Abstract
Genetic studies have examined body-shape measures adjusted for body mass index (BMI), while allometric indices are additionally adjusted for height. We performed the first genome-wide association study of A Body Shape Index (ABSI), Hip Index (HI) and the new Waist-to-Hip Index and compared these with traditional indices, using data from the UK Biobank Resource for 219,872 women and 186,825 men with white British ancestry and Bayesian linear mixed-models (BOLT-LMM). One to two thirds of the loci identified for allometric body-shape indices were novel. Most prominent was rs72959041 variant in RSPO3 gene, expressed in visceral adipose tissue and regulating adrenal cell renewal. Highly ranked were genes related to morphogenesis and organogenesis, previously additionally linked to cancer development and progression. Genetic associations were fewer in men compared to women. Prominent region-specific associations showed variants in loci VEGFA and HMGA1 for ABSI and KLF14 for HI in women, and C5orf67 and HOXC4/5 for ABSI and RSPO3, VEGFA and SLC30A10 for HI in men. Although more variants were associated with waist and hip circumference adjusted for BMI compared to ABSI and HI, associations with height had previously been reported for many of the additional variants, illustrating the importance of adjusting correctly for height.
Similar content being viewed by others
Introduction
The cardiometabolic complications of obesity are influenced by body shape, showing a positive association with abdominal size and an inverse association with gluteofemoral size1. The waist-to-hip ratio (WHR) and body mass index (BMI) are used correspondingly as an index of body shape and an index of general obesity2. The WHR, however, is moderately correlated with BMI and cannot differentiate abdominal from gluteofemoral size. Although waist (WC) and hip circumference (HC) are measures of specific body regions, they are both strongly correlated with BMI. To account for the correlation with BMI, genome-wide association studies (GWAS) have used traditionally residuals from linear models regressing each of WHR, WC, or HC on BMI, thus creating body-shape indices independent from BMI3,4. Nevertheless, the adjustment of WC or HC for BMI introduces a positive correlation with height, which is stronger than the association of WC or HC with height4.
An alternative approach to creating body-shape indices independent from body size and general obesity has been implemented in the development of A Body Shape Index (ABSI) and Hip Index (HI)5,6. Similarly to BMI, ABSI and HI are based on the principle of allometry, which accounts for the expansion of the dimensions of individual body parts relative to the total body size with log-linear rather than linear models7. Similar to the scaling of log-transformed weight to log-transformed height used for the development of BMI, log-transformed WC and HC have each been scaled to log-transformed weight and log-transformed height to develop ABSI and HI5,6. This scaling accounts for the expansion of body circumferences proportional to body size, as reflected in height, and additionally accounts for the proportional expansion of body circumferences with general adiposity, as reflected in body weight. Consequently, the allometric body-shape indices are independent by design from height, as well as from BMI. Strong associations of ABSI with mortality and cardio-metabolic risk factors have been reported5,8 and we have demonstrated that ABSI achieves better mortality risk stratification than alternative body-shape indices, which are correlated with BMI9.
There are, however, no studies to date examining the genetic associations of ABSI and HI, no allometric counterpart of WHR and no insight into the influence of the correlation of traditional body-shape indices with height. The aims of our study, therefore, were to perform the first GWAS of allometric body-shape indices and to compare allometric and traditional body-shape indices with respect to their genetic associations. Our GWAS provides novel information for unbiased genetic associations of body-shape indices.
Results
We used data from the UK Biobank for 219,872 women and 186,825 men with white British ancestry (Supplementary Table S1). For allometric body-shape indices, we used ABSI, HI and a waist-to-hip index (WHI) calibrated for UK Biobank participants (ABSIUKB, HIUKB and WHIUKB, see details in Methods). For traditional body-shape indices, we used WC, HC and WHR adjusted for BMI in linear models (WCadjBMI, HCadjBMI and WHRadjBMI). We examined women and men separately, as pronounced sexual dimorphisms have been reported for the genetic associations of traditional body-shape indices10.
Genetic variants associated with allometric body-shape indices
We determined independent significant single nucleotide polymorphisms (SNPs) and consolidated them in independent genomic risk loci, represented by a locus lead SNP, with Functional Mapping and Annotation (FUMA) (see details in Methods).
A larger number of independent genomic risk loci were associated with WHIUKB (282 in women, 97 in men) compared to ABSIUKB (200 in women, 65 in men) and HIUKB (171 in women, 75 in men) (Table 1). The highest-ranked lead SNPs associated with WHIUKB (rs72959041 in women and rs577721086 in men) were both in the RSPO3 locus and in very strong linkage disequilibrium (r2 > 0.99). In addition, rs72959041 was the second highest-ranked lead SNP for ABSIUKB in women and was the highest-ranked lead SNP for HIUKB in both sexes (Supplementary Table S2). This variant stood out with some of the largest effect sizes, with a positive sign of the regression coefficients for ABSIUKB and WHIUKB and a negative sign for HIUKB, reflecting the phenotype of the minor allele. Most of the independent variants associated with WHIUKB similarly showed regression coefficients with opposite signs for ABSIUKB and HIUKB (Supplementary Fig. S1). Nevertheless, some variants were associated preferentially with ABSIUKB or HIUKB. In women, the most outstanding examples of loci associated exclusively with ABSIUKB were VEGFA and HMGA1, while the most prominent locus associated exclusively with HIUKB was KLF14. In men, prominent loci with variants associated exclusively with ABSIUKB were AC022431.2 (C5orf67), RP11-115J16.1, RP5-859D4.3 (CASC20) and the region including loci HOXC4, RP11-834C11.14 and HOXC5, while the most prominent loci associated exclusively with HIUKB were RSPO3, VEGFA and RP11-95P13.2 (SLC30A10) (Supplementary Fig. S2).
Approximately two thirds of the independent significant SNPs but less than one fifth of the corresponding genomic risk loci associated in our study with WHIUKB were novel, i.e. they have not been reported in the NHGRI-EBI GWAS Catalog11 in association with WHR or WHRadjBMI (Table 1, Supplementary Table S3). Nevertheless, over two thirds of the independent significant SNPs and one third of the corresponding genomic risk loci associated with ABSIUKB were novel, i.e. not previously reported in association with WC or WCadjBMI. Further, most of the independent significant SNPs and over two thirds of the corresponding genomic risk loci associated with HIUKB were novel, i.e. not previously reported in association with HC or HCadjBMI. It was notable, however, that many of the novel highest-ranked lead SNPs associated with allometric body-shape indices were in strong linkage disequilibrium (LD at r2 ≥ 0.6) with variants previously reported in association with the corresponding traditional body-shape index, which in our study showed lower significance (Fig. 1). Exceptions were the novel highly ranked lead SNPs associated with HIUKB in men, rs998584 in the VEGFA locus and rs6066114 in the EYA2 locus, which did not include in their clumps variants previously reported in association with HC or HCadjBMI, while for rs113733630 in the TFAP4 locus, the previously reported variant was not in the same LD block (Supplementary Table S2).
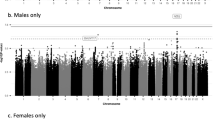
Miami plots of candidate SNPs identified for allometric body-shape indices in women and men. (a) GWAS of waist-to-hip index calibrated for UK Biobank participants (WHIUKB); (b) GWAS of a body shape index calibrated for UK Biobank participants (ABSIUKB); (c) GWAS of hip index calibrated for UK Biobank participants (HIUKB); P-P-values were derived from BOLT-LMM infinitesimal models; SNP-single nucleotide polymorphism; horizontal lines correspond to the genome-wide significance cut-off P = 5*10–8. Genomic risk loci with their corresponding locus lead SNPs were identified with FUMA v1.3.6a. All candidate SNPs are shown in grey, locus lead SNPs are colour-coded as follows: (grey circle) candidate SNPs; (dark red circle) novel genomic risk locus identified in the current study, with no previously reported candidate SNPs; (orange circle) genomic risk locus with a previously reported locus lead SNP; (yellow circle) genomic risk locus with a previously reported SNP in strong linkage disequilibrium (LD) with the locus lead SNP at r2 ≥ 0.6; (cyan circle) genomic risk locus with previously reported other candidate SNP. SNPs identified for allometric body-shape indices in the current study were matched against SNPs reported in the NHGRI-EBI GWAS Catalog11 (https://www.ebi.ac.uk/gwas/home, accessed on 07/04/2021) in association with the corresponding traditional body-shape index (with or without adjustment for body mass index, BMI), i.e. the waist-to-hip ratio for WHIUKB (catalogue sets EFO_0004343, EFO_0007788, EFO_0004302); waist circumference for ABSIUKB (EFO_0004342, EFO_0007789, EFO_0004302); hip circumference for HIUKB (EFO_0005093, EFO_0008039, EFO_0004302).
The deleteriousness Combined Annotation Dependent Depletion (CADD) score was above the recommended cut-off 12.3712 for 8% to 20% of the locus lead SNPs associated with allometric body-shape indices (Fig. 2). Nevertheless, almost half of all locus lead SNPs were in strong LD with a variant with genome-wide significance showing higher CADD than the cut-off.
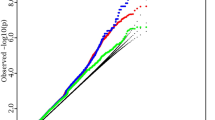
Deleteriousness (CADD) score of locus lead SNPs identified for allometric body-shape indices. (a) waist-to-hip index (WHIUKB) calibrated for UK Biobank women (n = 282 genomic risk loci with the corresponding locus lead SNPs); (b) a body shape index (ABSIUKB) calibrated for UK Biobank women (n = 200); (c) hip index (HIUKB) calibrated for UK Biobank women (n = 171); (d) WHIUKB for UK Biobank men (n = 97); (e) ABSIUKB for UK Biobank men (n = 65); (f) HIUKB for UK Biobank men (n = 75). CADD-Combined Annotation Dependent Depletion; LD-linkage disequilibrium; SNP-single nucleotide polymorphism; red vertical lines-recommended cut-off 12.37 for CADD (the higher the more deleterious)12; horizontal line-genome-wide significance cut-off (P = 5*10–8); left-hand side-CADD for the locus lead SNP of each genomic risk locus, with the proportion above the cut-off; right-hand side-candidate SNPs in strong LD with the locus lead SNP (colour-coded according to r2 ≥ 0.6), showing the maximum CADD within the LD block, plotted with the corresponding significance on the y-axis; PWHI / ABSI / HI-P-values for body-shape indices, derived from BOLT-LMM infinitesimal models; (black circle)-marks the locus lead SNP when this is showing the maximum CADD within the corresponding LD block; percentages (top corners)-percentage above the cut-offs for both, CADD and genome-wide significance (all differences between left-hand side and right-hand side proportions were significant at P < 0.0001 when compared with Fisher’s exact test, except for ABSIUKB in men (P = 0.0008).
Following recent reports of a fine-scale population structure in UK Biobank13, we checked matches with the SNPs reported in association with birth location. Only rs1805007 (MC1R), reported in association with height, was identified in our study as a candidate SNP and was included in the LD block of locus lead SNPs for WCadjBMI and HCadjBMI in women and men (lowest P = 4.4*10–11). Although rs9268556 (MHC region), reported in association with forced vital capacity, was additionally identified as a candidate SNP for WHIUKB, ABSIUKB and WHRadjBMI in women and HCadjBMI in men (lowest P = 2.4*10–9), this was not included in the LD block of a locus lead SNP.
Sexual dimorphism in the genetic associations of allometric body-shape indices
The genetic association patterns of allometric body-shape indices differed considerably between women and men. The heritability was larger in women compared to men, with up to three times more independent significant SNPs identified in women (Table 1), when the excess of women vs men in the dataset was only approximately 20%. Some 30% of the independent significant SNPs for ABSIUKB but 10% or less for HIUKB showed sex differences in effect size at psex < 5*10–6 (Table 1). Several highly ranked variants, however, were particularly affected (Supplementary Fig. S3). Thus, prominent associations with WHIUKB and ABSIUKB in women but not in men showed variants in loci COBLL1, RP11-95P13.1, ADAMTS9-AS2, CMIP and AC022431.2 (C5orf67) and variants in the region including loci ZNF664 and FAM101, while KLF14 was the most prominent locus associated with HIUKB in women but not in men (psex < 5*10–28 for all independent significant SNPs in these loci). In addition, although variants in RSPO3 locus were the highest-ranked associated with WHIUKB and HIUKB in both sexes, variants in the VEGFA and RSPO3 loci were associated with ABSIUKB almost exclusively only in women, while variants in the VEGFA locus were associated with HIUKB mainly in men (Supplementary Fig. S3).
Gene-level associations with allometric body-shape indices
We examined gene-level associations with Multi-marker Analysis of GenoMic Annotation (MAGMA) employed in FUMA, which uses the association statistics of all SNPs included in a gene (see Methods for details).
Similar to SNP-based analyses, a larger number of non-overlapping genomic risk regions were associated with WHIUKB (237 in women, 83 in men) compared to ABSIUKB (158 in women, 61 in men) and HIUKB (136 in women, 52 in men) and the proportions of novel gene-based regions were comparable to the proportions of novel SNP-based loci (Table 1). There were, however, differences in the lead genes (Fig. 3). The highest-ranked region for WHIUKB and ABSIUKB in women was FAM101A and for WHIUKB and HIUKB in men was DLEU1. Prominent novel genes with their corresponding genomic risk regions were XKR6 for ABSIUKB and ERI1 for WHIUKB and ABSIUKB in men, EYA2 for HIUKB in both women and men, SLC30A10 and SSPN for HIUKB in women and PEMP and AKR1C2 for HIUKB in men (Supplementary Table S4). Although few independent significant SNPs had previously been reported in association with cancer (or were in strong LD with previously reported SNPs, Supplementary Fig. S4), one third of the lead genes had previously been reported in association with cancer (Table 1), including the prominent lead genes DLEU1 and ZMIZ1 and a notable cluster of significant genes in the region of NEU1 on chromosome 6 (Supplementary Fig. S5).
Miami plots of significant genes identified for allometric body-shape indices in women and men. (a) waist-to-hip index calibrated for UK Biobank participants (WHIUKB); (b) a body shape index calibrated for UK Biobank participants (ABSIUKB); (c) hip index calibrated for UK Biobank participants (HIUKB); P-P-values were derived from MAGMA v1.08 employed in FUMA v1.3.6a and were adjusted with Bonferroni correction for 19,088 protein-coding genes; SNP-single nucleotide polymorphism; horizontal lines-correspond to P = 0.05 after Bonferroni correction. Significant genes within 250 kb window were consolidated in genomic risk regions represented by a lead gene. All significant genes are shown in grey, lead genes are colour-coded as follows: (grey circle)-significant genes; (dark red circle)-novel genomic risk region identified in the current study, with no previously reported significant gene; (orange circle)-genomic risk region with a previously reported lead gene; (cyan circle)-genomic risk region including a previously reported significant gene (not the lead gene). Genes identified for allometric body-shape indices in the current study were matched against genes reported in the NHGRI-EBI GWAS Catalog11 (https://www.ebi.ac.uk/gwas/home, accessed on 07/04/2021) in association with the corresponding traditional body-shape index (with or without adjustment for body mass index, BMI), i.e. the waist-to-hip ratio for WHIUKB (catalogue sets EFO_0004343, EFO_0007788, EFO_0004302); waist circumference for ABSIUKB (EFO_0004342, EFO_0007789, EFO_0004302); hip circumference for HIUKB (EFO_0005093, EFO_0008039, EFO_0004302).
Competitive gene-set analysis showed associations of allometric body-shape indices with gene sets related to embryonal morphogenesis and organogenesis, regulation of DNA binding, gene expression, biosynthetic processes and cell differentiation, transcription factor complexes, extracellular matrix component, circulatory system development, skeletal system development, ossification and chondrocyte differentiation and several cancers, with only a single gene set related to fat cell differentiation, which was associated with ABSIUKB in women (Fig. 4). A larger number of gene sets showed significant associations with WHIUKB and ABSIUKB in women (42 and 29, correspondingly) compared to men (16 and 2) but a similar number were associated with HIUKB (14 in women, 12 in men), although with only three overlapping.
Gene sets associated with allometric body-shape indices. ABSI-a body shape index calibrated for UK Biobank participants (ABSIUKB); Currated-curated gene set; GO-BP-gene ontology biological process; GO-CC-gene ontology cellular component; GO-MM-gene ontology molecular function; HI-hip index calibrated for UK Biobank participants (HIUKB); WHI-waist-to-hip index calibrated for UK Biobank participants (WHIUKB); #-reactome runx1 regulates transcription of genes involved in wnt signaling. The figure includes gene sets identified using competitive gene-set analysis with MAGMA v1.08 employed in FUMA v1.3.6a as significant for at least one of the allometric indices in women or men with adjusted Padj < 0.05, incorporating Bonferroni correction for multiple comparisons for 15,485 gene sets. Gene sets with Padj < 0.05 for WHIUKB in women were sorted in descending order of Padj, for WHIUKB in women, then the remaining gene sets with Padj < 0.05 for WHIUKB in men were sorted in descending order of Padj for WHIUKB in men, then the remaining gene sets with Padj < 0.05 for HIUKB in women were sorted in descending order of Padj for HIUKB in women and last, the remaining gene sets with Padj < 0.05 for HIUKB in men were sorted in descending order of Padj for HIUKB in men, leaving a single gene set associated only with ABSIUKB in women. The horizontal lines mark the start of the next sorting criterion. White cells are gene sets with Padj ≥ 0.05 for the corresponding body-shape index.
Associations with expression Quantitative Trait Loci (eQTL) were significant in both women and men for adipose tissue (subcutaneous and visceral omentum), arteries (tibial, coronary and aorta) and, unexpectedly, for female reproductive organs (breast mammary tissue, uterus, ectocervix, endocervix and fallopian tube) and, in women only, also for ovary and vagina (Fig. 5).
eQTLs associated with allometric body-shape indices. ABSI-a body shape index calibrated for UK Biobank participants (ABSIUKB); eQTLs-expression Quantitative Trait Loci; HI-hip index calibrated for UK Biobank participants (HIUKB); WHI-waist-to-hip index calibrated for UK Biobank participants (WHIUKB). The figure includes specific tissues from the Genotype-Tissue Expression (GTEx) v8.0 database, queried for associations with eQTLs with gene-property analysis, which were identified by MAGMA v1.08 employed in FUMA v1.3.6a as significant for at least one of the allometric indices in women or men, i.e. with adjusted Padj < 0.05, incorporating Bonferroni correction for multiple comparisons for 54 tissues. White cells are tissues with Padj ≥ 0.05 for the corresponding body-shape index.
Comparison between allometric and traditional body-shape indices
On the one hand, WHIUKB and WHRadjBMI were very strongly phenotypically correlated with each other and both were uncorrelated with height or BMI (Supplementary Table S5). Correspondingly, the ranking of independent significant SNPs associated with them was in excellent agreement, especially in women (Fig. 6a,d, Supplementary Fig. S6), with only some 5% showing differences in effect size at pdifference < 5 * 10–6 (Table 1, Supplementary Fig. S7 for women, Supplementary Fig. S8 for men), mainly for variants previously reported in association with height (Supplementary Fig. S9 for women, Supplementary Fig. S10 for men).
Significance ranking of independent significant SNPs: comparison between pairs of traditional and allometric body-shape indices. (a) independent significant SNPs (at r2 < 0.6 within 1 Mb window) for WHIUKB (n = 1,988) or WHRadjBMI (n = 1,959) in women; (b) independent significant SNPs for ABSIUKB (n = 1,273) or WCadjBMI (n = 1,586) in women; (c) independent significant SNPs for HIUKB (n = 814) or HCadjBMI (n = 2,008) in women; (d) independent significant SNPs for WHIUKB (n = 617) or WHRadjBMI (n = 571) in men; (e) independent significant SNPs for ABSIUKB (n = 266) or WCadjBMI (n = 842) in men; (f) independent significant SNPs for HIUKB (n = 222) or HCadjBMI (1,400) in men. Association statistics p-values were derived from BOLT-LMM infinitesimal models. ABSIUKB-a body shape index calibrated for UK Biobank participants; BMI-body mass index; HCadjBMI-hip circumference adjusted for BMI; HIUKB-hip index calibrated for UK Biobank participants; SNP-single nucleotide polymorphism; WCadjBMI-waist circumference adjusted for BMI; WHRadjBMI-waist-to-hip ratio adjusted for BMI; WHIUKB-waist-to-hip index calibrated for UK Biobank participants. Colour scale-colour-marked were only SNPs reported as associated with height in the NHGRI-EBI GWAS Catalog11 (https://www.ebi.ac.uk/gwas/home, accessed on 07/04/2021), i.e. included in catalogue sets EFO_0004339 or EFO_0004302; (navy circle)-independent significant SNP reported in association with height; (cyan circle)-independent significant SNP in strong LD (at r2 ≥ 0.6) with a SNP reported in association with height.
On the other hand, while the allometric indices ABSIUKB and HIUKB were phenotypically uncorrelated with height and BMI, the traditional indices WCadjBMI and HCadjBMI were uncorrelated only with BMI. The adjustment for BMI was apparently introducing a moderate positive correlation with height, as height was only weakly correlated with unadjusted WC and HC (Supplementary Table S5). Notably, a larger number of independent significant SNPs were associated with WCadjBMI and HCadjBMI, with half or more of them showing significant differences in effect size compared to their allometric counterparts ABSIUKB and HIUKB (Table 1, Supplementary Fig. S7, Supplementary Fig. S8), except for WCadjBMI and ABSIUKB in women, which showed more similarity (Fig. 6b). The main differences between WCadjBMI and ABSIUKB (Fig. 6b,e) and between HCadjBMI and HIUKB (Fig. 6c,f) concerned independent variants which were either previously reported in association with height or were in strong LD with variants reported in association with height (Supplementary Fig. S9, Supplementary Fig. S10). Half or more of the locus lead SNPs for WCadjBMI in men and for HCadjBMI in men and women had previously been associated with height, compared to less than one third for ABSIUKB and HIUKB (Table 1). More comparable proportions of lead genes (with gene-level significance) had previously been reported in association with height (less than half for allometric indices, Supplementary Fig. S11, and more than half for traditional indices), but the absolute number of lead genes identified for traditional indices was considerably larger (Table 1).
Discussion
Our study presents the first GWAS of allometric body-shape indices, performed separately in women and men with white British ancestry, and a comparison with traditional body-shape indices. One third of the genomic risk loci associated with ABSIUKB and over two thirds of the genomic risk loci associated with HIUKB in our study were novel. Genetic associations were sexually dimorphic, with fewer independent variants identified in men compared to women. The highest-ranked independent variant for WHIUKB, ABSIUKB in women and HIUKB was rs72959041 in the RSPO3 locus. WHIUKB, combining WC and HC, showed higher sensitivity to detect genetic associations compared to the regional indices ABSIUKB and HIUKB, based individually on WC or HC. The genetic association patterns of WHIUKB and WHRadjBMI were very similar. ABSIUKB and HIUKB showed fewer genetic associations compared correspondingly to WCadjBMI and HCadjBMI, which were associated with a larger proportion of variants previously reported in association with height.
Our study highlights the importance of the RSPO3 locus for body shape and fat distribution in individuals with white British ancestry and potentially in individuals with white ethnic background. RSPO3 protein, together with wingless-type WNT proteins, activate the canonical WNT/β-catenin pathway, which plays a depo-specific role in regulating locally adipocyte hyperplasia, hypertrophy and browning14. RSPO3 shows differential expression in adipose depots, with the highest expression in visceral, intermediate in abdominal subcutaneous and lowest in gluteal subcutaneous adipose tissue15. RSPO3 additionally promotes angioblast specification and vascular development16, plays a key role throughout life in maintaining the structural zonation and the replenishment of damaged cells in the adrenal glands17, regulates the renewal and differentiation of stem cells18, and contributes to cancer development and progression19,20. Genetic variants in the RSPO3 locus associated with abdominal obesity have additionally been associated with dyslipidaemia21, thus relating the RSPO3 gene to the metabolic syndrome. The highest-ranked RSPO3 variants, rs72959041 (in women) and rs577721086 (in men), which are in strong linkage disequilibrium, are specific to European populations22 and have previously been reported in association with WHRadjBMI3,4,23. The active variant, however, may be rs577721086, as this is located in the attachment site of CCCTC‑binding factor, which acts as a gene repressor, insulator or activator, but also regulates the chromatin structure and enables inter-chromosomal interactions24.
Although the genetic associations of body-shape indices are interpreted traditionally from the perspective of the adipose tissue and insulin resistance25, highly ranked variants associated with body shape are located in genes coding transcription factors, receptors and enzymes involved in morphogenesis, embryonal developmental, cell proliferation and cell survival, which have additionally been linked to various cancers. Thus, in addition to the RSPO3 proteins, HMGA1 and TFAP4 activate the WNT/β-catenin pathway, stimulate cell migration and invasion and promote cancer progression26,27. VEGFs, which are upregulated by hypoxia, are key factors for tumour-associated angiogenesis, tissue infiltration and metastasis28. PLXND1, which is involved in angiogenesis and is upregulated by VEGFs, has a constitutively low expression in adult tissues but is overexpressed in cancer tissues and their vasculature29. KLF14 is associated with insulin resistance, dyslipidaemia, type 2 diabetes and a female-specific shift of body fat from gynoid to abdominal stores, in agreement with the hip-specific association in women found in our study. In mice, adipocyte-specific deletion of Klf14 results in similar metabolic effects, while Klf14 knockout results in spontaneous tumorigenesis30. TBX15 belongs to a family of transcription factors regulating differentiation, proliferation, tissue integrity and epithelial-mesenchymal transition, which are relevant to cancer development and metastasis31. Overexpression, mutations and epigenetic silencing of ADAMTS genes have been reported in different tumours32. SLC30A10 is a zinc transporter maintaining zinc homeostasis, which when dysregulated can result in cancer initiation and progression33. FGFR4 plays a critical role in embryonic development, tissue repair, tumour angiogenesis and progression34. EYA1-4 proteins can influence tumour progression through several mechanisms35. Homeobox (HOX) genes code transcription factors with DNA-binding activity, which regulate the formation of axial patterns and body shape during embryogenesis and have been implicated in cancer development36. COBLL1 is the ancestor of a family of proteins involved in morphogenesis and embryonal patterning in organisms with axial symmetry and is upregulated in castrate-resistant prostate cancer with poor prognosis37. Upregulation of DLEU1 can also promote tumorigenesis38. A question, therefore, emerges whether excess abdominal fat mediates the development of cancers associated with abdominal obesity39, or abdominal obesity and cancer are parallel outcomes with shared genetic predisposition. Although studies exploring a causal relationship between body shape and cancer are limited, Mendelian randomisation based on traditional body-shape indices has already provided evidence for causal positive associations of WHR or WHRadjBMI with colorectal cancer40,41 and renal cell carcinoma42 but an inverse association with breast cancer43.
Allometric body-shape indices showed sexually dimorphic genetic association patterns, like traditional body-shape indices3,4,10,44, with an overall lower heritability in men. There were, however, fewer genetic associations and with lower significance in men then would correspond proportionally to the slightly smaller sample size compared to women, as noted for traditional indices3. In contrast height, which is also phenotypically dimorphic between sexes, shows considerably larger heritability than body-shape indices and a closer agreement of the genetic association patterns between women and men45. This raises the question, how women and men differ with respect to the regulatory factors determining fat distribution and body shape.
Sex steroid hormones play a central role in the regulation of fat distribution. Oestrogens counter fat accumulation in the abdominal area and favour fat accumulation in the gluteofemoral area46. Oestrogen levels in blood47 and the expression of ERα in the abdominal area48 decrease after the menopause, which permits a functional androgen dominance and the development of android type obesity in post-menopausal women49. Nevertheless, age-related differences in the genetic associations have been reported only for BMI and not for WHRadjBMI10. In contrast, testosterone levels in women remain lower than in men at all ages50 and are comparable between pre- and post-menopausal women, as ovarian androgen production is largely maintained long after the menopause51. The stress hormone cortisol, regulated by the hypothalamic–pituitary–adrenal (HPA) axis, is also closely involved in the regulation of fat distribution, with chronic cortisol excess resulting in the development of visceral adiposity and the metabolic syndrome52. Notably, the HPA axis shows a sexually dimorphic activity, with stronger responses to stimulation and suppression in women compared to men53,54 and increased responses to HPA stimulation in men after suppression of gonadal testosterone production55. Abdominal obesity also influences HPA responsiveness, with larger WHR associated with a stronger response to HPA stimulation56 and a weaker response to HPA suppression53. Animal studies provide further evidence for a sexually dimorphic adrenal function, suggesting that androgens rather than oestrogens are important for adrenal regulation57. Furthermore, absence of the androgen receptor in mice results in downregulation of the glucocorticoid receptor in the pituitary gland, adrenal hypertrophy and glucocorticoid overproduction58. Thus, the higher HPA responsiveness in women, enabled by constitutively lower testosterone levels, may engage a more complex regulatory network and present more opportunities for genetic polymorphisms to act as rate-limiting steps in fat distribution, potentially explaining the pronounced sexual dimorphism in the heritability and the genetic association patterns of body-shape indices4,45.
Despite the genetic and phenotypic differences in body-shape between women and men, the eQTL associations of genes associated with body-shape indices were in line with an involvement of adipose and vascular tissues, but also intriguingly of female reproductive organs in men as well as in women. This apparently paradoxical association in men may not be completely illogical, as female reproductive organs would be responsive to oestrogen-activated pathways and the formation of the Mullerian duct, from which the uterus and its adnexa are derived, is regulated by WNT signalling pathways59, which may have pleiotropic roles.
We have previously demonstrated the need to adjust body-shape indices for BMI prior to using them in statistical models examining phenotypic associations9. We have now shown that it is also essential to account correctly for height, in order to avoid an “over-adjustment” arising from constraining the relationship between weight and height to a fixed proportion in BMI. In the case of WCadjBMI and HCadjBMI, the adjustment for BMI does not reflect correctly the scaling of WC or HC with height and introduces phenotypic and genetic associations with height. When WC and HC are adjusted for height, in addition to BMI or weight, as in the allometric indices ABSIUKB and HIUKB, the relationship between weight and height is unconstrained and the correlation of WC and HC with height, as well as with BMI, is minimised. In the case of WHRadjBMI, the phenotypic and genetic association patterns were very similar to the allometric counterpart WHIUKB, despite the lack of additional adjustment for height, because the scaling coefficients for weight and height in individuals with white British ancestry were in proportion 1:2, similar to BMI. Nevertheless, given the large ethnic variability in body shape, this may not be universally applicable to other ethnicities. It would thus be advisable to use body-shape indices adjusted for height, as well as for BMI or weight, in order to minimise correlations with body size. Evaluating correctly the pleiotropic contributions is particularly important as many variants and genes associated with allometric body-shape indices are apparently related to growth and regulatory factors and a sizeable proportion of them have previously been reported in association with height. A mechanistic parallel with a pathway affecting either height or regional size when dysregulated in different periods of life could be drawn with growth hormone signalling. Growth hormone excess in adolescents is associated with increased height but an excess in adults, after epiphyseal closure, is associated with regional enlargement of the hands, feet and the face, i.e. acromegaly. Growth hormone signalling is further related to metabolic alterations and some cancers60. A similar principle could be relevant to other growth or regulatory factors.
Our study benefited from a large sample size, anthropometric measurements performed by trained personnel according to standardised protocols and access to established bioinformatics pipelines. There were, however, several limitations. It should first be acknowledged, that the UK Biobank cohort is not representative of the UK population at the time of recruitment and is subject to a healthy volunteer bias which should be considered when interpreting the findings61. In addition, we lacked a validation cohort of similar ethnicity to the discovery dataset, although this would affect mainly the validity of associations with lower and borderline significance, while the highest-ranked genetic polymorphisms showed very strong and convincing associations. We also lacked datasets of comparable size including individuals with different ethnic backgrounds, as less than 5% of UK Biobank participants reported black or Asian ethnic background. Further, there were no reliable imaging measures of body composition (i.e. from dual-emission X-ray absorptiometry scans or magnetic resonance imaging) with comparable sample size. Furthermore, we did not perform a meta-analysis of waist and hip indices, since there are no other GWAS of ABSI and HI and we have already argued that WCadjBMI and HCadjBMI are not reliable body-shape indices. It would be useful, however, to consider a multi-trait meta-analysis approach62 when studies of other ethnicities are available. We did not specifically adjust our analyses for geographical location, which has been highlighted as a source of residual confounding of associations with BMI, bioelectric impedance fat mass measurements and height in UK Biobank13, as we minimised the number of adjustment variables to avoid introducing collider bias. There were, however, no matches of the reported variants related to geographical location with the main variants identified in our study, hence a confounding from fine-scale population structure is unlikely. Further, we could not perform mechanistic investigations linking genetic polymorphisms to adrenal function or cancer, which were beyond the scope of our study. Lack of Mendelian randomisation analysis, which may support a causal effect of the identified variants and genes on cancer and may further clarify the relationships in the network of mediators63, is clearly a limitation. This, however, was also beyond the scope of the current study. Due to the large heterogeneity of cancers, associations would need to be consider separately for each individual cancer location and for each major histological type and the question of potential pleiotropy, which is very likely, would need to be addressed in detail. It should last be noted, that although examining ABSIUKB and HIUKB enabled the identification of variants associated exclusively with the abdominal or the gluteofemoral regions, these associations may be determined by features other than fat accumulation, e.g. the status of lean mass in the gluteofemoral region. It would therefore be important to examine the association patterns of genetic variants identified for anthropometric indices with measures of body compositions, which would become available in the future for a larger part of UK Biobank participants.
In conclusion, in the first GWAS of allometric body-shape indices, we have identified novel genetic variants and genes and have obtained unbiased association statistics for individuals with white British ancestry, which would inform future studies of fat distribution, body shape and the disorders associated with them. The highest-ranked genes associated with body-shape indices have previously been associated in mechanistic studies with adrenal cell renewal, vascularisation and cancer development and progression, in addition to their functions in adipose tissue. The comparison of allometric and traditional body-shape indices demonstrated that adjustment of body-shape indices for height, as well as for BMI or weight, is warranted to avoid associations with height, which were more pronounced for WCadjBMI and HCadjBMI. Differences between allometric and traditional body-shape indices with respect to their phenotypic and genetic associations were minimal when accounting correctly for height.
Methods
Study participants
UK Biobank is a prospective cohort with an ongoing follow-up, including 502,543 participants. Recruitment and data collection have previously been described64,65. We excluded in total 95,846 participants. Exclusions were determined by a lack of genetic data or withdrawn consent (n = 15,229); outliers for heterozygosity or missing genotyping rate, or sex chromosome aneuploidy, or a mismatch between genetic and self-reported sex (n = 843); age below 40 or above 70 years when attending an assessment centre at baseline (n = 14); missing weight, height, waist or hip circumference measurements (n = 2,097); or pregnancy at baseline (n = 115). We further restricted the selection to participants with white British ancestry, defined by UK Biobank according to their genetic characteristics (excluded n = 77,548). The final dataset comprised 406,697 participants.
Genotyping and imputation
Genotyping, imputation and quality control were performed centrally by UK Biobank and have previously been described65. Two genotyping arrays were used: Applied Biosystems UK BiLEVE Axiom Array (~ 50,000 participants) and a closely related Applied Biosystems UK Biobank Axiom Array (~ 450,000 participants).
Outcome measures and indices
We converted all anthropometric measurements to body-shape indices with adjustments either for BMI in linear models, or for weight and height in log-linear models. BMI was calculated by dividing weight (kg) by squared height (m).
For traditional body-shape indices, we used residuals of linear models regressing each of WC (cm), HC (cm) or WHR on BMI (WHRadjBMI, WCadjBMI, HCadjBMI). For the corresponding allometric body-shape indices, we used ABSI and HI, and created a new waist-to-hip index (WHI), in order to complete the set. The published formulas for ABSI and HI have previously been derived for participants in the National Health and Nutrition Examination Survey (NHANES)5,6:
To avoid phenotypic correlations between anthropometric indices arising from differences in the anthropometric characteristics of UK Biobank and NHANES participants and to enable comparability with traditional residual methods, we calibrated ABSI and HI, as well as WHI, for UK Biobank participants (ABSIUKB, HIUKB and WHIUKB). We derived the power coefficients from linear models regressing each of log-transformed WC (cm), HC (cm) or WHR on log-transformed weight (kg) and height (m) measured at baseline:
and generated allometric body-shape indices according to the general formula:
where β and γ are the regression coefficients for weight and height (Supplementary Table S6). We included ABSI and HI calculated with the published regression coefficients from NHANES only for the phenotypic comparisons.
The absolute values of the power coefficients for weight and height for WHRUKB were close to a ratio of 1:2 in both men and women, which corresponds to the relationship weight/height2 in BMI (Supplementary Table S6). Therefore, in analogy to ABSI, for which the published coefficients were derived by rounding the power coefficients obtained in NHANES to simple fractions5, we generated for comparison a simplified version of WHI with the formula:
As body-shape patterns show distinct differences between sexes10, we generated all indices and performed all statistical analyses separately for men and women. We used Blom’s method for inverse normal transformation of anthropometric indices (package RNOmni in R), as in66.
Association testing
We obtained Pearson’s coefficient for partial phenotypic correlation between anthropometric indices with function pcor (package ppcor in R), adjusting for age at baseline.
We used Bayesian linear mixed-model analysis BOLT-LMM v2.3 for genome-wide association testing67,68, which incorporates in the statistical algorithm a correction for population stratification and thus accounts for the relatedness between UK Biobank participants. To estimate the parameters of the LMM, we used linkage disequilibrium (LD) scores from the 1000 Genomes EUR samples from individuals with European ancestry69 and selected a coreset with high-quality genetic variants by restricting the list of variants released by UK Biobank after the centrally performed quality control to variants with missingness < 0.015, minor allele frequency MAF > 5% and Hardy–Weinberg exact test P > 1*10–6 (Supplementary Table S7). We performed the main analyses with variants with MAF ≥ 1% and imputation quality factor INFO > 0.1. We adjusted all models for age at baseline, age squared and a binary indicator of genotyping array. Supplementary Table S8 includes quality control parameters for BOLT-LMM. We obtained heritability estimates (h2g) from BOLT-LMM, based on the genetic relationship matrix. We report association statistics based on the BOLT-LMM infinitesimal models, which use a Gaussian single nucleotide polymorphism (SNP) effect prior.
Mapping, annotation and prioritisation of genetic variants
Individual genetic variants were defined with the chromosome, the genomic position in base pairs, the minor (alternative) allele and the major (reference) allele. For some SNPs there were more than one alternative alleles, hence the total number of genetic variants associated with a given body-shape index could be higher than the total number of SNPs. We used the SNP2GENE process of the web application Functional Mapping and Annotation (FUMA) v1.3.6a70 to perform positional mapping, clumping and annotation of genetic variants according to the p-values of the association statistics obtained from the corresponding BOLT-LMM infinitesimal model for variants with high imputation quality (INFO > 0.9). Independent significant SNP were defined as variants with genome-wide significance (P ≤ 5*10–8), which were in approximate linkage equilibrium with each other, at r2 < 0.6 within a 1 Mb window. Variants with nominal significance (P < 0.05) in LD with an independent significant SNP (r2 ≥ 0.6 within a 1 Mb window) formed the corresponding LD block of candidate SNPs. To map LD, we used the “UKB release2b 10 k White British” panel in FUMA. Independent significant SNPs in LD with each other (r2 ≥ 0.1 within 1 Mb window) were consolidated in a clump, represented by a lead SNP with the lowest p-value. Lead SNPs with corresponding clump boundaries less than 250 kb apart were merged into a genetic risk locus, represented by the locus lead SNP with the lowest p-value. All candidate SNPs were mapped to genes within a maximum distance of 1 kb (based on their genomic position) and for functional consequences (based on Ensemble genes v92) with ANNOVAR employed in FUMA71. We used as a measure of pathogenicity of a given variant the deleteriousness score (Combined Annotation Dependent Depletion (CADD) score) calculated in FUMA and evaluated this against the recommended cut-off of 12.3712. We additionally annotated the functionality of candidate SNPs with genome-wide significance with Ensembl Variant Effect Predictor (VEP) v9072. This maps each variant to the gene or location with the nearest protein-coding transcription start site within a window of 200 kb from the position of the examined variant.
Gene-based association analysis
For gene-level, gene-set and gene-property analysis, we used Multi-marker Analysis of GenoMic Annotation (MAGMA) v.1.0873, which is employed in FUMA v1.3.6a with default settings. The reference panel for LD and the Ensemble version for assigning SNPs to genes (within a symmetric window of 1 kb from both sides) were set as described for SNP annotation and prioritisation above.
Gene analysis was based on the summary SNP statistics of all variants with INFO > 0.9 (i.e. including the complete distribution). A SNP-wide mean model was used to calculate an association statistic and p-value for each gene. Significant genes were considered those with P < 0.05, after applying Bonferroni correction for multiple comparisons for 19,088 identified protein-coding genes. Significant genes with boundaries within 250 kb of each other were clumped in genomic risk regions, represented by a lead gene with the lowest P-value.
Competitive gene-set analysis was performed as a gene-level linear regression model to test whether the genes included in each gene-set (a binary indicator) showed stronger (positive) associations with the phenotype than other genes, generating a one-sided p-value. The model was conditioned by default on gene size, gene density (reflecting LD between SNPs in the gene), the inverse of the mean MAF in the gene (to account for potential power loss in very low MAF SNPs), and the log values of the three variables73. FUMA v1.3.6a uses gene sets obtained from the Molecular Signatures Database (MSigDB) v7.0, including curated gene sets from online pathway databases and gene ontology (GO) terms. The significance of associations with gene sets was evaluated at P < 0.05, after Bonferroni correction for 15,485 examined gene sets.
The Genotype-Tissue Expression (GTEx) v8.0 database was used to perform gene-property analysis, as implemented in FUMA v1.3.6a, which examines associations with expression Quantitative Trait Loci (eQTL). This uses a similar linear regression model to gene-set analysis, but with the average log2 transformed gene expression values per tissue (a continuous variable), to test the (positive) relationship between highly expressed genes in a specific tissue and genetic associations represented by gene-level statistics, generating a two-sided p-value. The significance of associations with eQTLs was evaluated at P < 0.05, after Bonferroni correction for 54 individual tissue types.
Match against published reports
We matched all candidate SNPs and the significant genes identified by MAGMA in FUMA against SNPs and genes previously reported in the NHGRI-EBI GWAS Catalog11 (accessed on 07/04/2021, https://www.ebi.ac.uk/gwas/home) in association with anthropometric indices. Novelty for a genomic risk locus was concluded if there was no match of any candidate SNP included in the locus with a SNP reported in the NHGRI-EBI GWAS Catalog in association with the corresponding traditional index. Similarly, novelty for a genomic risk region was concluded if there was no match of any significant gene included in the region with a gene reported in the NHGRI-EBI GWAS Catalog in association with the corresponding traditional index. Match with reported associations with height or cancer were compared at the level of independent significant SNPs (i.e. a match with any candidate SNP in the LD block) or at the level of an individual significant gene.
Difference analysis
To test for sex-dimorphic effects, we calculated a t-statistic as follows:
where β are the regression coefficients for a given variant in men or women, SE are the corresponding standard errors and rsex is the Spearman rank correlation coefficient between the regression coefficients in men and women for all examined genome-wide variants3,10. We extracted the corresponding p-values (psex) from a t-distribution with function pt in R. We used a similar formula to compare heritability between sexes, replacing the regression coefficients with heritability estimates in men and women.
To test for difference in effect sizes between allometric and traditional indices, we calculated, separately in women and men, t-statistics and the corresponding pdifference with a similar formula to the one used for sex dimorphisms:
where β are the regression coefficients for a given variant for the corresponding allometric and traditional index in a pair (i.e. WHIUKB and WHRadjBMI, ABSIUKB and WCadjBMI, HIUKB and HCadjBMI), SE are the corresponding standard errors and rallo-trad is the Spearman rank correlation coefficient between the regression coefficients for the allometric and traditional index for all examined genome-wide variants. To test for difference in effect sizes between the allometric waist and hip indices, the parameters for allometric and traditional indices were replaced with the corresponding parameters for ABSIUKB and HIUKB and rABSI-HI was calculated as above.
As differences in effect sizes were considered only for independent significant SNPs identified for at least one of the two compared sexes or indices and this would vary between comparisons, we evaluated differences relative to a single universal conservative cut-off P < 5 × 10–6, which is equivalent to a Bonferroni correction for 10,000 comparisons and reflects strong evidence for association.
We used R version 3.6.1. for the management of data and results74.
Ethical approval and consent to participate
This research was conducted according to the principles expressed in the Declaration of Helsinki. The UK Biobank cohort has been approved by the North West Multicenter Research Ethics Committee, UK (Ref: 16/NW/0274). Written informed consent has been obtained from all study participants. The current study was approved by the UK Biobank access management board under application 41952. Participants who had withdrawn consent by the time of the analysis were excluded from the dataset.
Data availability
The data supporting the findings of the study are available to bona fide researchers upon approval of an application to the UK Biobank (https://www.ukbiobank.ac.uk/researchers/) and a material transfer agreement. The results from FUMA with additional annotations are included in Supplementary Table S9 (candidate SNPs) and Supplementary Table S10 (all other analyses) and the gene-level analysis from MAGMA and a list of the sets and traits from the NHGRI-EBI GWAS Catalog used for matching are included in Supplementary Table S10. Summary statistics from BOLT-LMM infinitesimal models for all candidate SNPs identified in our study are included in Supplementary Table S11. The SNP2GENE output will also be made available upon publication on the FUMA website (https://fuma.ctglab.nl/) and locus lead SNPs will be uploaded to the NHGRI-EBI GWAS Catalog (https://www.ebi.ac.uk/gwas/home).
Abbreviations
- ABSI:
-
A body shape index
- ABSIUKB :
-
A body shape index calibrated for UK Biobank participants
- ADAMTS:
-
A disintegrin and metalloprotease domains with thrombospondins motifs
- AKR1C2:
-
Aldo-keto reductase family 1 member C2
- BMI:
-
Body mass index
- BOLT-LMM:
-
Bayesian mixed-model association method
- C5orf67:
-
Chromosome 5 open reading frame 67
- CADD:
-
Combined Annotation Dependent Depletion
- CASC20:
-
Cancer susceptibility 20
- CMIP:
-
C-Maf inducing protein
- COBLL1:
-
Cordon-bleu WH2 repeat protein like 1
- DLEU1:
-
Deleted in lymphocytic leukemia 1
- eQTL:
-
Expression Quantitative Trait Loci
- ERα:
-
Oestrogen receptor α
- ERI1:
-
Exoribonuclease 1
- EYA:
-
Eyes absent homolog
- FAM101A:
-
Filamin-interacting protein (readthrough region with ZNF664)
- FGFR4:
-
Fibroblast growth factor receptor 4
- FUMA:
-
Functional Mapping and Annotation
- GO:
-
Gene ontology
- GWAS:
-
Genome-wide association study
- HC:
-
Hip circumference
- HCadjBMI:
-
Hip circumference adjusted for body mass index
- HI:
-
Hip index
- HIUKB :
-
Hip index calibrated for UK Biobank participants
- HMGA1:
-
High mobility group AT-hook 1
- HPA:
-
Hypothalamic-pituitary-adrenal axis
- KLF14:
-
Kruppel-like factor 14
- LD:
-
Linkage disequilibrium
- MAF:
-
Minor allele frequency
- MAGMA:
-
Multi-marker Analysis of GenoMic Annotation
- MC1R:
-
Melanocortin 1 receptor
- NEU1:
-
Neutraminidase 1
- NHANES:
-
National Health and Nutrition Examination Survey
- PEMP:
-
Palmitoylated erythrocyte membrane protein
- PLXND1:
-
Plexin D1
- SLC30A10:
-
Solute carrier family 30 member 10
- SNP:
-
Single nucleotide polymorphism
- SSPN:
-
Sarcospan
- TBX15:
-
T-box transcription factor 15
- TFAP4:
-
Transcription factor AP-4
- VEGFA:
-
Vascular endothelial growth factor-A
- VEP:
-
Ensembl Variant Effect Predictor
- WC:
-
Waist circumference
- WCadjBMI:
-
Waist circumference adjusted for body mass index
- WHI:
-
Waist-to-hip index
- WHIUKB :
-
Waist-to-hip index calibrated for UK Biobank participants
- WHR:
-
Waist-to-hip ratio
- WHRadjBMI:
-
Waist-to-hip ratio adjusted for body mass index
- WNT:
-
Wingless-type
- XKR6:
-
XK related 6
- ZMIZ1:
-
Zinc-finger MIZ-type containing 1
- ZNF664:
-
Zinc finger protein 664
References
Hill, J. H., Solt, C. & Foster, M. T. Obesity associated disease risk: the role of inherent differences and location of adipose depots. Horm. Mol. Biol. Clin. Investig. https://doi.org/10.1515/hmbci-2018-0012 (2018).
Carmienke, S. et al. General and abdominal obesity parameters and their combination in relation to mortality: a systematic review and meta-regression analysis. Eur. J. Clin. Nutr. 67, 573–585 (2013).
Pulit, S. L. et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 28, 166–174 (2019).
Shungin, D. et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 518, 187–196 (2015).
Krakauer, N. Y. & Krakauer, J. C. A new body shape index predicts mortality hazard independently of body mass index. PLoS One 7, e39504. https://doi.org/10.1371/journal.pone.0039504 (2012).
Krakauer, N. Y. & Krakauer, J. C. An anthropometric risk index based on combining height, weight, waist, and hip measurements. J. Obes. 2016, 8094275. https://doi.org/10.1155/2016/8094275 (2016).
Stevens, C. F. Darwin and Huxley revisited: the origin of allometry. J. Biol. 8, 14. https://doi.org/10.1186/jbiol119 (2009).
Bertoli, S. et al. Association of Body Shape Index (ABSI) with cardio-metabolic risk factors: A cross-sectional study of 6081 Caucasian adults. PLoS One 12, e0185013. https://doi.org/10.1371/journal.pone.0185013 (2017).
Christakoudi, S. et al. A Body Shape Index (ABSI) achieves better mortality risk stratification than alternative indices of abdominal obesity: results from a large European cohort. Sci. Rep. 10, 14541. https://doi.org/10.1038/s41598-020-71302-5 (2020).
Winkler, T. W. et al. The influence of age and sex on genetic associations with adult body size and shape: a large-scale genome-wide interaction study. PLoS Genet. 11, e1005378. https://doi.org/10.1371/journal.pgen.1005378 (2015).
Buniello, A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Cook, J. P., Mahajan, A. & Morris, A. P. Fine-scale population structure in the UK Biobank: implications for genome-wide association studies. Hum. Mol. Genet. 29, 2803–2811 (2020).
Chen, N. & Wang, J. Wnt/beta-catenin signaling and obesity. Front. Physiol. 9, 792. https://doi.org/10.3389/fphys.2018.00792 (2018).
Loh, N. Y. et al. LRP5 regulates human body fat distribution by modulating adipose progenitor biology in a dose- and depot-specific fashion. Cell. Metab. 21, 262–273 (2015).
Kazanskaya, O. et al. The Wnt signaling regulator R-spondin 3 promotes angioblast and vascular development. Development 135, 3655–3664 (2008).
Vidal, V. et al. The adrenal capsule is a signaling center controlling cell renewal and zonation through Rspo3. Genes. Dev. 30, 1389–1394 (2016).
Raslan, A. A. & Yoon, J. K. R-spondins: multi-mode WNT signaling regulators in adult stem cells. Int. J. Biochem. Cell. Biol. 106, 26–34 (2019).
Chen, Z. et al. RSPO3 promotes the aggressiveness of bladder cancer via Wnt/beta-catenin and Hedgehog signaling pathways. Carcinogenesis 40, 360–369 (2019).
Tocci, J. M., Felcher, C. M., Garcia, S. M. E. & Kordon, E. C. R-spondin-mediated WNT signaling potentiation in mammary and breast cancer development. IUBMB Life 72, 1546–1559 (2020).
Hoffmann, T. J. et al. A large electronic-health-record-based genome-wide study of serum lipids. Nat. Genet. 50, 401–413 (2018).
Hunt, S. E. et al. Ensembl variation resources. Database 2018, 119. https://doi.org/10.1093/database/bay119 (2018).
Tachmazidou, I. et al. Whole-genome sequencing coupled to imputation discovers genetic signals for anthropometric traits. Am. J. Hum. Genet. 100, 865–884 (2017).
Kim, S., Yu, N. K. & Kaang, B. K. CTCF as a multifunctional protein in genome regulation and gene expression. Exp. Mol. Med. 47, 166. https://doi.org/10.1038/emm.2015.33 (2015).
Sulc, J., Winkler, T. W., Heid, I. M. & Kutalik, Z. Heterogeneity in obesity: genetic basis and metabolic consequences. Curr. Diab. Rep. 20, 1. https://doi.org/10.1007/s11892-020-1285-4 (2020).
Han, X., Cao, Y., Wang, K. & Zhu, G. HMGA1 facilitates tumor progression through regulating Wnt/beta-catenin pathway in endometrial cancer. Biomed. Pharmacother. 82, 312–318 (2016).
Song, J. et al. Transcription factor AP-4 promotes tumorigenic capability and activates the Wnt/β-catenin pathway in hepatocellular carcinoma. Theranostics 8, 3571–3583 (2018).
Ceci, C., Atzori, M. G., Lacal, P. M. & Graziani, G. Role of VEGFs/VEGFR-1 signaling and its inhibition in modulating tumor invasion: experimental evidence in different metastatic cancer models. Int. J. Mol. Sci. 21, 1388. https://doi.org/10.3390/ijms21041388 (2020).
Vivekanadhan, S. & Mukhopadhyay, D. Divergent roles of Plexin D1 in cancer. Biochim. Biophys. Acta Rev. Cancer 1872, 103–110 (2019).
Chen, X., Shi, W. & Zhang, H. The role of KLF14 in multiple disease processes. BioFactors 46, 276–282 (2020).
Papaioannou, V. E. The T-box gene family: emerging roles in development, stem cells and cancer. Development 141, 3819–3833 (2014).
Cal, S. & López-Otín, C. ADAMTS proteases and cancer. Matrix Biol. 44–46, 77–85 (2015).
Wang, J., Zhao, H., Xu, Z. & Cheng, X. Zinc dysregulation in cancers and its potential as a therapeutic target. Cancer Biol. Med. 17, 612–625 (2020).
Liu, Y. et al. Dissecting the role of the FGF19-FGFR4 signaling pathway in cancer development and progression. Front. Cell Dev. Biol. 8, 95. https://doi.org/10.3389/fcell.2020.00095 (2020).
Zhou, H., Zhang, L., Vartuli, R. L., Ford, H. L. & Zhao, R. The Eya phosphatase: its unique role in cancer. Int. J. Biochem. Cell. Biol. 96, 165–170 (2018).
Brotto, D. B. et al. Contributions of HOX genes to cancer hallmarks: enrichment pathway analysis and review. Tumour Biol. 42, 1010428320918050. https://doi.org/10.1177/1010428320918050 (2020).
Takayama, K. I., Suzuki, T., Fujimura, T., Takahashi, S. & Inoue, S. COBLL1 modulates cell morphology and facilitates androgen receptor genomic binding in advanced prostate cancer. Proc. Natl. Acad. Sci U S A 115, 4975–4980 (2018).
Pang, B. et al. Upregulation of DLEU1 expression by epigenetic modification promotes tumorigenesis in human cancer. J. Cell Physiol. 234, 17420–17432 (2019).
Silveira, E. A., Kliemann, N., Noll, M., Sarrafzadegan, N. & De Oliveira, C. Visceral obesity and incident cancer and cardiovascular disease: an integrative review of the epidemiological evidence. Obes. Rev. https://doi.org/10.1111/obr.13088 (2020).
Bull, C. J. et al. Adiposity, metabolites, and colorectal cancer risk: mendelian randomization study. BMC Med. 18, 396 (2020).
Jarvis, D. et al. Mendelian randomisation analysis strongly implicates adiposity with risk of developing colorectal cancer. Br. J. Cancer 115, 266–272 (2016).
Johansson, M. et al. The influence of obesity-related factors in the etiology of renal cell carcinoma-a mendelian randomization study. PLoS Med 16, e1002724 (2019).
Shu, X. et al. Associations of obesity and circulating insulin and glucose with breast cancer risk: a Mendelian randomization analysis. Int. J. Epidemiol. 48, 795–806 (2019).
Heid, I. M. et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 42, 949–960 (2010).
Zillikens, M. C. et al. Sex-specific genetic effects influence variation in body composition. Diabetologia 51, 2233–2241 (2008).
Palmer, B. F. & Clegg, D. J. The sexual dimorphism of obesity. Mol. Cell. Endocrinol. 402, 113–119 (2015).
Coburn, S. B. et al. Comparability of serum, plasma, and urinary estrogen and estrogen metabolite measurements by sex and menopausal status. Cancer Causes Control 30, 75–86 (2019).
Park, Y.-M., Erickson, C., Bessesen, D., Van Pelt, R. E. & Cox-York, K. Age- and menopause-related differences in subcutaneous adipose tissue estrogen receptor mRNA expression. Steroids121, 17–21 (2017).
Lizcano, F. & Guzman, G. Estrogen deficiency and the origin of obesity during menopause. Biomed. Res. Int. 2014, 757461. https://doi.org/10.1155/2014/757461 (2014).
Fanelli, F. et al. Revisiting hyper- and hypo-androgenism by tandem mass spectrometry. Rev. Endocr. Metab. Disord. 14, 185–205 (2013).
Fogle, R. H., Stanczyk, F. Z., Zhang, X. & Paulson, R. J. Ovarian androgen production in postmenopausal women. J. Clin. Endocrinol. Metab. 92, 3040–3043 (2007).
Pasquali, R., Vicennati, V., Cacciari, M. & Pagotto, U. The hypothalamic-pituitary-adrenal axis activity in obesity and the metabolic syndrome Ann. N.Y. Acad. Sci. 1083, 111–128 (2006).
Rutters, F., Nieuwenhuizen, A. G., Lemmens, S. G., Born, J. M. & Westerterp-Plantenga, M. S. Hypothalamic-pituitary-adrenal (HPA) axis functioning in relation to body fat distribution. Clin. Endocrinol. (Oxf.) 72, 738–743 (2010).
Vicennati, V. et al. Sex difference in the relationship between the hypothalamic-pituitary-adrenal axis and sex hormones in obesity Obesity (Silver Spring) 14, 235–243 (2006).
Rubinow, D. R. et al. Testosterone suppression of CRH-stimulated cortisol in men. Neuropsychopharmacology 30, 1906–1912 (2005).
Pasquali, R. et al. The hypothalamic-pituitary-adrenal axis in obese women with different patterns of body fat distribution. J. Clin. Endocrinol. Metab. 77, 341–346 (1993).
Grabek, A. et al. The adult adrenal cortex undergoes rapid tissue renewal in a sex-specific manner. Cell Stem Cell 25, 290-296.e292 (2019).
Miyamoto, J. et al. The pituitary function of androgen receptor constitutes a glucocorticoid production circuit. Mol. Cell. Biol. 27, 4807–4814 (2007).
Vainio, S., Heikkilä, M., Kispert, A., Chin, N. & Mcmahon, A. P. Female development in mammals is regulated by Wnt-4 signalling. Nature 397, 405–409 (1999).
Strous, G. J. et al. Growth hormone receptor regulation in cancer and chronic diseases. Front. Endocrinol (Lausanne) 11, 597573 (2020).
Keyes, K. M. & Westreich, D. UK Biobank, big data, and the consequences of non-representativeness. The Lancet 393, 1297 (2019).
Wu, Y. et al. Multi-trait analysis for genome-wide association study of five psychiatric disorders. Transl. Psychiatry 10, 209 (2020).
Hou, L. et al. Exploring the causal pathway from ischemic stroke to atrial fibrillation: a network Mendelian randomization study. Mol. Med. 26, 7 (2020).
UK Biobank Coordinating Centre: UK Biobank: Protocol for a large-scale prospective epidemiological resource. Protocol No: UKBB-PROT-09-06 (main phase, amendment one final). http://www.ukbiobank.ac.uk/wp-content/uploads/2011/11/UK-Biobank-Protocol.pdf (2007)
Bycroft, C. et al. Genome-wide genetic data on ~ 500,000 UK Biobank participants. bioRxiv https://doi.org/10.1101/166298 (2017).
Sinnott-Armstrong, N. et al. Genetics of 38 blood and urine biomarkers in the UK Biobank. bioRxiv https://doi.org/10.1101/660506 (2019).
Loh, P. R., Kichaev, G., Gazal, S., Schoech, A. P. & Price, A. L. Mixed-model association for biobank-scale datasets. Nat. Genet. 50, 906–908 (2018).
Loh, P.-R. et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47, 284–290 (2015).
Abecasis, G. R. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Watanabe, K., Taskesen, E., Van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Mclaren, W. et al. The ensembl variant effect predictor. Genome Biol. 17, 122. https://doi.org/10.1186/s13059-016-0974-4 (2016).
De Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. (2017).
Acknowledgements
This research has been conducted using the UK Biobank Resource (https://www.ukbiobank.ac.uk/about-biobank-uk/) under Application number 41952, granting access to the corresponding genetic and phenotype data (released 19 Nov 2018). The bioinformatics pipeline for GWAS BOLT-LMM analysis and variant annotation in the UK MED-BIO resources was created by Dr David Mosen-Ansorena and was further curated and maintained by Dr Paul Blakeley at Imperial College London (UK).
Funding
This work used the computing resources of the UK MEDical BIOinformatics partnership—aggregation, integration, visualisation and analysis of large, complex data (UK MED-BIO), which is supported by the Medical Research Council [Grant Number MR/L01632X/1]. Infrastructure support for the Department of Epidemiology and Biostatistics at Imperial College London (UK) was provided by the NIHR Imperial Biomedical Research Centre (BRC). The funders had no role in the design and conduct of the study, the collection, analysis, and interpretation of the data, or the preparation, review, and approval of the manuscript, or in the decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
S.C., K.K.T. and E.R. conceived and designed the study. K.K.T. and E.E. provided statistical advice. S.C. led the research and performed the statistical analysis. S.C. had full access to all of the data in this study and takes responsibility for the integrity of the data and the accuracy of the data analysis. S.C. drafted the paper with contributions from E.E, K.K.T and E.R. All authors, i.e. S.C., E.E., E.R. and K.K.T. were involved in the interpretation of the results, the critical revisions of the paper, and the approval of the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Christakoudi, S., Evangelou, E., Riboli, E. et al. GWAS of allometric body-shape indices in UK Biobank identifies loci suggesting associations with morphogenesis, organogenesis, adrenal cell renewal and cancer. Sci Rep 11, 10688 (2021). https://doi.org/10.1038/s41598-021-89176-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-89176-6
This article is cited by
-
Polymorphism rs143384 GDF5 reduces the risk of knee osteoarthritis development in obese individuals and increases the disease risk in non-obese population
Arthroplasty (2024)
-
Links between the genetic determinants of morning plasma cortisol and body shape: a two-sample Mendelian randomisation study
Scientific Reports (2024)
-
SMYD3: a new regulator of adipocyte precursor proliferation at the early steps of differentiation
International Journal of Obesity (2024)
-
An epigenome-wide association study of waist circumference in Chinese monozygotic twins
International Journal of Obesity (2024)
-
Allometric versus traditional body-shape indices and risk of colorectal cancer: a Mendelian randomization analysis
International Journal of Obesity (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.