik+/Y n = 8, 4930567H17Rik∆CDS/Y n = 11, Mageb5+/Y and Mageb5∆Arm∆CDS/Y n = 9 (C) Sperm counts. Each point represents a single individual where 2 technical replicates were counted and averaged. 4930567H17Rik+/Y n = 7, 4930567H17Rik∆CDS/Y n = 8, Mageb5+/Y and Mageb5∆Arm∆CDS/Y n = 9. (D) Percent motile sperm. 4930567H17Rik+/Y and 4930567H17Rik∆CDS/Y n = 5, Mageb5+/Y n = 7 and Mageb5∆Arm∆CDS/Y n = 8. (E) Pups/litter from mating three males from each genotype with CD1 females. 4930567H17Rik+/Y n = 18, 4930567H17Rik∆CDS/Y and Mageb5+/Y n = 17 and Mageb5∆Arm∆CDS/Y n = 18 litters. (F) Sex genotyping performed on pups from litters shown in (E). Number of male offspring are shown as percentage of total number of pups in parenthesis below. All comparisons in (B–F) were performed using an unpaired two-tailed student’s t-test between 4930567H17Rik+/Y versus 4930567H17Rik∆CDS/Y and Mageb5+/Y versus Mageb5∆Arm∆CDS/Y. All error bars represent mean with standard deviation.
We also assessed whether there were defects in total sperm or sperm motility in 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice. There were no differences in the number of sperm produced between 4930567H17Rik+/Y and 4930567H17Rik∆CDS/Y and, Mageb5+/Y and Mageb5∆Arm∆CDS/Y mice (4930567H17Rik∆CDS/Y = 15.6 ± 8.5 × 106/ml, 4930567H17Rik+/Y = 15.0 ± 4.4 × 106/ml, p = 0.88, n = 5; Mageb5∆Arm∆CDS/Y = 11.9 ± 5.4 × 106/ml, n = 8, Mageb5+/Y = 12.6 ± 3.3 × 106/ml, n = 7, p = 0.78) (Fig. 3C). Sperm from both 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice were > 50% motile (4930567H17Rik∆CDS/Y = 60 ± 2.6%, n = 2 and Mageb5+/Y = 67 ± 9.9% motile, n = 2). This motility was further examined via a swim-up assay and did not differ regardless of genotype (4930567H17Rik∆CDS/Y = 22.91 ± 17.99%, 4930567H17Rik+/Y = 24.95 ± 20.37%, p = 0.87; Mageb5∆Arm∆CDS/Y 49.28 ± 24.43%, Mageb5+/Y = 30.14 ± 16.09%, p = 0.10) (Fig. 3D).
4930567H17Rik ∆CDS/Y and Mageb5 ∆Arm∆CDS/Y mice exhibit wild-type levels of fertility, fecundity, and sex ratio
To assess if 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice sire fewer offspring, 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice were mated to female CD1 mice. Both 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice exhibited wild-type levels of fertility and fecundity, producing litters of equivalent size to wild-type controls (4930567H17Rik∆CDS/Y = 10.2 ± 3.2 pups/litter, n = 174, 4930567H17Rik+/Y = 10.8 ± 2.6 pups/litter, n = 195, p = 0.55; Mageb5∆Arm∆CDS/Y = 12.1 ± 2.3 pups/litter, n = 203, Mageb5+/Y = 11.9 ± 1.9 pups, n = 191, p = 0.77) (Fig. 3E). Previous studies have found X-palindrome-associated genes can skew the offspring sex ratio8, thus we genotyped the sex of all offspring. No sex ratio distortion was detected in the offspring of either 4930567H17Rik∆CDS/Y or Mageb5∆Arm∆CDS/Y mice (4930567H17Rik∆CDS/Y = 49% male offspring, 4930567H17Rik+/Y = 55% male offspring, p = 0.25; Mageb5∆Arm∆CDS/Y = 50% male offspring, Mageb5+/Y = 44% male offspring, p > 0.99, unpaired two-tailed t-test) (Fig. 3F), suggesting mouse 4930567H17Rik or Mageb5 do not influence sex ratio distortion, autonomously.
4930567H17Rik ∆CDS/Y and Mageb5 ∆Arm∆CDS/Y mice display altered sperm morphology
Defects in sperm morphology can still be present, despite wild-type levels of sperm production and motility, so we examined sperm morphology in 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice. We analyzed multiple attributes of sperm morphology, including area, size of hook, and overall width of the sperm heads, using a custom plugin in ImageJ software22. We find that sperm from Mageb5∆Arm∆CDS/Y mice were slightly more elongated than Mageb5+/Y sperm (0.24 versus 0.16 p = < 0.0001). Sperm from 4930567H17Rik∆CDS/Y mice are larger overall (4739 versus 4420 square pixels, p < 0.0001) and have slightly longer hooks (54 versus 52 pixels, p < 0.0001) than 4930567H17Rik+/Y sperm (Fig. 4A, Fig S6B). The only additional statistical trend observed in morphology was the aspect ratio, the inverse of ellipticity, which was statistically different in 4930567H17Rik∆CDS/Y (p < 0.02) and in Mageb5∆Arm∆CDS/Y mice (p < 0.001) (Fig S6). Despite these differences, 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice are still fertile under laboratory conditions, suggesting that these morphological differences play a minor role in overall fertility.
4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice display altered sperm morphology. (A) Sperm morphology characteristics calculated from assessment of DAPI images processed with a custom plugin to ImageJ22. All data were compared using an unpaired two-tailed t-test between 4930567H17Rik+/Y versus 4930567H17Rik∆CDS/Y and Mageb5+/Y versus Mageb5∆Arm∆CDS/Y. * p < 0.05 **p < 0.001, ***p < 0.0001. Additional parameters are shown in Fig S7. (B) Averages traces and shaded overlays of sperm head morphology from each genotype. Number of sperm assessed are shown inside each respective outline. Representative individual sperm nuclei are also shown in Fig S6B.
Discussion
Our study addresses whether gene families harbored within large singleton X-palindromes are required for male fertility and spermatogenesis in mice. While null mutants of the Slx and Slxl1 gene families harbored in X-palindrome arrays result in male infertility and defects in spermatogenesis8, null mutants of the 4930567H17Rik and Mageb5 gene families in singleton X-palindromes do not. The absence of an overt reproductive phenotype in male mice lacking 4930567H17Rik or Mageb5 may in part be due to genetic redundancy. 4930567H17Rik is related to heat shock transcription factors, which could have compensating gene family members. Indeed, Hsf1 and Hsf2 are expressed in post-meiotic cells23,24 and Hsf2 is known to regulate post-meiotic X- and Y-palindromic gene families24 suggesting Hsf1 and Hsf2 could compensate for the loss of 4930567H17Rik. The most likely candidate to compensate for loss of 4930567H17Rik is Hsf2, based on the robust spermatid expression of Hsf2, as compared to Hsf1 (Fig S8, left). Similarly, Mageb5 has eight X-linked3 and one autosomal Mageb gene family members expressed in the testis that could potentially compensate for the loss of the Mageb5 gene family. The most likely candidate to compensate for loss of Mageb5 is Mageb3, because Mageb3 is expressed at the highest level in spermatids, as compared to other Mageb gene family members (Fig. S8, right). To better understand the spermatogenic role of Mageb5, and Mageb family members, removal of multiple Mageb family members, particularly Mageb3, may be necessary. For both 4930567H17Rik and Mageb5, further studies investigating these possibly redundant genes could help elucidate the roles of 4930567H17Rik and Mageb5 in spermatogenesis. Furthermore, studies of 4930567H17Rik orthologs in rats or primates, that still possesses HSF DNA-binding domains, could shed light on the ancestral function of 4930567H17Rik.
Despite the lack of overt reproductive phenotypes in 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice, differences in sperm head morphology suggest 4930567H17Rik and Mageb5 play a role in sperm development. Sperm head morphology analysis uses DAPI stained images of sperm to detect chromatin. 4930567H17Rik+/Y and Mageb5∆Arm∆CDS/Y sperm had increased size and elongation, respectively, compared to wild type sperm (Fig. 4). This finding may represent that sperm from these mice have a reduced level of chromatin compaction. Thus, 4930567H17Rik and Mageb5 may alter chromatin compaction during spermiogenesis and epididymal transit, a time in development when sperm chromatin compaction is dynamic25. Supporting a potential role for 4930567H17Rik and Mageb5 in sperm chromatin compaction, both gene families exhibit increasing expression levels from late round spermatids to elongated spermatids (Fig. 1D), a developmental time for sperm chromatin compaction. Tracking the dynamics of sperm head morphology during development and epididymal transit may help further define the role of 4930567H17Rik and Mageb5 in sperm development.
Our current analyses of HSFX gene families and previous studies on 4930567H17Rik16 demonstrate that HSFX and 4930567H17Rik sequences are rapidly diverging, suggesting the 4930567H17Rik/HSFX gene family is under positive selection throughout mammals. The gene family’s presence within a X-linked palindrome may facilitate this rapid evolution in multiple ways. First, positive selection is known to be stronger for X-linked genes with male-beneficial functions, because of male sex chromosome hemizygosity26. Second, the second gene copy provide more substrate for new beneficial mutations upon which selection pressures can readily act27. Third, the second copy could relax constraint on palindromic genes and facilitate the acquisition of novel functions27. Fourth, any beneficial mutation arising in one gene copy could be readily spread to other gene copies in the palindrome through arm-to-arm gene conversion16. In the future, it will be important to connect how the rapid sequence divergence of HSFX and 4930567H17Rik relates to their spermatogenic functions.
Large palindromic regions are challenging to study in mice and thus are not a priority in large mouse knockout project consortiums28,29,30,31. CRISPR now enables the study of both X-palindromic structures and their associated gene families. Megabase-sized deletions of arrays of large palindromes demonstrate the necessity of large palindromes and their associated genes for male fertility8,9,10. However, these studies cannot resolve whether the palindrome structure or the associated gene families are responsible for male infertility. Our study demonstrates how CRISPR can generate specific deletions of a single palindrome-associated gene family, while keeping the palindrome structure largely intact. Our study also improves our understanding of large X-palindrome-associated gene function, by demonstrating that individual X-palindrome associated gene families are dispensable for male fertility. Future studies using CRISPR to genetically dissect the importance of palindrome structures versus associated gene families in reproduction will provide a more complete understating of the importance of these large genomic regions and their implications in male fertility.
Materials and methods
Generation of mice lacking 4930567H17Rik and Mageb5 palindrome-associated gene families
Mice lacking the X-linked palindrome-associated 4930567H17Rik and Mageb5 gene families were generated using a CRISPR Cas9 strategy. We selected single guide RNAs (sgRNAs) within the coding sequences of 4930567H17Rik or Mageb5 (Table S3). The Cas9 (ESPCAS9PRO, Sigma-Aldrich/Merck KGaA, Darmstadt, Germany) cleavage efficiency of individual sgRNAs was determined via injection of sgRNA (30 ng/ul)/Cas9 (50 ng/ul) complexes into mouse zygotes and screening for edits via PCR using primers flanking sgRNA cut sites (Table S4) and subsequent Sanger sequencing. We selected sgRNAs with cleavage efficiencies of > 30% to delete 4930567H17Rik and Mageb5 gene families.
To generate mice lacking the 4930567H17Rik gene family (4930567H17Rik∆CDS/Y), C57BL/6 J X SJL hybrid females were crossed with existing 4930567H17Rik+/Y mice12. Zygotes were injected with Cas9 protein (50 ng/μl), a single-stranded oligonucleotide donor (10 ng/μl), and dual sgRNAs (30 ng/μl) targeting each 4930567H17Rik gene copy to achieve a ~ 650 base pair deletion within each copy on both palindrome arms (Table S3). The deletion breakpoints were verified via PCR and subsequent Sanger sequencing (Fig S7A). An F1 male carrying a deletion of both 4930567H17Rik coding sequences in cis (4930567H17Rik∆CDS/Y) was bred to a C57BL/6 J female to generate 4930567H17Rik∆CDS/+ female mice. 4930567H17Rik∆CDS/+ females were backcrossed to C57BL/6 J males to generate 4930567H17Rik∆CDS/Y mice, which were used for all experiments. 4930567H17Rik∆CDS/Y mice used in the described experiments were backcrossed to C57BL/6 J for > 7 generations.
To generate mice lacking the Mageb5 gene family, zygotes from Mageb5∆Arm/+ females crossed to Mageb5∆Arm/Y mice12 were injected with Cas9 protein (50 ng/μl), an oligonucleotide donor (10 ng/μl), and dual sgRNAs (30 ng/μl) targeting a ~ 900 bp deletion of Mageb5 (Table S3). These injections resulted in two independent Mageb5∆Arm∆CDS/Y lines, “L1” carrying a 860 bp deletion and “L2” carrying a 400 bp deletion. The deletion breakpoints of the two lines were verified via PCR and Sanger sequencing (Fig S7B). F1 females with both the Mageb5 palindrome arm and coding sequence deleted in cis were bred to C57BL/6 J males to generate Mageb5∆Arm∆CDS/+ female mice. Mageb5∆Arm∆CDS/+ females were backcrossed to C57BL/6 J males for > 10 generations to generate Mageb5∆Arm∆CDS/Y mice, which were used for all experiments.
Both 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice transmitted the 4930567H17Rik and Mageb5 coding sequence deletions through the germline and no changes in overall health were observed due to off-target effects of CRISPR or as a consequence of the deletions. All mice used in these studies were between 3–7 months of age. 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice were directly compared to wild type littermates (4930567H17Rik+/Y and Mageb5+/Y mice) in all experiments allowing for the minimization of the effects of genetic background and age. If wild-type littermates were not available, then age-matched controls were used. Because both Mageb5∆Arm∆CDS/Y L1 and Mageb5∆Arm∆CDS/Y L2 mice were able to be maintained easily (had normal breeding), Mageb5∆Arm∆CDS/Y L1 were used for experiments presented in this work unless otherwise specified. Cages were kept on ventilated racks at 72°F, 30–70% humidity, on a 12 h:12 h light: dark cycle in a specific-pathogen free room. Cages were monitored daily by husbandry personnel and changed every two weeks. Mice were given water and fed Lab Diet 5008 food ad libitum. Adult mice were sacrificed by CO2 asphyxiation followed by cervical dislocation and pups were sacrificed by decapitation in compliance with ULAM standard procedures in euthanasia. The Institutional Animal Care and Use Committee of the University of Michigan approved all animal procedures (PRO00009403) and all experiments followed the National Institutes of Health Guidelines of the Care and Use of Experimental Animals and the ARRIVE guidelines.
Genotyping
Genotypes of 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice were determined via PCR on DNA samples collected from 1–2 mm tail snips. Tails were digested in 50 mM NaOH for 20 min at 95 °C and briefly vortexed to dissolve tissues. 50 µl of Tris HCl (pH 6.8) was added to neutralize NaOH and samples were centrifuged at 13,000 rpm for 30 s32. PCR was performed with Taq DNA polymerase (New England Biolabs) per manufactures instructions. To verify genotypes of 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y mice, we used primers flanking the coding sequence of each gene (primers 1–5 Table S4). For the Mageb5 lines, we used primers flanking the Mageb5 palindrome arm to verify the deletion of the palindrome arm, as previously described12 (primers 6,7 Table S4).
Reverse transcriptase-PCR
Total testis RNA was extracted using Trizol (Life Technologies) according to the manufacturer’s instructions. ~ 10 µg of total RNA was DNase treated using Turbo DNase (Life Technologies) and reverse transcribed using Superscript II (Invitrogen) using oligo (dT) primers to generate first-strand cDNA. RT-PCR was performed on adult testis cDNA preparations with primers residing in the single exon coding sequence of 4930567H17Rik (primers 3,8 Table S4), and with intron-spanning primers for Mageb5 (primers 9,10 Table S4). Primers to the round spermatid-specific gene Trim42 (primers 11,12 Table S4) served as a positive control8. To control for genomic DNA contamination, a reaction lacking reverse transcriptase was performed in parallel.
RNA-sequencing
Testis RNA was extracted from three 4930567H17Rik∆CDS/Y and three Mageb5∆Arm∆CDS/Y L1 mice, along with three wild-type littermate controls from each line, and DNase treated as described above. Total RNA quality was assessed using the Tapestation 4200 (Agilent) (minimum DV200 value of greater than 30% and a minimum concentration of 3.32 ng/µl). RNA used in this study had RIN (RNA integrity number) values ranging from 6.1–8.9. Ribo-minus (RNaseH-mediated) stranded RNA-seq libraries with indexed adaptors were generated (New England BioLabs). Final libraries were quantitated by Kapa qPCR using Kapa’s library quantification kit for Illumina sequencing platforms (Kapa Biosystems, catalog # KK4835). Pooled libraries were subjected to 150 bp paired-end sequencing according to the manufacturer’s protocol (Illumina NovaSeq6000) giving an average of 50 million reads per sample. Bcl2fastq2 Conversion Software (Illumina) was used to generate de-multiplexed Fastq files. RNA-seq reads were pseudoaligned to the NCBI RefSeq gene annotation for the Mus musculus C57BL/6 J (mm10) reference genome by Kallisto33, using the default settings. Transcript per million (TPM) numbers were generated by Kallisto. The estimated number of RNA-seq reads aligning to each gene, as provided by Kallisto, were used as input to DESeq34 to determine differentially expressed genes between 4930567H17Rik∆CDS/Y and Mageb5∆Arm∆CDS/Y and wild-type mice. All Illumina sequences can be found on NCBI’s sequence read archive under BioProject number: PRJNA748373 and accession numbers of SRR15198217 – SRR15198228.
Testis histology and staining
Testes were fixed overnight in Bouin’s solution (Ricca Chemical, Arlington, TX). Following fixation, testes were washed in 5–10 ml of 70% ethanol on a rotating tube holder at 4 °C for 6–48 h with three or more changes of ethanol to remove excess Bouin’s. Fixed testes were paraffin embedded, sectioned to 5 μm, and stained with Periodic-Acid Schiff (PAS) and Hematoxylin. Testis sections were imaged on an Olympus BX61 equipped with an Olympus DP73 color camera. Specific germ cell populations were identified by their location within a tubule, nuclear size, and the nuclear staining pattern of chromatin35.
Testis to body weight ratio
To calculate testis/body weight ratio, total testis mass was divided by the total body mass taken at the time of euthanasia.
Sperm counts, motility and swim-up assay
Following dissection from the body cavity, the two cauda epididymis were dissected and nicked three times to allow sperm to swim out. Cauda were placed in 1 ml of Human Tubal Fluid (HTF) (Millipore) at 37 °C, and rotated in a 37 °C incubator for 10 min. Cauda were removed and a 100 μl aliquot was used for pre-swim-up baseline sperm counts and motility assessment. For swim-up assays, the remaining portion of sperm in HTF was then removed and placed in a new tube using wide bore tips. Sperm were centrifuged for 5 min at 400xg and the supernatant discarded. The pellet was re-suspended in 1 ml of fresh 37 °C HTF and centrifuged for 5 min at 400×g. The supernatant was removed and 1 ml of fresh 37 °C HTF was carefully overlaid on top of the pellet. The tube was then placed at a 45° angle in a 37 °C incubator for 1 h; after which the top 800 μl containing motile sperm was removed and placed in a new tube. All aliquots of sperm used for counting were diluted 1:10 in H2O and counted using a hemocytometer. Counts were performed blind with four technical replicates per mouse. Sperm counts were calculated by taking the average number of sperm from each of the four technical replicates per mouse. Motility was assessed by counting ≈ 200 sperm for each replicate on a hemocytometer across at least 5 frames. Sperm were considered motile if they showed both progressive movement and signs of flagellar activity36,37. Percent motility for visual assessment was calculated by dividing the number of motile sperm by the total sperm counted and multiplying by 100. Motility assayed via swim up was calculated from dividing the post swim-up count by the pre-count and multiplying by 100. All analyses between groups were performed with an unpaired two-tailed student’s t-test, unless otherwise noted.
Fecundity and fertility
Three 4930567H17Rik∆CDS/Y and three Mageb5∆Arm∆CDS/Y mice, and equal numbers of wild-type litter mate controls, were each repeatedly mated with two CD1 females. Litter size was recorded, and the sex of each offspring was determined with sex-genotyping PCR primers (primers13,14 Table S4) specific to the X- and Y-linked gene Ube1 (Ubiquitin-like modifier activating enzyme, 1 as previously described8.
Sperm head morphology assessment
To assess sperm head morphology, 25 μl of the pre-swim-up sperm aliquot above was placed on a slide and allowed to dry. Cells were fixed for 10 min in 500 μl of 4% PFA (diluted in PBS). Slides were rinsed twice in PBS for 5 min and left to dry. Slides were stained with Vectashield with DAPI (Vector Laboratories) under a 22 × 40 mm cover slip and imaged using an Olympus UPlanSApo 100 × oil objective on an Olympus BX61 equipped with a Hamamatsu Orca-ER camera and Excelitas X-Cite 120LED fluorescence illuminator. Nucleus detection and morphology assessment was performed using the default settings of the custom plugin “Nuclear_Morphology_Analysis_1.20.0_standalone” to the image analysis software ImageJ22. ~ 100 sperm heads from each genotype were blindly selected, imaged, and input into the software. Default edge detection settings were used, and sperm heads were manually inspected to ensure all sperm were accurately detected and only sperm were selected by the software. Sperm from Mageb5∆Arm∆CDS/Y mice were not originally oriented correctly so the top and bottom vertical border tag was placed manually for the dataset38.
Data availability
The data underlying sections of this article are available in NCBI’s sequence read archive at https://www.ncbi.nlm.nih.gov/sra and can be accessed under BioProject number: PRJNA748373 and accession numbers of SRR15198217 – SRR15198228. The rest of the data underlying this article are available in the article and in the online supplementary material.
References
Warburton, P. E., Giordano, J., Cheung, F., Gelfand, Y. & Benson, G. Inverted repeat structure of the human genome: The X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 14, 1861–1869. https://doi.org/10.1101/gr.2542904 (2004).
Skaletsky, H. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837. https://doi.org/10.1038/nature01722 (2003).
Mueller, J. L. et al. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat. Genet. 40, 794–799. https://doi.org/10.1038/ng.126 (2008).
Mueller, J. L. et al. Independent specialization of the human and mouse X chromosomes for the male germ line. Nat. Genet. 45, 1083–1087. https://doi.org/10.1038/ng.2705 (2013).
Haldane, J. B. S. Sex ratio and unisexual sterility in hybrid animals. J. Genet. 12, 101–109. https://doi.org/10.1007/BF02983075 (1922).
Trombetta, B. & Cruciani, F. Y chromosome palindromes and gene conversion. Hum. Genet. 136, 605–619. https://doi.org/10.1007/s00439-017-1777-8 (2017).
Arlt, M. F., Brogley, M. A., Stark-Dykema, E. R., Hu, Y. C. & Mueller, J. L. Genomic structure, evolutionary origins, and reproductive function of a large amplified intrinsically disordered protein-coding gene on the X chromosome (Laidx) in mice. G3 10, 1997–2005. https://doi.org/10.1534/g3.120.401221 (2020).
Kruger, A. N. et al. A neofunctionalized X-linked ampliconic gene family is essential for male fertility and equal sex ratio in mice. Curr. Biol. 29, 3699-3706.e3695. https://doi.org/10.1016/j.cub.2019.08.057 (2019).
Zhou, J., McCarrey, J. R. & Wang, P. J. A 1.1-Mb segmental deletion on the X chromosome causes meiotic failure in male mice. Biol. Reprod. 88, 159. https://doi.org/10.1095/biolreprod.112.106963 (2013).
Kuroda-Kawaguchi, T. et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat. Genet. 29, 279–286. https://doi.org/10.1038/ng757 (2001).
Repping, S. et al. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am. J. Hum. Genet. 71, 906–922. https://doi.org/10.1086/342928 (2002).
Kruger, A. N., Ellison, Q., Brogley, M. A., Gerlinger, E. R. & Mueller, J. L. Male mice with large inversions or deletions of X-chromosome palindrome arms are fertile and express their associated genes during post-meiosis. Sci. Rep. 8, 8985. https://doi.org/10.1038/s41598-018-27360-x (2018).
Sun, J. et al. CRISPR/Cas9-based genome editing in mice uncovers 13 testis- or epididymis-enriched genes individually dispensable for male reproduction†. Biol. Reprod. 103, 183–194. https://doi.org/10.1093/biolre/ioaa083 (2020).
Park, S. et al. CRISPR/Cas9-mediated genome-edited mice reveal 10 testis-enriched genes are dispensable for male fecundity. Biol. Reprod. 103, 195–204. https://doi.org/10.1093/biolre/ioaa084 (2020).
Holcomb, R. J. et al. The testis-specific serine proteases PRSS44, PRSS46, and PRSS54 are dispensable for male mouse fertility†. Biol. Reprod. 102, 84–91. https://doi.org/10.1093/biolre/ioz158 (2019).
Swanepoel, C. M., Gerlinger, E. R. & Mueller, J. L. Large X-linked palindromes undergo arm-to-arm gene conversion across MUS lineages. Mol. Biol. Evol. 37, 1979–1985. https://doi.org/10.1093/molbev/msaa059 (2020).
Katayama, S. et al. Antisense transcription in the mammalian transcriptome. Science 309, 1564–1566. https://doi.org/10.1126/science.1112009 (2005).
Wang, Z. Y. et al. Transcriptome and translatome co-evolution in mammals. Nature 588, 642–647. https://doi.org/10.1038/s41586-020-2899-z (2020).
Green, C. D. et al. A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-seq. Dev. Cell 46, 651-667.e610. https://doi.org/10.1016/j.devcel.2018.07.025 (2018).
Xiao, T. Z. et al. MAGE I transcription factors regulate KAP1 and KRAB domain zinc finger transcription factor mediated gene repression. PLoS ONE 6, e23747. https://doi.org/10.1371/journal.pone.0023747 (2011).
Akerfelt, M., Morimoto, R. I. & Sistonen, L. Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11, 545–555. https://doi.org/10.1038/nrm2938 (2010).
Skinner, B. M. et al. A high-throughput method for unbiased quantitation and categorization of nuclear morphology†. Biol. Reprod. 100, 1250–1260. https://doi.org/10.1093/biolre/ioz013 (2019).
Korfanty, J. et al. Crosstalk between HSF1 and HSF2 during the heat shock response in mouse testes. Int. J. Biochem. Cell Biol. 57, 76–83. https://doi.org/10.1016/j.biocel.2014.10.006 (2014).
Akerfelt, M. et al. Promoter ChIP-chip analysis in mouse testis reveals Y chromosome occupancy by HSF2. Proc. Natl. Acad. Sci. U.S.A. 105, 11224–11229. https://doi.org/10.1073/pnas.0800620105 (2008).
Martins, M. C. et al. Sperm head morphometry and chromatin condensation are in constant change at seminiferous tubules, epididymis, and ductus deferens in bulls. Theriogenology 161, 200–209. https://doi.org/10.1016/j.theriogenology.2020.12.004 (2021).
Torgerson, D. G. & Singh, R. S. Enhanced adaptive evolution of sperm-expressed genes on the mammalian X chromosome. Heredity (Edinb) 96, 39–44. https://doi.org/10.1038/sj.hdy.6800749 (2006).
Conant, G. C. & Wolfe, K. H. Turning a hobby into a job: How duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. https://doi.org/10.1038/nrg2482 (2008).
Peterson, K. A. & Murray, S. A. Progress towards completing the mutant mouse null resource. Mamm. Genome https://doi.org/10.1007/s00335-021-09905-0 (2021).
Birling, M. C. et al. A resource of targeted mutant mouse lines for 5,061 genes. Nat. Genet. 53, 416–419. https://doi.org/10.1038/s41588-021-00825-y (2021).
Guan, C., Ye, C., Yang, X. & Gao, J. A review of current large-scale mouse knockout efforts. Genesis 48, 73–85. https://doi.org/10.1002/dvg.20594 (2010).
Bradley, A. et al. The mammalian gene function resource: The International Knockout Mouse Consortium. Mamm. Genome 23, 580–586. https://doi.org/10.1007/s00335-012-9422-2 (2012).
Nagy, A. Manipulating the mouse embryo : a laboratory manual. (2003).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. https://doi.org/10.1038/nbt.3519 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Russell, L. D., Ettlin, R. A., Hikim, A. P. S. & Clegg, E. D. Histological and histopathological evaluation of the testis. Int. J. Androl. 16, 83–83. https://doi.org/10.1111/j.1365-2605.1993.tb01156.x (1993).
Riel, J. M., Yamauchi, Y., Huang, T. T., Grove, J. & Ward, M. A. Short-term storage of human spermatozoa in electrolyte-free medium without freezing maintains sperm chromatin integrity better than cryopreservation. Biol. Reprod. 85, 536–547. https://doi.org/10.1095/biolreprod.111.091322 (2011).
Organisation, W. H. WHO Laboratory Manual for the Examination of Human Semen and Sperm-Cervical Mucus Interaction (Cambridge University Press, 1999).
Sugimoto, M. & Abe, K. X chromosome reactivation initiates in nascent primordial germ cells in mice. PLoS Genet. 3, e116. https://doi.org/10.1371/journal.pgen.0030116 (2007).
Acknowledgements
We thank M. Arlt, C. Swanepoel, I. Mier, A. Lawson, D. de Rooij for editorial comments as well as D. de Rooij and M. Brogley for technical assistance. Additionally, we would like to thank the PATH-TMPRS, the Rogel Cancer Center Tissue and molecular Pathology Shared Resource (funding support NIH P30 CA04659229) for help with tissue sectioning and staining. We also would like to thank the transgenic animal model core at the University of Michigan for generation of our mutant mice, and the advanced genomics core at the University of Michigan for Sanger sequencing and RNA-seq.
Author information
Authors and Affiliations
Contributions
E.S.-D. and E.D. contributed equally to this work. E.S.-D., E.D. and J.M. designed the experiments. E.S.-D., E.D., E.G. and J.M. performed the experiments and analyzed the data. E.S.-D., E.D. and J.M. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Stark-Dykema, E.R., Dulka, E.A., Gerlinger, E.R. et al. X-linked palindromic gene families 4930567H17Rik and Mageb5 are dispensable for male mouse fertility. Sci Rep 12, 8554 (2022). https://doi.org/10.1038/s41598-022-12433-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-12433-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.