Abstract
Glomerular filtration rate (GFR), or the rate of primary urine formation, is the key indicator of renal function. Studies have demonstrated that GFR exhibits significant circadian rhythmicity and, that these rhythms are disrupted in a number of pathologies. Here, we tested a hypothesis that the circadian rhythm of GFR is driven by intrinsic glomerular circadian clocks. We used mice lacking the circadian clock protein BMAL1 specifically in podocytes, highly specialized glomerular cells critically involved in the process of glomerular filtration (Bmal1lox/lox/Nphs2-rtTA/LC1 or, cKO mice). Circadian transcriptome profiling performed on isolated glomeruli from control and cKO mice revealed that the circadian clock controls expression of multiple genes encoding proteins essential for normal podocyte function. Direct assessment of glomerular filtration by inulin clearance demonstrated that circadian rhythmicity in GFR was lost in cKO mice that displayed an ultradian rhythm of GFR with 12-h periodicity. The disruption of circadian rhythmicity in GFR was paralleled by significant changes in circadian patterns of urinary creatinine, sodium, potassium and water excretion and by alteration in the diurnal pattern of plasma aldosterone levels. Collectively, these results indicate that the intrinsic circadian clock in podocytes participate in circadian rhythmicity of GFR.
Similar content being viewed by others
Introduction
The kidney plays a pivotal role in maintaining homeostasis. The homeostatic equilibrium is continuously challenged by a variety of external and internal factors, including those affected by circadian rhythms. Accordingly, it has long been recognized that homeostatic control over extracellular fluids requires continuous adjustment of diverse specific renal functions throughout the circadian cycle. For instance, renal blood flow (RBF), renal tissue oxygenation, renal cortico-medullary osmotic gradient, glomerular filtration rate (GFR) and tubular reabsorption/secretion of water and major electrolytes have been shown to display circadian rhythms with apparently similar kinetics characterized by a trough in the middle of the biological night and peak values in the middle of the active phase (reviewed in1,2). It has been hypothesized that these rhythms are driven, at least in part, by the circadian clock, a molecular timer formed by several transcriptional-translational feedback loops that oscillate with the periodicity of ~24 hours in a self-sustained and cell-autonomous manner. The core of the circadian clock imposes functional rhythms via the temporal control over transcription, translation and posttranslational modifications (reviewed in3).
Numerous studies have examined the role of the circadian clock in renal function. Analyses of mouse models with null mutations in core clock genes revealed that the circadian clock controls a large number of genes involved in diverse homeostatic renal functions4,5. Functionally, disruption of the circadian clock leads to dramatic changes in circadian patterns of urinary excretion of water and major electrolytes, impairment in cortico-medullary osmotic gradient and, loss of blood pressure (BP) control6,7,8,9. Recent research has addressed the role of intrinsic renal circadian clocks. Nikolaeva et al. have shown that conditional disruption of the essential circadian transcriptional activator Bmal1 in the renal tubule results in dysregulation of a wide range of intrarenal and systemic metabolic processes and, in impairment in renal xenobiotic elimination5. Tokonami et al. have demonstrated that ablation of the circadian clock specifically in renin-secreting granular cells resulted in a complex renal phenotype characterized by impaired handling of sodium, water, calcium and magnesium, a significantly modified circadian pattern of plasma aldosterone levels and, decreased BP10. Collectively, these studies have clearly established an important role for the circadian clock in renal tubular function.
The role of the circadian clock in the process of glomerular filtration remains much less understood. It has been shown that GFR display circadian oscillations with an amplitude of 20–40% when measured by inulin clearance11,12,13. The GFR depends on the difference between glomerular capillary and Bowman’s space hydrostatic pressure, the transcapillary oncotic pressure and the ultrafiltration coefficient (Kf) which, in turn, depends on the hydraulic permeability of the glomerular filter and on the effective filtration surface. Studies have shown that circadian oscillations in GFR are independent of circadian rhythms in systemic blood pressure14 and, of the sympathetic renal innervation15. Moreover, Koopman et al., have shown that the GFR rhythm is not due to differences in posture and food intake over the circadian cycle16. Plasma albumin and total protein levels exhibit similar with GFR circadian patterns therefore acting as factors opposing GFR oscillations17,18,19. Altogether, these data raised the possibility that circadian rhythm in GFR is driven, at least in part, by intrinsic circadian clocks located in glomerular cells and/or juxtaglomerular apparatus. Here, we addressed the role of the circadian clock in podocytes, which are highly specialized epithelial cells that cover the outer surface of glomerular capillaries and that have been proposed to actively participate in the control of Kf20,21.
Results
The Bmal1 expression in podocytes
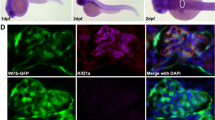
Although it is generally assumed that the circadian clock machinery is present in nearly all eukaryotic cells, the expression and abundance of core clock components in podocytes has not been tested. To address this question, we performed RNAscope in situ hybridization of mouse kidneys with probes specific to Bmal1 and to Nphs2 (podocin, a critical element of the filtration slit specifically expressed in podocytes). As shown in Fig. 1, Bmal1 and Nphs2 mRNAs are co-localized within a subset of glomerular cells, thereby confirming the expression of Bmal1 in podocytes. The Bmal1 mRNA content measured by image quantitation revealed that 16.2 ± 2.4% (n = 14 glomeruli (SEM)) of glomerular Bmal1 mRNA molecules are expressed in podocytes, while podocytes represent ~30% of all glomerular cells, according to current estimates22,23. These results suggested that podocytes exhibit lower Bmal1 expression compared to the other glomerular cell types.
The Bmal1 lox/lox/Nphs2-rtTA/LC1 conditional knockout (cKO) model
To study the role of the circadian clock in podocytes we generated mice with podocyte-specific doxycycline (DOX)-inducible inactivation of the Arntl (Bmal1) gene encoding BMAL1, an indispensable transcriptional activator in the circadian clock machinery. To this aim, mice bearing floxed Bmal1 alleles24 were crossed with Nphs2-rtTA/Cre transgenic mice commonly used for podocyte-specific conditional gene targeting25. Inactivation of the Bmal1 gene was induced by 2-weeks treatment with DOX (2 mg/ml in drinking water) of 8-weeks old Bmal1lox/lox/Nphs2-rtTA/LC1 mice (hereafter referred to as conditional knockout mice or cKO mice). The same DOX treatment was provided to their littermate controls (Bmal1lox/lox mice, hereafter referred to as Control mice). All experiments were performed 1 month after the end of DOX treatment in order to avoid potential side effects of DOX on renal function. The model was validated by demonstration of Cre-mediated genomic excision of floxed Bmal1 alleles in the kidney and in isolated glomeruli from cKO mice (Supplementary Fig. 1A) and, by RT-PCR analysis performed on RNAs extracted from glomeruli isolated at different circadian time-points from kidneys of Control and cKO mice (Supplementary Fig. 1B). Body weights, 24-hour food and water intake (Supplementary Table 1) as well as physical activity rhythms (Supplementary Fig. 2) were not different between Control and cKO mice. The cKO mice did not show any obvious morphological and ultrastructural abnormalities and did not develop proteinuria (Supplementary Fig. 3).
Transcriptome profiling of glomeruli isolated from Control and cKO mice
To identify molecular pathways controlled by the circadian clock in podocytes we performed transcriptome profiling of glomeruli isolated from kidneys of Control and cKO mice at six different circadian time-points (ZT0, ZT4, ZT8, ZT12, ZT16 and ZT20; ZT – Zeitgeber or circadian time; ZT0 is the time of light-on and ZT12 is the time of light-off). Transcriptome analysis revealed multiple genes critically involved in a variety of podocyte-specific processes and exhibiting differential expression levels in glomeruli of Control and cKO mice (Fig. 2 and Supplementary Table 2). A significant reduction in expression levels (at one or more circadian time-points) was observed, e.g., for: Tcf21, a transcriptional factor playing a key role in podocyte differentiation and maintenance26; N-Ethylmaleimide Sensitive Factor (Nsf) and G Protein Subunit Alpha 12 (Gna12), genes involved in the organization and integrity of podocyte cytoskeleton27,28; Uncoupling protein 2 (UCP2) and Farnesoid X-Activated Receptor (FXR or Nr1h4), key proteins controlling renal metabolism29,30; and Rho GTPase Activating Protein 24 (Arhgap24), an enzyme critical for podocyte interaction with the basement membrane31. An increased expression was found, e.g., in: Cathepsin L (Ctsl), a protease highly expressed in the foot processes and important for normal podocyte architecture32, sulfatase 2 (Sulf2), an enzyme that controls paracrine interglomerular communication between podocytes and endothelial cells33 and Gprc5a, a highly podocyte-specific orphan G protein-coupled receptor which controls the thickness of the glomerular basement membrane34. Interestingly, among the circadian core clock genes, only Cry1, Npas2, Rora and Rorc displayed significant changes in the expression levels in glomeruli of cKO mice. The heterogeneous cellular composition of glomeruli and lower than the mean expression levels of Bmal1 in podocytes (see above) are possible explanations for these results.
Analysis of glomeruli transcriptome revealed altered expression of genes involved in diverse podocyte-specific processes or being part of the circadian clock core. mRNA expression profiles of Tcf21, Nsf, Arhgap24, Ctsl, Sulf2, Gprc5a, Aff3, Cry1 and Npas2 in Control (black) and cKO (red) glomeruli. Values are means ± SEM. Statistical analyses were performed with the R package limma48. Contrasts between cKO and Control at each time point were combined into one F-test. *Time point with difference after post-hoc classification of significant genes (false discovery rate <5%). n = 6 mice/time-point/genotype.
cKO mice lose circadian rhythmicity in GFR
To determine whether the intrinsic circadian clock in podocytes participates in the control of GFR we compared circadian patterns of inulin clearance in Control and cKO mice. As shown in Fig. 3A, GFR in Control mice exhibited significant circadian rhythm (circadian fit p-value = 0.023) with an acrophase at ZT16 and a trough during the light (rest) phase. This circadian pattern in GFR was disrupted in cKO mice (circadian fit p-value = 0.336) that displayed an ultradian rhythm (12-hours period fit p-value = 0.003) with two peaks, the first at ZT4 and the second at ZT16 (Fig. 3B). Importantly, the integrated 24-hour GFR was not different between Control and cKO mice (p = 0.985, ANOVA). Because alterations in GFR are expected to elicit reciprocal adjustments in renin-angiotensin-aldosterone system (RAAS), which is a part of the tubulo-glomerular feedback (TGF) mechanism, we examined the circadian pattern of plasma aldosterone levels in Control and cKO mice. As shown in Fig. 3C, this pattern was strikingly altered in cKO mice (p = 0.0095, ANOVA). Of note, maximal values of GFR in cKO mice (ZT4 and ZT16) negatively correlated with minimal values in plasma aldosterone levels (ZT4 and ZT20), as could be expected from the general concept of TGF. As another independent approach to corroborate these results, we analyzed circadian profiles of urinary creatinine excretion in hourly collected urines. As shown in Fig. 4A, at baseline (before DOX treatment), both Control and cKO mice displayed similar patterns of urinary creatinine excretion with the percentage of creatinine excreted during the light phase (ZT0 to ZT11) not different between the two genotypes. One month after the end of DOX treatment, the percentage of creatinine excreted during the light phase was significantly higher in cKO mice compared to Control mice, thereby confirming the role of the circadian clock in podocytes in the control of GFR (Fig. 4B). A similar disruption of urinary excretory rhythms in cKO mice was observed for sodium, potassium and urine volume (Supplementary Fig. 4). However, the total 24-hour excretory rates for creatinine, sodium, potassium and water (Supplementary Table 1) as well as basic plasma parameters (Supplementary Table 1) and BP (Supplementary Fig. 5) were not different between Control and cKO mice.
Circadian pattern of GFR is disrupted in cKO mice. Temporal profiles of GFR in (A) Control and (B) cKO mice. (C) Temporal profile of plasma aldosterone levels in Control (black) and cKO (red) mice. Values are means ± SEM. *p < 0.05 (unpaired t test). Circadian fit were analyzed with a linear model of a pair of cosine curves with a period of 24 hours. (A,B) Sin and Cos coefficients were combined into one contrast with the ‘glht’ (generalized linear hypothesis test) function of the R package ‘multcomp’49. Effect on aldosterone levels was tested with two-way Anova with genotypes and time points as factors. n = 6 mice/time-point/genotype, except n = 5 for cKO mice at ZT16.
Urinary creatinine excretion during the inactive (light) phase (ZT0-ZT11) is higher in cKO mice. Profiles of every-hour urinary creatinine excretion in Control (black) and cKO (red) mice in (A) baseline (before DOX treatment) and (B) one month after the end of the DOX treatment. Bar plots represent the percentage of 24-hour urinary creatinine excretion excreted during the inactive (light) phase (ZT0-ZT11). *p < 0.05 (unpaired t test). n = 10 for cKO mice after DOX treatment, n = 11 in all other groups.
Discussion
The cellular origin and molecular mechanisms underlying the circadian rhythm of GFR have intrigued physiologists and clinicians for decades. Interest into this field was stimulated by clinical observations that the amplitude of circadian oscillations in GFR is reduced with age35 and, that the circadian rhythm in GFR is dampened or inverted in patients with the nephrotic syndrome14, heart failure36 or cirrhosis12. Early studies demonstrating that the circadian rhythm in GFR is independent on circadian oscillations in BP, cardiac output and sympathetic regulation suggested that an intrarenal self-autonomous mechanism(s) might be involved14,15. Here, we hypothesized that circadian oscillations in GFR are driven, at least in part, by the intrinsic circadian clock in podocytes, the highly specialized terminally differentiated glomerular cells that are essential for the formation of the glomerular filtration barrier and that are actively involved in control of GFR by regulating slit diaphragm and by influencing properties of glomerular endothelial cells via paracrine factors20,21. This hypothesis was tested at the transcriptional and functional levels.
Current estimates suggest that 10 to 50% of all cellular transcripts are rhythmic37. It has been shown that a majority of oscillating transcripts (with the exception of transcripts encoding proteins of the core clock machinery) exhibit tissue-specific expression patterns and/or display tissue-specific oscillation amplitudes and phases37. Accordingly, it has been proposed that the main role of peripheral circadian clocks consists in the dynamic adjustment of tissue-specific physiological functions. Recent studies demonstrating that the kidney exhibits one of the highest levels of circadian transcripts across mammalian tissues have underscored the potential importance of circadian clocks in renal cells37. Here, we showed that inactivation of the intrinsic circadian clock in podocytes led to significant alterations in expression patterns of multiple genes that have been implicated in diverse cellular processes critical for podocyte function, including podocyte differentiation, metabolism, cytoskeleton organization and adhesion. Polymorphic variants in several genes have been associated with human glomerular disease. For instance, single nucleotide polymorphisms (SNPs) in Ucp2 have been linked to decreased GFR in type 1 diabetic patients (T1D)38, SNPs in the Arhgap24 gene are associated with the focal segmental glomerulosclerosis (FSGS)31,39, SNPs in the AF4/FMR2 Family Member 3 (Aff3) gene are associated with diabetic end stage renal disease (ESRD) in T1D patients40 and, Ctsl levels show strong positive correlation with proteinuria in chronic kidney disease (CKD)32. Collectively, these results suggested that the intrinsic circadian clock is significantly involved in the control of podocyte transcriptome and that transcriptional changes in cKO mice may result in functional alterations.
We further provided three lines of functional evidence demonstrating that the intrinsic circadian clock in podocytes plays an essential role in the maintenance of the circadian rhythm of GFR. First, disruption of circadian rhythms of GFR in cKO mice was revealed through the direct assessment of the circadian pattern of GFR using inulin clearance. Second, inactivation of the circadian clock in podocytes was shown to increase the rate of urinary creatinine excretion in cKO mice during the inactive phase. Third, loss in circadian rhythmicity of GFR was paralleled by alterations in the circadian pattern of plasma aldosterone levels. Importantly, the circadian clock deficiency did not result in proteinuria and did not affect the integrated 24-hour GFR. This indicates that the main role of the circadian clock in podocytes consists in the maintenance of the circadian rhythm in GFR and not, or to a lesser extent, in the control of other specific cellular functions in podocyte (at least in unstressed conditions). These results, together with evidence from another recent study that addressed the role of intrinsic circadian clocks in the renal tubule5, support the idea that circadian clocks serve very specialized renal functions. Indeed, Nikolaeva et al. have shown that conditional ablation of the circadian clock in tubular cells resulted in impaired secretion of organic anions, but tubular handling of water and major electrolytes as well as the glomerular function were fully preserved in this model5. Our study also provides additional evidence for the intrinsic glomerular origin of circadian oscillations in GFR since circadian rhythm in BP was fully maintained in cKO mice. Collectively, these results demonstrate that intrinsic renal circadian clocks drive circadian oscillations in a number of essential renal functions, including glomerular filtration.
The cause(s) of disruption of circadian rhythms in GFR in patients with cardiovascular, renal or liver diseases remains unknown. However, it is well established that intrinsic circadian clocks in peripheral tissues are orchestrated by external circadian cues, of which systemic circulating factors (hormones, food, food metabolites) are probably most important for renal cells. Accordingly one may hypothesize that profound changes in the circulating metabolome that are characteristic for the aforementioned disorders may cause dysfunction in circadian clocks within podocytes and/or other glomerular cells.
Methods
Animals
The procedures used to generate, and the characterization of, Bmal1lox/lox and LC-1 Cre mice were described previously41. The Nphs2lox/+ mice were obtained from The Jackson Laboratory. The three mouse lines used in this study are inbred strains, bred on the genetic background of C57BL/6J mice. The animals were maintained ad libitum on the standard laboratory chow diet (KLIBA NAFAG diet 3800). Before all experiments, mice were adapted to a 12-hour light/ 12 hour dark cycle for two weeks. All experiments were performed on male mice.
PCR for recombined Bmal1 allele
Excision of floxed Bmal1 allele was assessed on cDNA or gDNA using the following primers; F_5′-TGG ACA CAG ACA AAG ATG ACC CTC A-3′ and R_5′-TCC CTC GGT CAC ATC CTA CGA CA-3′ (for cDNA) and F_5′-AGG GAC AGG CCA AAA GTC TG-3′ and R_5′-GGC ACA TGT CTT AAT CTA CCC-3′ (for gDNA).
Metabolic cages
Mice were individually housed in metabolic cages (Tecniplast) and subjected to 3 days of adaptation before urine collection. The every hour urine collection was performed using a 12-chanel peristaltic pump, as previously described by Nikolaeva et al.8.
GFR
GFR measurements were performed on anesthetized animals with inulin-FITC as previously described42. Briefly, ~2.5% FITC-inulin (2 μl/g BW) was injected into the retro-orbital plexus of Control or cKO mice. Blood collection was performed 3, 7, 10, 15, 40 and 60 minutes post-injection and inulin clearance was calculated using a two phase exponential decay curve model43. A total of 36 Control mice and 36 cKO mice were used for GFR measurement (6 mice/time-point/genotype).
Glomeruli isolation and RNA extraction
Isolation of mouse glomeruli was performed as previously described by Takemoto et al.44. Briefly, mice were anesthetized by intraperitoneal injection of Ketamine:Xylazine (1:1) and Dynabeads M-450 Tosylactivated (Invitrogen) perfused through the abdominal aorta. After perfusion, the 2 kidneys were harvested and minced into small pieces, before to be digested with collagenase at 30 °C for 30 minutes. The digested tissue was filtered and cell suspension centrifuged. Dynabeads containing-glomeruli were separated with a magnetic particle concentrator and washed. This method allows to recover ~10,000 glomeruli per kidney. Representative image of isolated glomeruli is shown in Supplementary Fig. 6. RNA from isolated glomeruli was directly extracted using RNAeasy MiniElute Spin Column (Qiagen). and 200 ng of purified RNA were used for RNA sequencing. A total of 36 Control mice and 36 cKO mice were used for the transcriptome experiments (6 mice/time-point/genotype).
Plasma aldosterone
Heparinized plasma was collected from anesthetized mouse at different ZT and aldosterone level measured by radioimmunoassay (DPC).
Arterial blood pressure
Blood pressure was measured with telemetry system (DSI) in freely moving mice.
Locomotor activity
The circadian patterns of general locomotor activity were measured by using the Mouse-E-Motion system (Infra-E-Motion Gmbh). This system allows real-time measurements of general motion activity in mice housed in normal laboratory cages.
RNA scope
The RNAscope analysis was performed according to the manufacturer’s protocol on kidneys harvested at ZT4.
RNA-seq
RNA-seq libraries were prepared using 200 ng of total RNA as described in Nikolaeva et al.5. Illumina TruSeq SR Cluster Kit v4 reagents were used. Sequencing data were processed as described in45 using Mus musculus.GRCm38.86 gene annotation. Statistical analysis was performed in R (version 3.4.0). Genes with low counts were filtered out according to the rule of 1 count per million (cpm) in at least 1 sample. Library sizes were scaled using TMM normalization and log-transformed into counts per million (CPM) using voom46.
Principal component analysis showed a large variability between replicate samples. Two factors of unwanted variation were removed using the RUVs function (R package RUVSeq v. 1.10.047). Differential expression between knock-out and wild-type was computed using limma48. A linear model with a factor for each combination of time point and genotyope was used. Factors to correct for batch effect and unwanted variation were also added in the design matrix. Differences in gene expression levels between KO and WT animals at each time points were combined into one F-test. Genes with a false discovery rate <5% were considered significant. The limma function ‘classifyTestsF’ was used to classify time point as significant or not for the selected genes.
Statistics
All data are expressed as mean ± SEM. Statistical differences between two groups were determined by Student’s 2-tailed t test. Differences between groups containing 2 variables were assessed by 2-way ANOVA. For repeated measurements, 2-way ANOVA was used. P < 0.05 was considered significant.
Study approval
All experiments with animals were performed in accordance with the Swiss guidelines for animal care, which conform to the National Institutes of Health animal care guidelines. All animal protocols were approved by Swiss veterinary authorities (authorisation #27750).
References
Firsov, D. & Bonny, O. Circadian rhythms and the kidney. Nature reviews. Nephrology (2018).
Solocinski, K. & Gumz, M. L. The Circadian Clock in the Regulation of Renal Rhythms. Journal of biological rhythms 30, 470–486 (2015).
Reinke, H. & Asher, G. Crosstalk between metabolism and circadian clocks. Nature reviews. Molecular cell biology (2019).
Zuber, A. M. et al. Molecular clock is involved in predictive circadian adjustment of renal function. Proc Natl Acad Sci USA 106, 16523–16528 (2009).
Nikolaeva, S. et al. Nephron-Specific Deletion of Circadian Clock Gene Bmal1 Alters the Plasma and Renal Metabolome and Impairs Drug Disposition. J Am Soc Nephrol 27, 2997–3004 (2016).
Doi, M. et al. Salt-sensitive hypertension in circadian clock-deficient Cry-null mice involves dysregulated adrenal Hsd3b6. Nat Med 16, 67–74 (2009).
Gumz, M. L. et al. The circadian clock protein Period 1 regulates expression of the renal epithelial sodium channel in mice. J Clin Invest 119, 2423–2434 (2009).
Nikolaeva, S. et al. The circadian clock modulates renal sodium handling. J Am Soc Nephrol 23, 1019–1026 (2012).
Hara, M. et al. Robust circadian clock oscillation and osmotic rhythms in inner medulla reflecting cortico-medullary osmotic gradient rhythm in rodent kidney. Scientific reports 7, 7306 (2017).
Tokonami, N. et al. Local renal circadian clocks control fluid-electrolyte homeostasis and BP. J Am Soc Nephrol 25, 1430–1439 (2014).
van Acker, B. A., Koomen, G. C., Koopman, M. G., Krediet, R. T. & Arisz, L. Discrepancy between circadian rhythms of inulin and creatinine clearance. J Lab Clin Med 120, 400–410 (1992).
Jones, R. A., Mc, D. G. & Last, J. H. Reversal of diurnal variation in renal function in cases of cirrhosis with ascites. J Clin Invest 31, 326–334 (1952).
Sirota, J. H., Baldwin, D. S. & Villarreal, H. Diurnal variations of renal function in man. J Clin Invest 29, 187–192 (1950).
Voogel, A. J., Koopman, M. G., Hart, A. A., van Montfrans, G. A. & Arisz, L. Circadian rhythms in systemic hemodynamics and renal function in healthy subjects and patients with nephrotic syndrome. Kidney Int 59, 1873–1880 (2001).
Buijsen, J. G., van Acker, B. A., Koomen, G. C., Koopman, M. G. & Arisz, L. Circadian rhythm of glomerular filtration rate in patients after kidney transplantation. Nephrol Dial Transplant 9, 1330–1333 (1994).
Koopman, M. G. et al. Circadian rhythm of glomerular filtration rate in normal individuals. Clin Sci (Lond) 77, 105–111 (1989).
Jubiz, W., Canterbury, J. M., Reiss, E. & Tyler, F. H. Circadian rhythm in serum parathyroid hormone concentration in human subjects: correlation with serum calcium, phosphate, albumin, and growth hormone levels. J Clin Invest 51, 2040–2046 (1972).
Mauvoisin, D. et al. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc Natl Acad Sci USA 111, 167–172 (2014).
Scheving, L. E., Pauly, J. E. & Tsai, T. H. Circadian fluctuation in plasma proteins of the rat. Am J Physiol 215, 1096–1101 (1968).
Pavenstadt, H. Roles of the podocyte in glomerular function. Am J Physiol Renal Physiol 278, F173–179 (2000).
Grahammer, F., Schell, C. & Huber, T. B. The podocyte slit diaphragm–from a thin grey line to a complex signalling hub. Nature reviews. Nephrology 9, 587–598 (2013).
Steffes, M. W., Schmidt, D., McCrery, R. & Basgen, J. M. Glomerular cell number in normal subjects and in type 1 diabetic patients. Kidney Int 59, 2104–2113 (2001).
Blutke, A., Schneider, M. R., Wolf, E. & Wanke, R. Growth hormone (GH)-transgenic insulin-like growth factor 1 (IGF1)-deficient mice allow dissociation of excess GH and IGF1 effects on glomerular and tubular growth. Physiological reports 4 (2016).
Storch, K. F. et al. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell 130, 730–741 (2007).
Shigehara, T. et al. Inducible podocyte-specific gene expression in transgenic mice. J Am Soc Nephrol 14, 1998–2003 (2003).
Maezawa, Y. et al. Loss of the podocyte-expressed transcription factor Tcf21/Pod1 results in podocyte differentiation defects and FSGS. J Am Soc Nephrol 25, 2459–2470 (2014).
Lu, Y. et al. Genome-wide identification of genes essential for podocyte cytoskeletons based on single-cell RNA sequencing. Kidney Int 92, 1119–1129 (2017).
Boucher, I. et al. Galpha12 activation in podocytes leads to cumulative changes in glomerular collagen expression, proteinuria and glomerulosclerosis. Laboratory investigation; a journal of technical methods and pathology 92, 662–675 (2012).
Zhou, Y. et al. UCP2 attenuates apoptosis of tubular epithelial cells in renal ischemia-reperfusion injury. Am J Physiol Renal Physiol 313, F926–f937 (2017).
Wang, X. X. et al. The farnesoid X receptor modulates renal lipid metabolism and diet-induced renal inflammation, fibrosis, and proteinuria. Am J Physiol Renal Physiol 297, F1587–1596 (2009).
Akilesh, S. et al. Arhgap24 inactivates Rac1 in mouse podocytes, and a mutant form is associated with familial focal segmental glomerulosclerosis. J Clin Invest 121, 4127–4137 (2011).
Sever, S. et al. Proteolytic processing of dynamin by cytoplasmic cathepsin L is a mechanism for proteinuric kidney disease. J Clin Invest 117, 2095–2104 (2007).
Schumacher, V. A. et al. WT1-dependent sulfatase expression maintains the normal glomerular filtration barrier. J Am Soc Nephrol 22, 1286–1296 (2011).
Ma, X. et al. Depletion of Gprc5a Promotes Development of Diabetic Nephropathy. J Am Soc Nephrol 29, 1679–1689 (2018).
Sunaga, K., Sudoh, T. & Fujimura, A. Lack of diurnal variation in glomerular filtration rates in the elderly. Journal of clinical pharmacology 36, 203–205 (1996).
Baldwin, D. S., Sirota, J. H. & Villarreal, H. Diurnal variations of renal function in congestive heart failure. Proceedings of the Society for Experimental Biology and Medicine. Society for Experimental Biology and Medicine (New York, N.Y.) 74, 578–581 (1950).
Zhang, R., Lahens, N. F., Ballance, H. I., Hughes, M. E. & Hogenesch, J. B. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci USA 111, 16219–16224 (2014).
de Souza, B. M. et al. Polymorphisms of the UCP2 Gene Are Associated with Glomerular Filtration Rate in Type 2 Diabetic Patients and with Decreased UCP2 Gene Expression in Human Kidney. PLoS One 10, e0132938 (2015).
Guan, M. et al. Association of kidney structure-related gene variants with type 2 diabetes-attributed end-stage kidney disease in African Americans. Human genetics 135, 1251–1262 (2016).
Sandholm, N. et al. New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS genetics 8, e1002921 (2012).
Traykova-Brauch, M. et al. An efficient and versatile system for acute and chronic modulation of renal tubular function in transgenic mice. Nat Med 14, 979–984 (2008).
Eisner, C. et al. Major contribution of tubular secretion to creatinine clearance in mice. Kidney Int 77, 519–526 (2010).
Qi, Z. et al. Serial determination of glomerular filtration rate in conscious mice using FITC-inulin clearance. Am J Physiol Renal Physiol 286, F590–596 (2004).
Takemoto, M. et al. Large-scale identification of genes implicated in kidney glomerulus development and function. Embo j 25, 1160–1174 (2006).
Siddiqui, I. et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity 50, 195–211.e110 (2019).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29 (2014).
Harrisson, H. E. A modification of the diphenylamine method for determination of inulin. Proc.Soc.Exp.Biol.Med. 49, 111–114 (1942).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015).
Hothorn, T., Bretz, F. & Westfall, P. Simultaneous inference in general parametric models. Biometrical journal. Biometrische Zeitschrift 50, 346–363 (2008).
Acknowledgements
This work was supported by Swiss National Science Foundation Research Grant 31003A-169493 (to D.F.) and by the Russian Federal Agency of Scientific Organizations Grant АААА-А18-118012290371-3 (to S.N.). Selected data in this manuscript were previously presented at the Experimental Biology 2019 meeting.
Author information
Authors and Affiliations
Contributions
C.A. and D.F. designed experiments and interpreted results. C.A. and G.C. carried out most of the experimental work with the help of S.N. and M.P.M. S.P. performed all statistical analyses. C.A., S.P. and D.F. analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ansermet, C., Centeno, G., Nikolaeva, S. et al. The intrinsic circadian clock in podocytes controls glomerular filtration rate. Sci Rep 9, 16089 (2019). https://doi.org/10.1038/s41598-019-52682-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-52682-9
This article is cited by
-
Significant impact of time-of-day variation on metformin pharmacokinetics
Diabetologia (2023)
-
Circadian Regulation of Blood Pressure: of Mice and Men
Current Hypertension Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.