Abstract
MmsR (33.3 kDa) is a putative LysR-type transcriptional activator of Pseudomonas denitrificans. With the help of 3-hydroxypropionic acid (3-HP), an important platform chemical, MmsR positively regulates the expression of mmsA, which encodes methylmalonylsemialdehyde dehydrogenase, the enzyme involved in valine degradation. In the present study, the cellular function of MmsR and its binding to the regulatory DNA sequence of mmsA expression were investigated both in vivo and in vitro. Transcription of the mmsA was enhanced >140-fold in the presence of 3-HP. In the MmsR-responsive promoter region, two operators showing dyad symmetry, designated O1 and O2 and centered at the −79 and −28 positions, respectively, were present upstream of the mmsA transcription start site. An electrophoretic mobility shift assay indicated that MmsR binds to both operator sites for transcription activation, probably in cooperative manner. When either O1 or O2 or both regions were mutated, the inducibility by the MmsR-3-HP complex was significantly reduced or completely removed, indicating that both sites are required for transcription activation. A 3-HP sensor was developed by connecting the activation of MmsR to a green fluorescent readout. A more than 50-fold induction by 25 mM 3-HP was observed.
Similar content being viewed by others
Introduction
3-Hydroxypropionic acid (3-HP) is a commercially important platform chemical that can be converted to acrylic acid, malonic acid, acrylamide, 1,3-propanediol and other valuable chemicals. Biologically, 3-HP can be produced from glucose or glycerol1. With glucose as the carbon source, 3-HP can be synthesized via malonyl-CoA or β-alanine. With glycerol as the carbon source, 3-HP is synthesized by two enzymatic reactions, catalyzed by the coenzyme B12-dependent glycerol dehydratase converting glycerol to 3-hydroxypropionaldehyde (3-HPA) and the 3-HPA-specific aldehyde dehydrogenase oxidizing 3-HPA to 3-HP, respectively. Several microorganisms, including Escherichia coli, Klebsiella pneumoniae, Pseudomonas denitrificans and Saccharomyces cerevisiae, have been developed as hosts2,3. Among them, K. pneumoniae and P. denitrificans are advantageous when glycerol is used as the carbon source because they can naturally synthesize coenzyme B124,5.
In the course of the development of P. denitrificans as the 3-HP production host, we observed that 3-HP was actively degraded6. Several enzymes, namely putative 3-hydroxyisobutyrate dehydrogenase IV (HbdH-4), 3-hydroxypropionate dehydrogenase (HpdH) and/or methylmalonate semialdehyde dehydrogenase (MmsA), were involved in 3-HP degradation7 (see Fig. 1a for the gene arrangement of mmsA and hbdH4; hpdH is present in a separate operon). Furthermore, we found that the transcription of the mmsA gene encoding the MmsA enzyme was up-regulated at a high level (>100-fold) but only in the presence of 3-HP8. This suggests that there exists positive regulation on the expression of the enzyme, where 3-HP acts as an inducer. Analysis of gene arrangement near mmsA revealed the presence of a putative LysR-family transcriptional regulator protein that could be transcribed in the opposite direction of mmsA. It was speculated that the transcriptional regulator protein, when complexed with 3-HP, activates the transcription of mmsA. Considering the nature of this promoter (inducible by 3-HP) and its high induction efficiency (over 100-fold), the promoter should be useful for the expression of pathway enzymes for 3-HP synthesis or development of 3-HP-responsive biosensors. However, the biochemical characteristics of the transcriptional activator protein, including its binding sequences, have not yet been revealed.
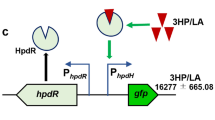
Regulation of gene expression by the LysR-type transcriptional activator protein, MmsR, in 3-hydroxypropionate degrading pathway of Pseudomonas denitrificans. (a) Hypothetical schematic representation of MmsR-based regulation. hpdH, 3-hydroxypropionate dehydrogenase; mmsA, methylmalonate semialdehyde dehydrogenase; hbdH4, 3-hydroxyisobutyrate dehydrogenase IV. (b) Relative mRNA abundance of mmsA in the presence of various candidate inducer molecules (25 mM). The amount of transcript was determined by real-time PCR analysis and ΔΔCT method as described previously25. The data were normalized against the un-induced mRNA expression level of mmsA in the wildtype strain, which was ~10-fold lower than that of the reference gene, rpoD. (c) Structures of chemical compounds tested for inducer specificity. 3-HP (1), 3-hydroxypropionic acid; 3-HIB (9), 3-hydroxyisobutyrate; 3-HB (5), 3-hydroxybutyrate; L-val (8), L-valine; LAC (2), L-lactate; AcOH (3), acetate; PA (4), propionaldehyde; 1,3-PDO (6), 1,3-propanediol; and 2,3-BDO (7), 2,3-butanediol.
This study aimed to elucidate the 3-HP-inducible gene regulation necessary for the expression of mmsA at the cellular and biochemical levels. At the cellular level, the specificity and/or spectrum of the small-molecule inducer(s) was studied using various acids structurally similar to 3-HP or the intermediates appearing in the L-valine degradation pathway and central carbon metabolism. The role of MmsR, the putative transcriptional activator protein, was confirmed by the removal of mmsR from the chromosome and subsequent complementation. Additionally, the promoter region that contains two operator sites (O1 and O2) was characterized by site-directed mutagenesis on each or both sites. At the biochemical level, the interactions of MmsR protein, 3-HP and the promoter region of mmsA were investigated by an electrophoretic mobility shift assay and DNase I footprinting using purified recombinant MmsR. Finally, a biosensor for the detection of 3-HP was developed and tested by expressing a green fluorescent protein (GFP) under the promoter activated by MmsR. This study should be useful for developing efficient 3-HP synthetic pathways and 3-HP sensors in many microorganisms.
Results and Discussion
Specificity of small-molecule inducers for MmsR transcriptional activator protein
According to our previous study, the putative transcription activator protein MmsR (previously designated C4-LysR) is composed of the N-terminal helix-turn-helix domain for DNA binding, the C-terminal domain for inducer binding that responds to 3-HP, and a linker connecting the two domains8. The spectrum of the inducer molecules for MmsR was studied using various acids and alcohols, such as L-lactic acid (LAC (2)), acetic acid (AcOH (3)), propionic acid (PA (4)), 3-hydroxybutyrate (3-HB (5)), 1,3-propanediol (1,3-PDO (6)) and 2,3-butanediol (2,3-BDO (7)), as well as L-valine (L-val (8)) and its degradation intermediate 3-hydroxyisobutyrate (3-HIB (9)). Most of the acids and alcohols tested were chosen mainly due to their similarity to 3-HP in size and/or structure. On the other hand, L-val and 3-HIB were chosen because mmsA, whose transcription was regulated by MmsR, encodes methylmalonylsemialdehyde dehydrogenase, an enzyme involved in L-val degradation. P. denitrificans was cultured on minimal medium with and without each compound to be tested, and transcription of mmsA was measured by quantitative RT-PCR (Fig. 1b). The housekeeping gene rpoD, encoding sigma factor 70, was used as a reference. Transcription of the mmsA gene was enhanced markedly upon exposure of P. denitrificans to chemicals compared to basal expression level in non-induction condition, such as 3-HIB (9) (154-fold), 3-HB (5) (38-fold) and L-val (8) (72-fold), in addition to 3-HP (1) (134-fold). By contrast, limited or no induction was observed after exposure to LAC (2), AcOH (3), PA (4), 1,3-PDO (6) and 2,3-BDO (7). As with all β-hydroxy acids, the chemicals 3-HP, 3-HB and 3-HIB are structurally similar (Fig. 1c), and the structural feature of having both the carboxyl and β-hydroxy groups seems to be integral to the ability to bind to MmsR. L-val, structurally much different from these three compounds, is converted to 3-HIB, and the induction by L-val is attributable to L-val-derived 3-HIB rather than to L-val itself (Fig. 1a).
We also examined the possibility that malonate semialdehyde (MSA, derived from 3-HP) and methylmalonate semialdehyde (MMSA, derived from L-val and 3-HIB) rather than 3-HP and/or 3-HIB are the physiological inducers (see Figs 1a and 2a). In general, aldehydes are toxic, and aldehyde-degrading genes are often up-regulated when aldehydes are accumulated9. According to our previous study6, 3-HP was degraded by the enzymes encoded by hpdH, hbdH4 and hbdH1, and the deletion mutant P. denitrificans ΔhpdHΔhbdH4ΔhbdH1 did not degrade 3-HP at an appreciable rate. As shown in Fig. 2a, in the triple mutant, which should not produce MSA from 3-HP, transcription of mmsA was nonetheless up-regulated by 3-HP. This indicates that 3-HP can activate MmsR protein without being converted to MSA. Similarly, transcription of mmsA was up-regulated by 3-HIB in the triple mutant, suggesting that 3-HIB is also a real inducer of MmsR.
Transcriptional analysis of mmsA in P. denitrificans with various genetic backgrounds. (a) Transcription of mmsA in the wildtype (WT) and mutant P. denitrificansΔhpdHΔhbdH4ΔhbdH1 (ΔΔΔ) strains. 3-HP and 3-HIB (each at 25 mM) were used as inducers. (b) Deletion and complementation of transcriptional activator, MmsR. ΔmmsR, P. denitrificansΔmmsR; and mmsR-C, P. denitrificansΔmmsR but with mmsR-complementation from a plasmid. The data were normalized against the un-induced mRNA expression level of mmsA in the wildtype strain, which was ~10-fold lower than that of the reference gene, rpoD.
To confirm the physiological role of MmsR as a transcriptional activator, deletion and subsequent complementation experiments were conducted (Fig. 2b). In the deletion mutant (ΔmmsR), transcription of mmsA was low, and the level was not affected by 3-HP. On the other hand, when mmsR was re-introduced to the ΔmmsR mutant by a plasmid, up-regulation of mmsA by 3-HP was fully restored. These results confirm that MmsR is a real transcriptional activator protein for the expression of mmsA.
In vivo characterization of mmsA promoter region
The transcriptional start site (TSS) of mmsA was identified by 5′ rapid amplification of cDNA ends (RACE)10. It was located at the G residue, −15 relative to the translation start of the mmsA structural gene (Supplementary Fig. S1). The putative −35 box (TGTTAG) and −10 box (AAAAAG) were predicted based on the TSS, which were spaced by 17 bp.
Importance of −10 and −35 boxes was studied by 5′ end mapping and site-directed mutagenesis (Fig. 3). For the mapping experiment, the PmmsA promoter region including the putative −10 and −35 regions were sequentially deleted and their strength was determined using GFP as reporter. P. denitrificans either lacking or expressing the mmsR gene was used as a host because MmsR considerably affects the promoter strength. In the ΔmmsR strain (un-induced condition), when the promoter region between −115 and −58 (P_Δ1) or −115 and −35 (P_Δ2) was deleted, the gfp expression decreased by approximately 16% compared to that of the control (P_wt), which contains the whole-length promoter. In comparison, when a longer upstream sequence before −24 (P_Δ3) or −14 (P_Δ4) was deleted, the promoter strength decreased more considerably by 39 or 58%, respectively (see Fig. 3B). These results suggest that the sequences deleted, from −35 to −14 relative to the TSS, in P_Δ3 and P_Δ4, should contain important elements for transcription, most likely the −10 or −35 regions. In addition, site-directed mutagenesis in the putative −10 (P-10mut) and −35 (P-35mut) regions reduced the promoter strength by 52% in comparison to that of P_wt (see Fig. 3B). The site-directed mutagenesis experiments have been repeated for the mmsR+ strain. Under the induced conditions with 3-HP, GFP intensity was high at 82 × 103 AU/OD600. In comparison, GFP intensity after mutation of either the putative −10 box (P-10mut) or −35 (P-35mut) box was only 3–5 × 103 AU/OD600 which is ~5% of that by P_wt (see Fig. 3B,C). When either the putative −10 or −35 box was deleted, presence of MmsR and 3-HP did not affect the gfp expression level (see Fig. 3C). Altogether, these results reveal the importance of the −10 and −35 regions which positioned from −34 to −29 and from −11 to −6 relative to the TSS, respectively, in PmmsA.
(A) Analysis of the mmsR-mmsA intergenic region for identification of the PmmsA promoter. A series of deletion (P_Δ1 − P_Δ4) and site-directed mutagenesis (P-35mut and P-10mut) are shown. Promoter elements (−10, −35) and TSS (+1) of PmmsA promoter were underlined. Change in gene expression of basal level (B) and inducibility (C) of the mmsA promoter after deletion and/or site-directed mutagenesis. GFP intensity (AU/OD600; AU, arbitrary unit) was measured in the absence (−) of 3-HP and MmsR (black bar) or presence (+) of 3-HP and MmsR (green bar). 3-HP was supplemented at 25 mM.
In vivo characterization of mmsA operator region
Before experimental characterization, in silico analysis of the operator region was briefly conducted11. Analysis by mfold web server12 identified the presence of a good stem-loop structure formed from dyad symmetry, often appearing as cis-acting element in many prokaryotic operators, with a spacer of 15 bp (see Supplementary Fig. S2a). This palindromic region was hypothesized as O1 site (from −95 to −63 relative to the TSS) and used as query for searching the second operator site O2, which is known to be less stringent, pseudo-palindrome. Two best candidates with good matches and spaced by 15 bp, AAAAATGCA-N15-CAGACAACAA (candidate 1, locating from −44 to −12 relative to the TSS) and TAGGATCGA-N15-AGCCACAGG (candidate 2, locating from −31 to +2 relative to the TSS) were identified (see Supplementary Fig. S2b). Furthermore, based on the typical location of O2 site (ABS) in relation to the promoter region in other LTTR systems13,14,15,16,17 (see Supplementary Fig. S2c), the first candidate, which was overlapped with the −35 element only, was suggested as the better candidate than the other one. The analysis shows that the dyad in the putative O1 site (from −95 to −63 relative to the TSS) is highly symmetrical, with only a single mismatch, while the inverted repeat in the putative O2 site (from −44 to −12 relative to the TSS) has 7 bases mismatched among the 9 (Fig. 4A). Alignment of the four palindromic fragments of the putative O1 and O2 operators showed the existence of the consensus sequence AATGTGCAA (Fig. 4B). Two bases T and G at positions 5 and 6 (bold), respectively, were fully conserved in all the fragments, and the three bases A, C, A at positions 2, 7 and 8 (underlined), respectively, were highly conserved in three fragments.
(A) Analysis of MmsR binding sites in the mmsR-mmsA intergenic region. A series of deletion (O_Δ1 – O_Δ4) and site-directed mutagenesis (O1mut, O2mut and O1mutO2mut) are shown. The symbols O1 and O2 denote the probable tandem operator sequences corresponding to palindromic (red letters) and pseudo-palindromic (blue letters) regions recognized by the MmsR protein. Promoter elements (−10, −35) and TSS (+1) of PmmsA promoter were underlined. F12, F12M, F1M2 and F1M2M are DNA fragments used for in vitro electromobility shift assay (EMSA). F12 includes the complete intergenic region, where both O1 and O2 are intact; F12M contains the intact O1 and mutant O2; F1M2 contains mutant O1 and intact O2; F1M2M contains mutant O1 and mutant O2. (B) Consensus sequence between O1 and O2 operator sequences of mmsR-mmsA intergenic region. The 9 bp of each inverted half-site is shown in 5′ to 3′ direction. The lower letters indicate the mismatch of repeat with the consensus sequence. The residues in boldface in consensus sequence are converted in each repeat sequence. The residues in under lined in consensus sequence are highly conserved in three fragments. (C) Change in gene expression after deletion and/or site-directed mutagenesis of the mmsA operator region. GFP intensity (AU/OD600; AU, arbitrary unit) was measured in the absence (black bar) and presence (green bar) of 25 mM 3-HP. MmsR was constitutively expressed from plasmid under the weak promoter, Pc1.
The 5′ end mapping and site-directed mutagenesis experiments were performed using GFP as reporter (Fig. 4A,C). To account for the possibility that mmsR is auto-regulated by itself and the auto-regulation interferes with the gfp transcription, mmsR was constitutively expressed from a plasmid under the control of the weak Pc1 promoter (3hibdH-I promoter18) (see Materials and Methods). The sequences of O_wt (intact intergenic region) and O_Δ1 (downstream from position −98 relative to the TSS) that contain both the putative operators (O1 and O2) exhibited high 3-HP induction up to ~34.6-fold. In comparison, when removing sequence upstream from position −79 containing the first half site of the putative O1 site (O_Δ2) or further eliminating additional sequences down to −48 (O_Δ3) and −21 (O_Δ4) containing the other half site of O1 and O2 regions, respectively; inducibility was completely lost (see Fig. 4C). It is noticed that the basal level under MmsR (over)expression (O_Δ series) has been significantly decreased compared to that lacking MmsR (P_Δ series shown in Fig. 3) even though their sequence features are nearly identical. This suggests that the inducer-free MmsR protein represses the mmsA expression. Once the complete O1 site and half site of O2 are removed, this repression effect is abolished, as depicted by similar expression levels for O_Δ4 and P_Δ3. Effect of mutation (site-directed mutagenesis) in the O1 and/or O2 region on the promoter strength was also investigated. Here, the operator sequences were carefully modified to destroy the dyad symmetry without affecting the −35 region. The palindromic region of O1 site (AATGTGCAA-N15-TTGCACATC) was changed to ATCCTTGCT-N15-CGCCCAGGC in O1mut or O1mutO2mut mutations; while the sequence ATCCTTGCT-N15-CAGGCACAA was used to replace O2 site region (AAAATGCAC-N15-AGACAACAA) in O2mut or O1mutO2mut mutations, with the underlined letters indicated the modified nucleotides (see Supplementary Table S2 for whole sequences). As expected, mutation of O1 (O1mut) or both O1 and O2 (O1mutO2mut) completely abolished the inducibility by 3-HP. When the putative O2 region alone was mutated (O2mut), promoter strength in the absence of 3-HP greatly increased (~10-fold) compared to that of wild-type O_wt. When 3-HP is present, the strength was reduced by 37% (compared to that of wild type O_wt) but still remained at a relatively high level. This suggests that the O2 site has an important role in regulating PmmsA function and strength, although its role or association with the O1 site is yet to be further elucidated. Collectively, the in vivo studies on the operator regions can be summarized as follows: (i) O1 alone can activate the transcription (with the help of 3-HP-MmsR complex), although less efficiently than when both O1 and O2 are present; (ii) binding of 3-HP-free MmsR to the O2 operator can suppress transcription from the PmmsA promoter; and (iii) the high strength of the promoter of the O2 mutant still requires the presence of the O1 site. It is most likely that the PmmsA promoter is regulated positively (in the presence of 3-HP) as well as negatively (in the absence of 3-HP).
The LTTR proteins and their cis-acting elements have been studied in several microorganisms. In the two symmetrical operator regions, termed RBS (Regulatory Binding Site) and ABS (Activator Binding Site), RBS was reported to have a better symmetry than ABS. In addition, the full sites of RBS and ABS were known to be often embraced by T-N11-A motifs. In E. coli and Salmonella typhimurium LT2, the IlvY protein (a LysR-type transcription regulatory protein) controlled the expression of ilvC gene (encoding acetohydroxy acid isomerase; EC 1.1.1.86)19. Transcription of the ilvC gene was induced by the substrates of acetohydroxy acid isomerase, acetohydroxybutyrate or acetolactate, and the induction was mediated by the IlvY protein. Similar to the PmmsA promoter under study, the promoter region of the ilvY and ilvC genes had two operators, O1 and O2, and each operator was composed of 9-bp-long inverted repeats. The repeats shared homologous sequences, A(T)CATTGCAA in E. coli and AT(A)GTTGC(A)GN in S. typhimurium LT2, respectively. Moreover, similar to the case in P. denitrificans, symmetry between the two dyads in these strains was more stringent in O1 (a single mismatch) than in O2 (six mismatches). Another LysR-type activator AtzR, which is present in Pseudomonas sp., has been reported to show the dissimilarity in the ABS operator region. AtzR activates the expression of divergently transcribed cyanuric acid degradative operon atzDEF. However, ABS contained three motifs, designated ABS-1, ABS-2 and ABS-3, in which individual subsites had distinct roles in the activation process. In vivo mutational analysis showed that ABS-1 and ABS-2 subsites were required for full activation of the PaztDEF promoter. In contrast, ABS-3 functioned as a ‘subunit trap’ leading to inactivation of PaztDEF when AtzR was shifted to the ABS-2 and ABS-3 subsites20. Similar dual functions of the ABS operator region have been reported with CbbR17 and DntR21. With the PmmsA under study, O2 site also seems to have the dual functions for both activation and repression. However, neither the presence of such multiple ABS’s nor shift of MmsR protein has been identified. For other LysR proteins such as IlvY14, CatR16, ClcR15 and HpaR22, the dual functions of O2 site has also been reported but without multiple ABS shown in AtzR. Further investigations are required to clarify the location of the O2 site(s) in the PmmsA promoter and the mechanism for the dual functions, if present.
Production of MmsR protein and its binding to operator sites in vitro
For biochemical characterization in vitro, a recombinant MmsR protein tagged with six histidine residues at the C-terminus of the protein was produced and purified from recombinant E. coli. The His-tag MmsR protein at either the C- or N-terminus showed the same performance as the native MmsR protein in the complementation experiments described in Fig. 2b (see Supplementary Fig. S3 for details); thus, one recombinant MmsR, C-His-tagged only, was studied in vitro. After careful optimization of the culture conditions (temperature, culture medium, IPTG concentration, harvest time and 3-HP concentration as well as co-expression with various chaperones, such as GroEL-ES, DnaKJ-GrpE and trigger factors; see Supplementary Fig. S4), the recombinant MmsR could be expressed in soluble form at a high level in E. coli by co-expression of mmsR-expressing plasmid and pGro7 containing groES-groEL chaperones (see Supplementary Fig. S4b) and purified by affinity chromatography (see Supplementary Fig. S5a). The size of the His-tag-fused MmsR protein was estimated to be 34.4 kDa, which is in good agreement with the size estimate based on the 6x his-mmsR gene sequence. According to native PAGE and/or gel filtration analyses, the MmsR protein was monomeric at the low concentration of 65 nM, while dimeric at the high concentration of 550 nM (see Supplementary Figs S5b and S7).
The binding ability of MmsR to the mmsR-mmsA intergenic region was studied in vitro by EMSA (Fig. 5 and Supplementary Fig. S6). A total of 40 nM of the complete 130 bp DNA fragment containing the sequence from −115 to +16 relative to the TSS was used as a probe (designated the F12 fragment; see Fig. 4A), and the purified MmsR protein at varying concentration at 0–48.5 nM was incubated with the DNA probe in both the presence and absence of 3-HP as an inducer. Similar to in vivo experiment, the sequences, ATCCTTGCT-N15-CGCCCAGGC and ATCCTTGCT-N15-CAGGCACAA, were applied to destroy the dyad symmetry in O1 and O2 sites, respectively, for generation of F1M2, F12M, and F1M2M fragments (see Fig. 4A and Supplementary Table S2 for sequence information). The DNA fragment mutated in both O1 and O2 (F1M2M) was used as a negative control. In electrophoresis, band shift appeared for the intact 130 bp DNA fragment when MmsR was added, and the ratio of the shifted DNA to un-shifted DNA increased upon amplifying the concentration of the MmsR protein during incubation. In contrast, no such mobility retardation was observed with the control DNA fragment up to 72.7 nM of MmsR protein (data not shown). This indicates that MmsR has a strong binding affinity to the native PmmsA, and they form a binding complex in vitro. At high concentrations of the MmsR protein, multiple bands traveling shorter distances appeared (see Fig. 5). It may attribute to the formation of diverse oligomeric complexes among the DNA fragments and protein molecules. The DNA probe has two binding sites (O1 and O2) for MmsR, and MmsR binds to the DNA as a dimer; thus, it is highly probable that DNA and protein form diverse oligomeric complexes when their concentrations are high. Figure 5 also shows the effect of 3-HP on binding between MmsR and the PmmsA promoter. In the presence of 3-HP, intensity of the shifted band increased at lower MmsR concentrations (for a clear comparison, see lane 4 in the panels of F12). This suggests that 3-HP promotes the binding affinity between MmsR and the PmmsA promoter. The EMSA experiments were also conducted with two DNA fragments having mutation (site-directed mutagenesis) in the palindromic regions of either O1 (F1M2) or O2 (F12M) operator site (see Fig. 4A and Supplementary Table S2 for sequence information). Under the same incubation condition, more intensive shifted-bands were observed with F12Mcompared to that with F1M2 in either absence or presence of 3-HP. This suggests higher affinity of MmsR toward O1 operator than O2 operator. Interestingly, the addition of 3-HP greatly reduced the binding of MmsR to the O2 region (see the panels for F1M2 in Fig. 5). This suggests that, along with the DNase I footprinting results (see below), ABS region should become less occupied by MmsR in the presence of 3-HP. Rhee et al. also studied the binding affinity of E. coli IlvY protein to DNA fragments containing either the tandem operators (O1O2) or the O1 or O2 operator alone. The binding affinity was highest with the fragment containing the tandem O1O2 operator and lowest with the O2 operator. However, in their system, the inducer molecule did not affect the binding of IlvY protein to the operators14.
In vitro electromobility shift assay (EMSA) to study binding of the MmsR protein to operator sites. Three DNA fragments containing intact O1and O2 (F12), or mutated in either O2 site (F12M) or O1 site (F1M2) were used as probe. The experiments were conducted in the absence (upper panel) or presence (lower panel) of 3-HP (at 25 mM). Lanes 1–10, increasing amounts of the MmsR protein at 0, 0.36, 0.73, 1.45, 2.9, 5.8, 11.6, 14.5, 24.2 and 48.5 nM. Lane 11–13 and 14–15, increasing amounts of the MmsR protein at 2.9, 11.6 and 48.5 nM. The red arrows indicate the shifted complexes. The concentration of DNA fragments was fixed at 40 nM. Figures for EMSA experiments of F12M and F1M2 were cropped from full-length gels shown in Supplementary Fig. S6.
DNAase I footprinting on operator-promoter region
The MmsR binding regions was also studied by DNase I footprinting based on capillary electrophoresis (Fig. 6). The longer 169 bp DNA fragment that includes the complete 130 bp intergenic region (employed in the previous EMSA experiments) was used as a probe (Supplementary Table S2). DNA concentration was fixed at 230 nM, while MmsR concentration varied in 0–1.2 μM. The footprinting results exhibit the presence of two protected regions by MmsR, which should correspond to operator O1 (centered at position −79) and O2 (centered at position −28), respectively. Protection became more evident as MmsR concentration increased. However, it was difficult to pinpoint the boundary of protection at the base-pair level of the DNA sequence. Between the O1 and O2 operators, the former was more protected than the latter and this confirms the higher affinity of the O1 site than the O2 site toward MmsR. Addition of 3-HP barely affected the DNase I footprinting for the O1 operator. However, for the O2 operator, as indicated by appearance of more unprotected peaks (at the high MmsR concentration of 1.2 μM), a weaker protection was observed when 3-HP was added (Fig. 6). This result agrees with EMSA experiment, i.e., in the presence of 3-HP, the O2 region is less susceptible to MmsR.
DNase I footprinting experiments on the mmsR-mmsA intergenic region. FAM-labeled mmsR-mmsA intergenic region was digested by DNase I in the absence (left) and presence (right) of 3-HP and analyzed using capillary electrophoresis. Sequence positions (x-axis) are marked with reference to the ROX 500 internal size-standard (M). The y-axis indicates relative fluorescent units (ABI 3130). MmsR concentration increased from 0 to 1.2 µM (concentrations were indicated on the left hand side). Dotted two black boxes (putative O1 and O2) in both panels indicate that DNase I digestion is inhibited by MmsR in dose-dependent manner. Red arrows highlight the change in protection by added 3-HP.
A relaxation of DNA bending in O2 or ABS region, as studied with AtzR13, IlvY19 and CatR15, was suggested as a common mechanism for transcription activation of LysR-type regulator. Two major mechanisms of LTTR-mediated transcription activation have been suggested; (i) ‘sliding dimer mechanism’ where a LysR protein, when bound with a inducer molecule, shifts from one ABS (called ‘downstream ABS’) to another ABS (‘upstream ABS’) leading to free −35 region, as studied with AtzR13, CbbR17 and DntR23, and (ii) ‘different-side binding model’ where RNAP and LysR can contact different sides of DNA helix, as studies with IlvY19, CatR15, and ClcR15. According to our study on the mmsR-mmsA intergenic region, no extra ABS is present and the PmmsA promoter functions better when the −35 box was not or less occupied by MmsR. This eliminate the first mechanism, although does not provide evidences to support the second mechanism.
Use of 3-HP-inducible promoter as 3-HP biosensor
The current dynamic promoter responding to 3-HP could be developed as a 3-HP sensor. The performance as a biosensor was briefly examined using the regulatory region containing both O1 and O2 sites (F12 fragment) with GFP as the reporter. MmsR protein was produced by the constitutive and weak promoters Pc1 of P. denitrificans and from the same plasmid as GFP. When measured at 12 h, the fluorescent signal was enhanced as the 3-HP concentration increased to 25 mM, and the fold difference compared with the control (for which no 3-HP was added) was >50 (Fig. 7A). The derived time-course profile (Fig. 7B) shows the fluorescent signal increasing with time and 3-HP concentration. The response of the biosensor to 25 mM 3-HP started immediately and did not reach the point of saturation until after 12 h. It was noted that the background fluorescence of the control (without 3-HP) was low and was not different from that of the host P. denitrificans without the recombinant plasmid. This indicates that the transcription of gfp is blocked almost completely and that leaky expression is negligible when 3-HP is absent. We attributed this tight control of gene expression to the repressive function of 3-HP-free MmsR protein when bound to the O2 operator.
(A) The 3-HP triggered fluorescence in cells containing the biosensor (green bar) in a dose-dependent manner up to 25 mM of 3-HP when measured at 12 h. The host cells without the biosensor (black bar) did not respond to 3-HP. (B) The response of the biosensor to 25 mM 3-HP (red downward triangle) started immediately and continued to increase up to 12 h after induction. Basal induction in the host cells (black circle) or in the biosensor without 3-HP (blue circle) was low. Three independent measurements for each were conducted and the results were averaged.
Recently, Rogers and Church reported development of 3-HP biosensors based on helper enzymes that convert 3-HP to the acuR-binding compound acrylate or prpR-binding 2-methylcitrate24. Both sensors could detect 3-HP successfully but only after conversion to acrylate or 2-methylcitrate. In comparison, the current MmsR biosensor directly recognizes 3-HP and does not require any helper enzymes. Furthermore, it generally has a better sensitivity and dynamic range. On this basis, we believe that the current biosensor is more convenient to use.
Conclusions
MmsR in P. denitrificans functions as a transcriptional regulator activating the expression of the mmsA gene from its responsive promoter PmmsA. The fold increase in the transcription of mmsA by 3-HP induction was >140. The responsive promoter PmmsA had two operator regions, designated O1 and O2, positioned at –79 and –28, respectively, upstream of the transcription start site of the mmsA gene. Binding of MmsR to O1 and O2 did not require an inducer molecule, but transcription activation strictly required it. Each of the O1 and O2 operators was composed of two 9-bp-long inverted repeats, and a mutation in the inverted repeats resulted in the reduction or complete loss of their binding capability to MmsR. The inducible promoter was developed as a biosensor for detection of 3-HP. With GFP as the reporter, the sensor showed a good response up to 25 mM 3-HP. This study will prove useful for the biotechnological application of the PmmsA promoter.
Materials and Methods
Materials
P. denitrificans ATCC 13867 was purchased from ATCC (America). The primers were synthesized by Macrogen Co. Ltd (Seoul, Korea). 3-HP was purchased from Tokyo Kasei Kogyo Co. Ltd. (Tokyo, Japan) and TCI America (Portland). Unless indicated otherwise, all other chemicals and enzymes were purchased from Sigma-Aldrich (St. Louis, MO).
Development of P. denitrificans ΔmmsR
To construct the deletion mutant P. denitrificansΔmmsR, the previously developed sacB-based method was used5. The pQSAK plasmid, which has a sacB-Km cassette, was used to knock off the mmsR gene from the chromosome (Supplementary Table S1). An engineered fragment containing the ~600–700 bp upstream (QS-mmsR-US) and downstream (QS-mmsR-DS) regions of the mmsR gene was generated by PCR from P. denitrificans genomic DNA. The fragment was cloned into the pQSAK vector using the GeneArt seamless cloning and assembly kit (Invitrogen, USA) and confirmed for sequence. The recombinant plasmid subsequently was used for homologous recombination.
Complementation of ΔmmsR mutation and effect of His-tag on MmsR function
The ORF of the mmsR gene was PCR-amplified from P. denitrificans genomic DNA and overlapped with a native constitutive Pc1 promoter (controlling expression of the 3hibdh-I gene18) of P. denitrificans. The overlapped fragment was then cloned into the shuttle vector pUCPK’, which had been modified from the previously developed plasmid pUCPK’5. The resultant plasmid was designated pUCPK’/Pc1-mmsR, and the recombinant plasmid was introduced by electroporation into the P. denitrificansΔmmsR mutant to develop the complementation strain, mmsR-C. In order to evaluate the effect of His-tag on MmsR function, plasmids overexpressing the His-tagged MmsR at the N- or C-terminus were constructed. The resultant plasmids and recombinant strains were designated pUCPK’/Pc1-N6xhis-mmsR, pUCPK’/Pc1-mmsR-C6xhis and N-his-mmsR-C, C-his-mmsR-C.Details of primers were given in Supplementary Table S3.
RNA extraction and Real-time PCR
The P. denitrificans strains were grown in M9-minimal medium containing 5 g/L sodium gluconate. The cells were cultivated under the aerobic condition at 37 °C and 200 rpm in an orbital incubator shaker. 3-HP and/or various chemicals (3-hydroxyisobutyrate, 3-hydroxybutyrate, L-valine, lactic acid, acetic acid, propionic acid, 1,3-propanediol and 2,3-butanediol) at 25 mM was supplemented at the OD600 of ~0.4–0.5. After a further 2 h cultivation, approximately 5 × 108 cells were collected and centrifuged at 5,000 × g for 10 min. The cell pellets were immediately resuspended in 500 μL of RNA solution (Ambion, UK) and RNA was extracted using a total RNA isolation kit (Macherey-Nagel, Germany). One microgram of total RNA was employed to synthesize the first-strand cDNA in a 20 µL reaction using the SuperScript III first-strand synthesis system (Invitrogen, USA). A real-time PCR analysis was performed, according to the SYBR green method, in a 20 µL reaction volume using the StepOne Real Time PCR system (Applied Biosystems, USA). The PCR efficiencies of all of the primers were experimentally determined and found to be suitable for reliable copy-number quantification. The mRNA quantity was estimated based on ΔΔCT value, as described previously25. The assays were performed in duplicate, and a template-less reaction was used as a negative control.
Construction plasmids
Supplementary Table S1 lists the bacterial strains and plasmids used in this study. E. coli BL21 (DE3) served as the host for protein production, and E. coli Top10 was used for routine cloning and plasmid maintenance. The LB medium was used for the growth of E. coli. Ampicillin at 100 mg/L was added to the culture medium when needed. Gene manipulation was carried out using standard methods7. The plasmid pET30b(+) was used to clone the mmsR gene in E. coli BL21(DE3). The mmsR gene was amplified from P. denitrificans genomic DNA by PCR and the fragment was ligated into the pET30b(+) vector using a seamless cloning and assembly kit (Invitrogen). The resulting plasmid, pET30b(+)/mmsR, which contained the ET-mmsR gene with the His-tag at the C-terminus, was sequenced by Macrogen Co., Ltd., Korea, and introduced into E. coli BL21(DE3).
The plasmid pUCPK was used to develop3-HP biosensor in P. denitrificans. The sequences for UC-Pc1, UC-mmsR and UC-PmmsA were amplified from P. denitrificans genomic DNA by PCR, while the UC-gfp gene, encoding the green fluorescence protein (GFP), was amplified from pPro24-gfp (Addgene). Four DNA fragments of UC-Pc1, UC-mmsR, UC-PmmsA and UC-gfp were overlapped and ligated into the pET30b(+) vector using a seamless cloning and assembly kit (Invitrogen). The resulting plasmid, pUCPK-Pc1-mmsR-PmmsA-gfp, was sequenced by Macrogen Co., Ltd., Korea, and introduced into P. denitrificans. Details of primers are given in Supplementary Table S3.
Plasmids used in deletion analysis and site-directed mutagenesis were constructed by amplifying various target fragments, such as P_wt-gfp, P_Δ1-gfp, P_Δ2-gfp, P_Δ3-gfp, P_Δ4-gfp, P-10mut-gfp, P-35 mut-gfp, O_wt-gfp, O_ Δ1-gfp, O_ Δ2-gfp, O_ Δ3-gfp, O_ Δ4-gfp, O1 mut-gfp, O2mut-gfp and O1mutO2mut-gfp, using the recombinant pUCPK-Pc1-mmsR-PmmsA-gfp plasmid as template with the primers shown in Supplementary Table S3. The amplified PCR fragments were ligated into the pUCPK or pUCPK-Pc1-mmsR vectors (Supplementary Table S3) using a seamless cloning and assembly kit (Invitrogen). The resulting plasmids was sequenced by Macrogen Co., Ltd., Korea, and introduced into P. denitrificansWT or P. denitrificansΔmmsR.
Transcription start site identification
To determine the 5′ end of the mmsA transcript, 5′ rapid amplification of cDNA ends (RACE) PCR was performed using a SMARTer® RACE 5′/3′ kit (Clontech, USA). The RACE experiment was carried out according to the manufacturer’s instruction. Synthesis of the first strand cDNA was performed on 1 μg of the DNase-treated total RNA of P. denitrificans (wild-type) grown under induction condition as described before. RACE fragments were purified from agarose gels using the Qiagen gel-extraction kit and cloned into the linearized pRACE vector with In-Fusion HD Cloning (Takara, Japan). DNA from seven colonies of each RACE synthesis was sequenced using M13 forward and reverse primers. Primer extension assays were performed as previously described (Zeng et al. 2002), except that 50 µg of total RNA, derived from transfected 293 T cells, was used. The sequence of gene-specific primer (mmsA-GSP5-RP) used for RACE was: ′5-GATTACGCCAAGCTTCGGCCCGAGGTCGCCCAGCTTGGAGCCGCG-3′.
Expression and purification of MmsR protein
For the production of soluble His-tagged MmsR, pET30b(+)/mmsR was co-expressed with various chaperone plasmids, i.e. pG-KJE8, pGRO7, pKJE7, pG-TF2, and pTF-16 (Takara, Japan), in E. coli BL21(DE3). The recombinant BL21(DE3) strains were grown in LB medium supplemented with 50 mg/L kanamycin, 25 mg/L chloramphenicol and/or 0.5 mg/mL L-arabinose. The cells were grown aerobically in 1 L Erlenmeyer flasks containing 350 mL of medium at 30 °C and 200 rpm in an orbital incubator shaker. The cells were induced at ~0.6 OD600 with 0.1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG) and grown at 25 °C, 150 rpm for 10 h. The cells were then harvested and centrifuged at 10,000 × g for 10 min. The pellets were washed with 100 mM potassium phosphate buffer (pH 7.0) and resuspended in the binding buffer (20 mM sodium phosphate buffer containing 0.5 M NaCl and 20 mM imidazole). The resuspended cells were disrupted using the French Pressure Cell (FA-078A, Thermo Electron Corp.; Waltham, MA, USA) at 1,250 psi. The cell lysate was centrifuged at 15,000 × g at 4 °C for 30 min to remove the particulate fraction and unbroken cells. The soluble fraction was subjected to purification under non-denaturing conditions by Ni-affinity chromatography using a Ni-NTA-HP resin column (17-5248-01; GE Healthcare, Sweden). The eluents from the column were pooled and dialyzed using a 10 kDa cut-off membrane to remove the salts. The resulting enzyme extract was electrophoresed under denaturing and non-denaturing conditions, as described by Laemmli, and the extract was stored at −80 °C in 20% glycerol.
DNase I footprinting analysis of the mmsA promoter
DNase I footprinting analysis was conducted using an automated capillary sequencer26. FAM-labeled fragments of the mmsA promoter were synthesized by a standard PCR method using a FAM-labeled oligonucleotide (Macrogen, Korea) and P. denitrificans genomic DNA as template, purified on agarose gel and extracted from the gel (Qiagen). The 250 ng DNA fragment (169 bp) was incubated with the recombinant MmsR (0.15, 0.6 and 1.2 µM) for protection at room temperature for 40 sec in 10 µl 1X binding buffer which contains 50 mM Tris (pH 7.4), 750 mM KCl, 0.5 mM dithiothreitol and 0.5 mM EDTA. Protected DNA fragments were digested with 0.05 units of DNase I at room temperature for 30 sec in the presence of 1X DNase I buffer, and the digestion reaction was stopped by adding the stop solution containing EDTA (pH 8.0) to a final concentration of 250 mM. Afterwards, the sample was heated to 70 °C and rapidly cooled to 4 °C. Denatured DNA fragments were separated on an ABI 3130xl Genetic analyzer (36 cm capillary array and POP4 polymer; Applied Biosystems Inc., Foster City, CA, USA). A 1.5 µL of assay product was mixed with 8.3 µL of deionized formamide (Applied Biosystems) and 0.2 µL of ROX 500 size-standard (Applied Biosystems Inc.), the latter of which wasdenatured at 95 °C for 5 min and quenched at 4 °C for 3 min. The mixture was injected into capillaries by applying 1.6 KV for 15 sec, and then electrophoresed under 10 kV and an operating temperature of 60 °C. Data analysis was conducted by GeneMapper software (version 4.1, Applied Biosystems). DNA fragments labeled with ROX were used as internal standard (Invitrogen, USA).
Electrophoretic Mobility Shift Assay (EMSA)
The 130 bp-long DNA fragments, designated F12, F12M, F1M2 and F1M2M, were PCR-amplified (Supplementary Table S2) and purified using a glass fiber column (PCR purification kit, Qiagen). EMSA was performed using Molecular Probes’ Fluorescence-based Electrophoretic Mobility Shift Assay (fluorescence-EMSA) kit (Invitrogen, E33075), following the standard protocols supplied by the manufacturer. Binding reactions were assembled on ice with increasing amounts of purified MmsR protein (34.4 kDa) added separately to a fixed concentration (40 nM) of the DNA fragments (F12, F12M, F1M2 and F1M2M) in a final 12 µl volume of 1X binding buffer (50 mM Tris (pH 7.4), 750 mM KCl, 0.5 mM dithiothreitol, 0.5 mM EDTA). The reaction was incubated at room temperature for 30 min before loading onto 6% non-denaturating polyacrylamide gel. The gel was run at 200 V in pre-chilled 0.5X TBE buffer (44.5 mM Tris base, 44.5 mM boric acid, 0.5 mM EDTA; pH~8) for 30 min. The gel was fixed and stained with SYBR Green EMSA stain for DNA observation or SYPRO Ruby EMSA stain for protein observation, before quantification on a gel documentation system (Bio-Rad systems).
Statistical analysis
Values form RNA transcription level or fluorescence (AU/OD600) were confirmed by one-way analysis of variance (ANOVA) with tukey post hoc comparisons for each dependent variable at 95% of confidence interval.
References
Kumar, V., Ashok, S. & Park, S. Recent advances in biological production of 3-hydroxypropionic acid. Biotechnol. Adv. 31, 945–961 (2013).
Rathnasingh, C., Raj, S. M., Jo, J. E. & Park, S. Development and evaluation of efficient recombinant Escherichia coli strains for the production of 3-hydroxypropionic acid from glycerol. Biotechnol. Bioeng. 104, 729–739 (2009).
Pfleger, B. F., Gossing, M. & Nielsen, J. Metabolic engineering strategies for microbial synthesis of oleochemicals. Metab. Eng. 29, 1–11 (2015).
Ashok, S. et al. Production of 3-hydroxypropionic acid from glycerol by recombinant Klebsiella pneumoniae ΔdhaTΔyqhD which can produce vitamin B12 naturally. Biotechnol. Bioeng. 110, 511–524 (2013).
Zhou, S., Catherine, C., Rathnasingh, C., Somasundar, A. & Park, S. Production of 3-hydroxypropionic acid from glycerol by recombinant Pseudomonas denitrificans. Biotechnol. Bioeng. 110, 3177–3187 (2013).
Zhou, S., Ashok, S., Ko, Y., Kim, D. M. & Park, S. Development of a deletion mutant of Pseudomonas denitrificans that does not degrade 3-hydroxypropionic acid. Appl. Microbiol. Biotechnol. 98, 4389–4398 (2014).
Lee, P. et al. 3-hydroxyisobutyrate dehydrogenase-I from Pseudomonas denitrificans ATCC 13867 degrades 3-hydroxypropionic acid. Biotechnol. Bioprocess. Eng. 19, 1–7 (2014).
Zhou, S., Ainala, S. K., Seol, E., Nguyen, T. T. & Park, S. Inducible gene expression system by 3-hydroxypropionic acid. Biotechnol. Biofuels. 8, 1–8 (2015).
Singh, S. et al. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic Biol. Med. 56, 89–101 (2013).
Schramm, G., Bruchhaus, I. & Roeder, T. 5′ end cDNA amplification using RACE approach. Nat. Protoc. 1, 2555–2562 (2006).
Reese, M. G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Biol Chem. 26, 51–56 (2001).
Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31, 3406–3415 (2003).
Porrúa, O., García-Jaramillo, M., Santero, E. & Govantes, F. The LysR-type regulator AtzR binding site: DNA sequences involved in activation, repression and cyanuric acid-dependent repositioning. Mol. Microbiol. 66, 410–427 (2007).
Rhee, K. Y., Senear, D. F. & Hatfield, G. W. Activation of gene expression by a ligand-induced conformational change of a protein-DNA complex. J. Biol. Chem. 273, 11257–11266 (1998).
McFall, S. M., Chugani, S. A. & Chakrabarty, A. M. Transcriptional activation of the catechol and chlorocatechol operons: variations on a theme. Gene. 223, 257–267 (1998).
Chugani, S. A., Parsek, M. R. & Chakrabarty, A. M. Transcriptional repression mediated by LysR-type regulator CatR bound at multiple binding sites. J. Bacteriol. 180, 2367–2372 (1998).
Keulen, G., Ridder, A., Dijkhuizen, L. & Meijer, W. G. Analysis of DNA binding and transcriptional activation by the LysR-type transcriptional regulator CbbR of Xanthobacter flavus. J. Bacteriol. 185, 1245–1252 (2003).
Zhou, S. et al. Cloning, expression and characterization of 3-hydroxyisobytyrate dehydrogenase from Pseudomonas denitrificans ATCC 13867. PloS One 8, e62666 (2013).
Wek, R. C. & Hatfield, G. W. Transcriptional activation at adjacent operators in the divergent-overlapping ilvY and ilvC promoters of Escherichia coli. J. Mol. Biol. 203, 643–663 (1988).
Porrúa, O. et al. Complex interplay between the LysR-type regulator AtzR and its binding site mediates atzDEF activation in response to two distinct signals. Mol. microbiol. 76, 331–347 (2010).
Lerche, M. et al. The solution configurations of inactive and activated DntR have implications for the sliding dimer mechanism of LysR transcription factors. Scientific reports 6, 19988 (2016).
Galán, B., Kolb, A., Sanz, J. M., García, J. L. & Prieto, M. A. Molecular determinants of the hpa regulatory system of Escherichia coli: the HpaR repressor. Nucleic Acids Res 31, 6598–6609 (2003).
Devesse, L. et al. Crystal structures of DntR inducer binding domains in complex with salicylate offer insights into the activation of LysR-type transcriptional regulators. Mol. Microbiol. 81, 354–367 (2011).
Rogers, J. K. & Church, G. M. Genetically encoded sensors enable real-time observation of metabolite production. Proc. Natl. Acad. Sci. USA 113, 2388–2393 (2016).
Yim, S. H. et al. Copy number variations in East-Asian population and their evolutionary and functional implications. Hum. Mol. Gen. ddp564 (2009).
Yindeeyoungyeon, W. & Schell, M. A. Footprinting with an automated capillary DNA sequencer. Biotechniques. 29, 1034–1036 (2000).
Acknowledgements
This study was supported financially by the Korean Ministry of Science, ICT and Future Planning through the Advanced Biomass R&D Center (Global Frontier Project, Grant No. 2010-0029799), KAIST, Korea. In addition, we highly appreciate Prof. Gyoo Yeol Jung and Dr. Jeongkyeong Na at POSTECH, Korea for helping us in DNase I footprinting experiment using capillary electrophoresis. This study was also supported by the BK21 Plus Program for Advanced Chemical Technology at Pusan National University.
Author information
Authors and Affiliations
Contributions
N.H.N., S.K.A., S.Z. and S.P. designed research; N.H.N., S.Z. performed research; N.H.N., S.K.A. and S.P. analyzed data; N.H.N. and S.P. wrote the paper. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nguyen, N.H., Ainala, S.K., Zhou, S. et al. A novel 3-hydroxypropionic acid-inducible promoter regulated by the LysR-type transcriptional activator protein MmsR of Pseudomonas denitrificans. Sci Rep 9, 5333 (2019). https://doi.org/10.1038/s41598-019-41785-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-41785-y
This article is cited by
-
Engineering of the Substrate Pocket of α-ketoglutaric Semialdehyde Dehydrogenase for Improving the Activity toward 3-hydroxypropanal
Biotechnology and Bioprocess Engineering (2023)
-
In vivo Characterization of the Inducible Promoter System of 3-hydroxypropionic Dehydrogenase in Pseudomonas denitrificans
Biotechnology and Bioprocess Engineering (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.