Abstract
The elongases of very long-chain fatty acids (Elovls) are responsible for the rate-limiting elongation process in long-chain polyunsaturated fatty acid (LC-PUFA) biosynthesis. The transcription factor, PPARα, regulates lipid metabolism in mammals; however, the detailed mechanism whereby PPARαb regulates Elovls remains largely unknown in fish. In the present study, we report the full length cDNA sequence of Trachinotus ovatus Elovl4a (ToElovl4a), which encodes a 320 amino acid polypeptide that possesses five putative membrane-spanning domains, a conserved HXXHH histidine motif and an ER retrieval signal. Phylogenetic analysis revealed that the deduced protein of ToElovl4a is highly conserved with the Oreochromis niloticus corresponding homologue. Moreover, functional characterization by heterologous expression in yeast indicated that ToElovl4a can elongate C18 up to C20 polyunsaturated fatty acids. A nutritional study showed that the protein expressions of ToElovl4a in the brain and liver were not significantly affected among the different treatments. The region from PGL3-basic-Elovl4a-5 (−148 bp to +258 bp) is defined as the core promoter via a progressive deletion mutation of ToElovl4a. The results from promoter activity assays suggest that ToElovl4a transcription is positively regulated by PPARαb. Mutation analyses indicated that the M2 binding site of PPARαb is functionally important for protein binding, and transcriptional activity of the ToElovl4a promoter significantly decreased after targeted mutation. Furthermore, PPARαb RNA interference reduced ToPPARαb and ToElovl4a expression at the protein levels in a time-dependent manner. In summary, PPARαb may promote the biosynthesis of LC-PUFA by regulating ToElovl4a expression in fish.
Similar content being viewed by others
Introduction
Long-chain polyunsaturated fatty acids (LC-PUFA) are involved in numerous biological processes and are major components of complex lipid molecules1. In vertebrates, two LC-PUFA biosynthetic pathways are defined: the “∆6 pathway” (∆6 desaturation-elongation-∆5 desaturation) and the “∆8 pathway” (elongation-∆8 desaturation-∆5 desaturation); these are initiated from α-linolenic (18:3n-3) and linoleic (18:2n-6) acids, respectively1,2,3,4,5. Two sets of enzymes, the elongases of very long-chain fatty acids (Elovls) and fatty acyl desaturases (Fads), are involved in these pathways6. The Elovls protein family include seven isozymes (Elovl1-7) in vertebrates7. In vitro FA elongation assays, knockdown and knockout (KO) of Elovl1-7 genes revealed that Elovl1-7 exhibits substrate specificity; each isozyme prefers acyl-CoAs with specific chain-lengths and/or a degree of saturation8. The over-expression of fish elongases has also elevated the endogenous production of LC-PUFA in fish species, such as transgenic Danio rerio and Miichthys miiuy9,10. The Elovl4 enzyme has been widely studied in teleosts, especially in marine species11,12,13,14,15. Yeast heterologous expression systems revealed that Elovl4 is mainly involved in the elongation of C20–22 LC-PUFA, producing polyenes of up to 36 carbons in the biosynthetic pathway of LC-PUFAs1,12,16.
Peroxisome proliferator-activated receptor alpha (PPARα) is a member of the steroid receptor superfamily of ligand-activated nuclear transcription factors and is known to regulate lipid and glucose metabolism17,18. Furthermore, PPARα stimulates the expression of target genes via direct binding to PPAR response elements (PPREs) in the promoter region of target genes19,20. It has been shown that PPARα upregulates Fads2 promoter activity in fish and avians21,22. Both PPARα1 and PPARα2 were found to activate the promoter activity of Fads2 in Lateolabrax japonicas; however, no such regulatory activity was detected for Larimichthys crocea22.
The golden pompano Trachinotus ovatus (Linnaeus 1758), Carangidae, and Perciformes are found in the Asia-Pacific region and are considered important aquaculture fish in China because of their economic value23,24. Furthermore, the T. ovatus muscle has been found to be rich in PUFAs (such as eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA)) after feeding without PUFAs (EPA and DHA)25, showing that it has the ability to endogenously compound PUFAs. Consequently, T. ovatus provides an exceptional model for the investigation of regulatory mechanisms in LC-PUFA biosynthesis in teleosts. To investigate the underlying function of T. ovatus Elovl4a (ToElovl4a) and the regulation of Elovl4a by PPARαb during LC-PUFA biosynthesis, the present study focused on clarifying the importance of PPARαb in regulating ToElovl4a transcriptional activity. First, a functional characterization of the ToElovl4a gene was performed using heterologous expression in yeast. Second, promoter activity assays via the mutation of potential PPARαb binding sites were performed to identify the key element in the ToElovl4a promoter. Finally, the suppression of expression (RNAi) of PPARαb was used to elucidate the transcriptional regulation of PPARαb with respect to ToElovl4a. These approaches have contributed to the identification of ToElovl4a function and showed that PPARαb performs a vital function in the regulation of Elovl4a expression.
Results
Molecular cloning and phylogenetics of T. ovatus Elovl4a
The T. ovatus putative elongase full length cDNA was 1,606 bp and included an ORF of 963 bp. This nucleotide sequence translated to a peptide sequence of 320 amino acids (Accession no. MG674424) (Supplementary Fig. 1). A BLAST analysis revealed that the ToElovl4a) protein sequence shared high sequence identity with Elovl4a sequences from other teleosts, including tilapia (Oreochromis niloticus, 96%, Ensembl No. ENSONIP00000009094.1) and zebrafish (Danio rerio, 81%, Ensembl No. ENSDARG00000006773), shared low sequence identity with chicken (Gallus gallus, 68%, Ensembl No. ENSGALG00000015876) and humans (Homo sapiens, 64%, Ensembl No. ENSG00000118402).
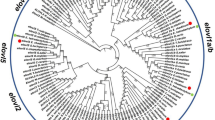
Interestingly, comparisons of the amino acid sequences for the Elovl4a protein from the above four species showed three conserved domains, which contained five putative membrane-spanning domains with a conserved HXXHH histidine motif and an ER retrieval signal (Fig. 1). The phylogenetic tree analysis indicated that ToElovl4a clustered with several other Elovl4a sequences from other osteichthyes, and more distantly, with avian (G. gallus) and mammalian (H. sapiens) Elovl4 (Fig. 2). ToElovl4a was grouped together with perciformes, such as O. niloticus.
Comparison of the deduced amino acid (AA) sequence of the Trachinotus ovatus Elovl4a with those of O. niloticus, D. rerio, H. sapiens and G. gallus Elvol4. Five (I–V) putative membrane-spanning domains are indicated by yellow colour. The conserved HXXHH histidine motif and ER retrieval signal are indicated by yellow and blue boxes, respectively. Dashes represent gaps created to maximize the degree of identity among all compared sequences. The accession numbers of the sequences used are from Supplementary Table 3.
Phylogenetic relationship of T. ovatus Elovl4a/b amino acid sequences with their counterparts from other species. The main topology was produced by MEGA 6 software with the maximum likelihood (ML) method with 1000 bootstrap replicates. The accession numbers of the sequences used are from Supplementary Table 3.
Heterologous expression of the elongase ORF in Saccharomyces cerevisiae
The function of ToElovl4a was characterized by determining the FA profiles in S. cerevisiae, was transformed with pYES2-Elovl4a and was grown in the presence of potential FA substrates, including C18 (18:2n-6, 18:3n-3, 18:3n-6 and 18:4n-3), C20 (20:4n-6 and 20:5n-3) and C22 (22:5n-3, 22:4n-6 and 22:6n-3) substrates. In yeast transformed with pYES2-Elovl4a and grown in the presence of 18:3n-6 (Fig. 3A), however, an additional FA peak was identified as 20:3n-6 (Fig. 3B) based on the gas chromatography (GC) retention times. Therefore, from this data, it was concluded that the ToElovl4a can efficiently elongate C18 up to C20. The conversion rates of 18:3n-6 to 20:3n-6 were calculated to be approximately 1.05% (Table 1). Moreover, upon comparison with the gas mass spectrometry database, our results indicated that no other FA mass spectrometry structures were detected except for the 20:3n-6 structure.
Functional characterization of the putative Elovl4a in transgenic yeast. (A and B) represent adding polyunsaturated fatty acid (FA) substrate of C18:3n−6. FAs were extracted from yeast transformed with the pYES2 vector, including the ORF of the putative Elovl4a cDNA as an insert. Peaks 1–4 represent the main endogenous FAs of T. ovatus, namely, C16:0, C16:1 isomers, C18:0 and C18:1n -9, respectively. Based in the retention times, additional peaks were identified as 20: 3n-6 (B). Vertical axis, FID response; horizontal axis, retention time.
Tissue distribution of ToElovl4a
Tissue distributions of ToElovl4a were delineated by qRT-PCR. The highest ToElovl4a mRNA levels were detected in the brain, followed by the stomach and intestine, whereas relatively low ToElovl4a expression levels were observed in the liver and spleen (Fig. 4). Notably, the expression of ToElovl4a in the brain was much higher than in other tissues (P < 0.05).
Gene transcriptions of Elovl4a in various tissues of T. ovatus. The twelve tissues are brain (Br), stomach (St), intestine (In), male gonad (Mg) and female gonad (Fg), fin (Fi), blood (Bl), kidney (Ki), gill (Gi), white muscle (Wm), liver (Li), and spleen (Sp). Significant differences at P < 0.05 are labelled with different letters, and mean ± SEM of each mRNA quantity is shown for each tissues tested.
Nutritional regulation of ToElovl4a
The protein expression of ToElovl4a in the liver and brain fed with different levels of LNA or LA (18:3n-3 or 18:2n-6) through the diet was determined by a western blot. The GAPDH was used as an internal control for normalization. The express pattern of ToElovl4a protein levels in the liver and brain were uncorrelated with the fatty acid compositions (Fig. 5) (also Supplementary Fig. S1).
Western blot analysis of Elovl4a proteins in livers and brains after eight dietary treatments in T. ovatus. The left side show the western blot result, and the right side shows the corresponding ratio of grey values of Elovl4a proteins. Dietary lipid sources: 1, FO; 2, KO; 3, SO; 4, CO; 5, 1:1 FO-SO; 6, 1:1 FO-CO; 7, 1:1 KO-SO; 8, 1:1 KO-CO. FO, fish oil; KO, krill oil; SO, soybean oil; CO, corn oil; FO-SO, fish oil-soybean oil; FO-CO, fish oil-corn oil; KO-SO, krill oil-soybean oil; KO-CO, krill oil-corn oil. Full-length blots are shown in Supplementary Fig. S2.
Promoter analysis of PPARαb regulation
The cloned candidate ToElovl4a promoter (1,057 bp) was an upstream non-transcribed sequence. To determine the binding region of PPARαb in the ToElovl4a promoter, a full length candidate promoter and several truncated mutants were constructed with a promoterless luciferase reporter vector, pGL3-basic. The promoter construct, Elovl4a-p5 (−148 bp to +258 bp), exhibited the highest promoter activity with PPARαb, suggesting that this region of the Elovl4a-p5 promoter sequence contained the PPARαb binding site (Fig. 6A).
Promoter activity analysis of ToElovl4a gene. A. The structure and transcriptional activity of the ToElovl4a promoters. Five recombinant plasmids, denoted as Elovl4a-1 (−148 to +56), Elovl4a-2 (−500 to +56), Elovl4a-3 (−1001 to +56), Elovl4a-4 (−148 to +155) and Elovl4a-5 (−148 to +258), were constructed and transfected with transcription factor PPARαb into HEK 293 T cells. B. Dual luciferase activity driven by the ToElovl4a-5 core promoter upon the transfection of pcDNA3.1-PPAR-α and pcDNA3.1 in HEK 293 T cells. All values are presented as the means ± SD (n = 3). Asterisks indicate that the values are memorably different from the individual controls (*p < 0.05 and **p < 0.01). Bars on the same group with different letters are statistically significant from one another.
To further confirm the interaction of ToPPARαb with ToElovl4a, the influence of ToPPARαb overexpression on ToElovl4a transcription was determined. PPARαb overexpression increased the promoter activity of ToElovl4a-5 at all tested time points in heterologous HEK 293 T cells, and the maximum difference occurred at 12 h posttransfection, which was 5.6-fold higher in the PPARαb-overexpressing cells than that in the controls (Fig. 6B). These results indicated that constitutively expressed PPARαb positively regulated ToElovl4a expression in HEK 293 T cells.
To identify the PPARαb binding sites in the Elovl4a promoter, the predicted binding sites were mutated (Fig. 7, Table 2). The effects on promoter activity were investigated in 293 T cells that were transfected with each mutant and PPARαb. The results revealed that mutation of the M2 binding site (+209 bp to +223 bp) caused significant reduction in promoter activity (Fig. 7), showing that M2 was the PPARαb binding site in the Elovl4a promoter. Notably, three other predicted binding sites did not induce luciferase activity with PPARαb, suggesting that these three sites were not required for triggering ToElovl4a expression with PPARαb.
The nucleotide sequence and predicted binding sites for transcription factors in the core region of the ToElovl4a promoter (A). Effects of transcription factor mutations on ToElovl4a-5 promoter activity (B). Binding sites are shown with boxes. Mutations of promoter sequences are listed in Table 2. All values are presented as the means ± SD (n = 3). Asterisks indicate that the values are memorably different from the individual controls (*p < 0.05 and **p < 0.01). Bars on the same group with different letters are statistically significant from one another.
Transcriptional regulation of ToElovl4a by PPARαb
Protein levels of ToPPARαb were considerably decreased in a time-dependent manner by the RNAi of PPARαb, suggesting effective knockdown of ToPPARαb expression in T. ovatus caudal fin cells (TOCF) (Fig. 8A) (also Supplementary Fig. S2A). When ToPPARαb expression was reduced, the protein levels of ToElovl4a were considerably depleted compared with the control group at corresponding time points (Fig. 8B) (also Supplementary Fig. S2B). These results suggested an active regulatory role of ToPPARαb on ToElovl4a expression in the TOCF cells.
ToPPARαb upregulates ToElovl4a expression. Western blot analysis was used to detect the expression of ToPPARαb (A) and ToElovl4a (B) after the transfection of either control RNA (Control) or siRNA (RNAi), respectively. Full-length blots are presented in Supplementary Fig. S3.
Discussion
The present study sought to gain insights into the mechanisms underlying the transcriptional regulation of LC-PUFA biosynthesis in T. ovatus. To achieve this, sequence and functional characterization, tissue expression patterns and transcriptional regulation of ToElovl4a were investigated. The ToElovl4a ORF encodes a protein that is 81%–96% identical to Elovl4 proteins from other teleosts. These isolated ToElovl4a proteins contain three classic structural motifs, including transmembrane domains, a conserved histidine box (HXXHH), and an ER retrieval signal (RXKXX) in the canonical C-terminal, indicating its specific role is in LC-PUFA biosynthesis26. These three conserved boxes were also found to be present in other species Elovl4 proteins11,12,13,14,15. The ToElovl4 sequence is positioned within the teleost Elovl4a clade together with O. niloticus, and the teleost Elovl4 clade is outgrouped by the tetrapod Elovl4 clade containing the Elovl4 sequences from avians and mammals.
Three members of the fatty acid elongases protein family, Elovl2, Elovl4 and Elovl5, have been described as crucial enzymes involved in the biosynthetic pathway of LC-PUFA in teleosts1,27. For marine fish, yeast heterologous expression systems indicated that Elovl5 can effectively elongate both C18 and C20 PUFA, whereas Elovl4 is mainly involved in the elongation of C20–22 LC-PUFA producing polyenes up to 36 carbons1,13. However, it was also revealed that Elovl4 proteins are able to utilize all assayed C18–22 PUFA substrates13,14,15. In this study, the functional characteristics of ToElovl4a via heterologous expression in S. cerevisiae showed that the T. ovatus putative elongase is Elovl4a, which can only elongate C18 (18:3n-6) substrates to C20 (20:3n-6) PUFA. In agreement with the functional data obtained for some marine fish, such as the 7.6% low activity in Scatophagus argus13, the 4.6% in Acanthopagrus schlegelii14, and the 6.1% in Larimichthys crocea15, ToElovl4 also showed low activity (1.05%) towards PUFA substrates, which confirmed its role in the biosynthesis of VLC-PUFA. However, until now, unlike the present study, Elovl4a was also found to effectively convert C18-C22 PUFA to longer polyenoic products up to C36 in other carnivorous fish1,11,12,13,14,15, suggesting that marine fish Elovl4 exhibited high elongation efficiency towards C18-C22 PUFA substrates, except ToElovl4a. It is inferred that ToElovl4a solely elongates omega-6 C18 fatty acids, and this has been hypothesized as an adaptive strategy to supplement for Elovl5 in T. ovatus27. For T. ovatus, yeast heterologous expression systems showed that Elovl5 can effectively transform C18-C20 PUFA and ToFads6 that possess Δ4/Δ5/Δ8 Fad desaturation activity27,28. In addition to the present study, thus far, the complete classical pathways of LC-PUFA biosynthesis have not been elucidated for T. ovatus29.
In the present study, the highest ToElovl4a mRNA expression was detected in the brain, showing that essential fatty acid metabolism occurs in the brain30. However, relatively moderate ToElovl4a mRNA expression levels were detected in the stomach, intestine and gonad. Interestingly, these are the first tissues exposed to dietary lipids, and they are the main lipid metabolism tissues in the body30. Moreover, the liver is the main site for LC-PUFA synthesis31. These studies indicate that lower levels of hepatic Elovl4a transcripts in carnivorous marine fish, like T. ovatus, may correlate with their limited LC-PUFA biosynthetic abilities3.
Previous studies have indicated that Fad enzymatic activity and gene expression vary with dietary LNA/LA (18:3n-3/18:2n-6) ratio13,32. Upregulation of Δ6 Fads2 gene expression was detected in Siganus fuscescens, Maccullochella peelii, Oncorhynchus mykiss and Scatophagus argus that were fed high dietary ratios of LNA/LA32,33,34,35. Unlike desaturases, there is a lack of data on the influence of dietary LNA/LA ratio on elongase expression. Xie et al.13 showed that the expression of Elovl4 and Elovl5 is significantly affected by dietary fatty acid composition, and they showed the highest expression of mRNA in the liver and eye of fish fed a diet a LNA/LA ratio of 1.7:1 in Scatophagus argus. Unfortunately, in the present nutritional experiment, no pattern was found between the expression of ToElovl4a and fatty acid composition.
In general, mRNA levels of some genes in eukaryotic cells are dependent on transcription factors and RNA polymerases binding to specific sequences in gene promoters36. Consequently, the integrity and activity of a promoter can affect the gene expression. Moreover, PPARs are ligand-activated transcription factors that are necessary for regulating gene expression in the PUFA biosynthesis pathway18. Dual luciferase reporter assays were conducted to clarify regulatory mechanisms whereby PPARαb is believed to modulate Elovl4a expression. Analysis of the truncated mutants indicated that ToElovl4a reporter activity was induced by the overexpression of PPARαb. The core binding region in the ToElovl4a promoter is −148 bp to +258 bp (Fig. 6A). This was the first evidence showing that the transcription of Elovl4a may be upregulated by PPARαb. In a previous study, PPARαb interacted with the binding site of the ToElovl5 and ToFads6 promoter region to positively regulate ToElovl5 and ToFads6 transcription, respectively27,28. Obviously, PPARαb plays a key regulatory role in the LC-PUFA biosynthesis in T. ovatus. Furthermore, the deletion of the PPARαb M2 binding site (+209 bp to +223 bp) results in significantly reduced promoter activity (Fig. 7). To further confirm whether PPARαb is a transcription factor implicated in ToElovl4a function, the effects of PPARαb knockdown on ToElovl4a protein expression were investigated by western blotting in TOCF cells. These data showed that PPARαb upregulated ToElovl4a protein levels.
In summary, the functional studies presented here show that ToElovl4a may effectively extend 18:3n-6 substrates. Moreover, the proposed synthesis pathway of LC-PUFA was for T. ovatus27,28. Furthermore, we demonstrated clear associations between PPARαb and the ToElovl4a promoter and the positive regulatory functions of PPARαb in ToElovl4a transcription. These results provide new insights into the regulation and function of Elovl4a in fish and further reveal the complexity of the associated regulatory mechanisms.
Materials and Methods
Ethics statement
All experiments in this study were approved by the Animal Care and Use Committee of South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences (No. SCSFRI96-253) and were performed according to the regulations and guidelines established by this committee. To minimize suffering of the fish, all surgeries were implemented with 0.01% 2-phenoxyethanol (Sigma-Aldrich) anaesthesia.
Diets, fish, feeding trial and sampling
Eight isonitrogenous and iso-lipidic diets were formulated with 45% crude protein and 12% crude lipid with different lipid sources (Supplementary Table 1). Diet 1 contained fish oil (FO) as the control, and diets 2–8 contained different proportions of fish oil, krill oil, soybean oil and corn oil. The dietary formulations, proximate and fatty acid compositions are shown in Supplementary Table 1.
T. ovatus juvenile fish (body weight: 82.9 ± 2.4 g) were collected from Linshui Marine Fish Farm in Hainan Province, China. The fish were raised on commercial feed (Hengxin, crude protein >37%, crude fat >7%) according to standard feeding schemes 2 weeks before the feeding trial and maintained in fresh seawater at 29 ± 1 °C, a salinity of 35‰, and with dissolved oxygen >6 mg/L in a recirculating aquaculture system. The feeding experiment was conducted in 32 cages (1 m × 1 m × 1.5 m) in a corresponding environment with each cage including 20 fish that were randomly allocated. The fish were anaesthetized using MS222 (0.1 g/L; Sigma, Alcobendas, Spain); then, the liver and brain were sampled, flash frozen in liquid nitrogen, and stored at −80 °C until further use.
To determine the tissue expression profile of ToElovl4a, healthy fish tissue (n = 6) containing small intestine, liver, white muscle, brain, spleen, fin, gill, head kidney, stomach, blood, males and female gonads were sampled, flash frozen in liquid nitrogen, and stored at −80 °C until further use.
Gene cloning and bioinformatics of ToElovl4a
Total RNA (1 μg) was extracted from T. ovatus brain by TRIzol Reagent (Takara, Japan). The quality and quantity (concentration) of isolated RNA were determined using a NANODROP 2000 spectrophotometer (Thermo Scientific). Subsequently, cDNA was synthesized using the PrimeScriptTM RT reagent kit (Takara, Kyoto, Japan), according to the manufacturer’s instructions. A putative ToElovl4a number was derived from the annotation file of T. ovatus. Subsequently, a putative ToElovl4a sequence was obtained based on CDS data of T. ovatus. (https://doi.org/10.6084/m9.figshare.7570727.v1 (2019)). To determine the veracity of the putative Elovl4a sequence, gene-specific primers were designed (Supplementary Table 2). The PCR protocol used has been previously described37. The amplified products were purified by a DNA purification kit (Tiangen, China), ligated into the pEASY-T1 vector (TransGen Biotech, China), and sequenced (Invitrogen, Guagnzhou, China). Validated plasmids were transformed into competent Trans1-T1 cells (TransGen Biotech, China). A Blast search on the putative Elovl4a ORF sequence further confirmed the accuracy and validity.
The deduced amino acid sequence of the cloned ToElovl4a open reading frame (ORF) was aligned with other Elovl4 orthologue ORFs (Supplementary Table 3). Multiple sequence alignments were conducted using ClustalX version 2.0 with default parameters38. Phylogenetic analyses for all Elovl4 amino acid sequences were performed using maximum likelihood (ML) methods (LG + G model, bootstrap 1000) with MEGA 6.039. All available Elovl4 genome sequences were downloaded from public databases of Ensembl (http://asia.ensembl.org/) and Genome Browser (http://genome.ucsc.edu/cgi-bin/hgBlat). The phylogenetic tree was embellished using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) and Adobe PhotoShop CS6 (Adobe, San Jose, CA).
Functional characterization of the ToElovl4a elongases
PCR products corresponding to the ToElovl4a ORF were amplified from the T. ovatus brain cDNA using high fidelity Pfu DNA polymerase (Promega, USA) with primers incorporating Kpn I and Xho I enzyme restriction sites (Supplementary Table 2). The PCR products were digested with the above restriction endonucleases (Takara, Japan) and ligated into a similarly digested pYES2 yeast expression vector (Invitrogen, USA). The recombinant plasmid (pYES2-Elovl4a) was transformed into Saccharomyces cerevisiae competent cells (S.c. EasyComp Transformation Kit, Invitrogen). The selection of recombinant yeast colonies and subsequent yeast culture was prepared according to previously published methods40,41. Fatty acids, including 18:2n-6 (Anandamide), 18:3n-3 (α-linolenic acid), 18:3n-6 (γ-linolenic acid), 18:4n-3 (Stearidonic acid), 20:4n-6 (Arachidonic acid, ARA), 20:5n-3 (Eicosapentaenoic acid, EPA), 22:5n-3 (Docosapentaenoic acid, DPA), 22:4n-6 (Adrenic Acid), and 22:6n-3 (Docosahexaneoic acid, DHA), were used as substrates for detecting the elongase activity of ToElovl4a. The final concentrations of the FA substrates varied according to their fatty acyl chain lengths and were 0.5 mM (C18) and 0.75 mM (C20). Yeast cultures were incubated for two days at 30 °C, harvested, and washed twice, as previously described8. As a control, the yeast were transformed with pYES2 only (no insert) and similarly treated. Fatty acid methyl esters (FAMEs) were prepared, extracted, purified and analysed via thin-layer chromatography (TLC) and gas chromatography (GC2010-plus; Shimadzu, Japan) as previously described42. The proportion of substrate fatty acids converted to elongated FA products was calculated as follows: [product area/(product area + substrate area) × 1008.
Real-time quantitative PCR analysis
Specific primers for real-time quantitative PCR (qRT-PCR) were designed by Primer Premier 5.0 (Premier Biosoft, USA) based on cloned nucleotide sequences (Supplementary Table 2). Translation elongation factor 1-alpha (EF1α) was verified and used as a reference gene43. The qRT-PCR amplifications were performed in a quantitative thermal cycler (Mastercycler EP Realplex, Eppendorf, Germany). The programme parameters were 95 °C for 2 min, followed by 40 cycles of 95 °C for 10 s, 56 °C for 10 s, and 72 °C for 20 s. Amplification efficiencies of the target and reference genes were observed from the slope of the log-linear portion of the calibration curve with PCR efficiency = 10(−1/Slope)−1. Expression levels of target genes were calculated using the 2−ΔΔCt method44.
Preparation of the Elovl4a polyclonal antibody and western blotting analysis
To prepare the polyclonal anti-Elovl4a antibody, a specific domain (Elovl4a aa92–106) of Elovl4a was compounded from Genecreate (Wuhan, China). The resulting PCR product was inserted into the pET-B2M vector using Nde I/Xho I sites. To express recombinant T. ovatus Elovl4a protein (rToElovl4a), the recombinant plasmid was transformed into Escherichia coli BL21 (DE3) (Novagen, Germany). The rToElovl4a was purified as previously described45. To generate a polyclonal antibody, purified rToElovl4a protein was injected into white New Zealand rabbits using standard methods46. Once generated, the polyclonal antibody was pre-adsorbed using E. coli lysate supernatants to eliminate inhomogeneous antibodies and was depurated on a HiTrapTM Protein A HP column on a AKTAprime™ Plus system (GE Healthcare, USA).
To confirm specificity of the rabbit anti-Elovl4a antibody, human embryonic kidney (HEK293T) cells were transfected with pcDNA3.1 and pcDNA3.1-Elovl4a for 48 h. After this period, cells were harvested by centrifugation at 160 g for 10 min at 4 °C. The total protein was extracted using ProteoPrep® Total Extraction Sample Kit (Sigma-Aldrich). Then, the total protein were electrophoresed on 12% SDS-PAGE and electrophoretically transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, USA) using the PierceG2 Fast Blotter (25 V for 10 min, Pierce, Rockford, IL, USA). Western blotting analyses was executed according to a previously described protocol47.
To observe the endogenous Elovl4a expression, T. ovatus caudal fin (TOCF) cells were cultured in six-well plates at a density of 2.5 × 106 cells/well. After the TOCF cells were transfected with PPARαb siRNA, cells were harvested and lysed as described above. Then, the total protein was incubated with/without calf intestinal alkaline phosphatase (CIAP) (20 U) at 37 °C for 30 min, separated by 12% SDS-PAGE and transferred to PVDF membranes using the PierceG2 Fast Blotter (25 V for 10 min; Pierce, Rockford, IL, USA). Primary antibodies [anti-Elovl4a, murine anti-Flag (Sigma-Aldrich, St. Louis, MO, USA) and the loading control, the anti-glyceraldehyde 3-phosphate dehydrogenase antibody (GAPDH; Sigma-Aldrich), 1:1000] were incubated with the PVDF membrane in 1% (w/v) non-fat milk in Tris-buffered saline and Tween 20 (TBST) buffer (0.1% Tween 20) for 3 h. Horse radish peroxidase-(HRP)-conjugated goat anti-rabbit antibody (1:3000) was used as a secondary antibody (Sigma-Aldrich). The results were observed using an electrochemiluminescence (ECL) system.
Cloning of the Elovl4a promoter and construction of deletion mutants
Genomic DNA was extracted from the muscle tissue of T. ovatus as described previously48 and used as a template for candidate promoter cloning. The sequence upstream of the Elovl4a gene was obtained from genomic sequencing data of T. ovatus. To identify the role of PPARαb in the transcriptional regulation of ToElovl4a, five different promoter regions of ToElovl4a were amplified by specific primers (Supplementary Table 2) and subcloned into the Kpn I and Xho I restriction sites of the pGL3-basic luciferase reporter plasmid (Promega, USA). Five recombinant plasmids, denoted pGL3-basic-Elovl4a-1 (−148 to +56), pGL3-basic-Elovl4a-2 (−500 to +56), pGL3-basic-Elovl4a-3 (−1001 to +56), pGL3-basic-Elovl4a-4 (−148 to +155) and pGL3-basic-Elovl4a-5 (−148 to +258), were constructed (Fig. 6A). The truncated mutants were amplified using PrimeSTAR Master Mix (Takara, Japan). The programme parameters were 95 °C for 4 min, followed by 30 cycles of 95 °C for 40 s, 56 °C for 40 s, and 72 °C for 1 min. A general DNA purification kit (Tiangen, China) was used to purify the PCR products. All purified PCR products and the pGL3-basic (Promega, USA) vector were digested with Kpn I and Xho I and concatenated by T4 DNA ligase (Takara, Japan) overnight at 16 °C. Recombinant plasmids were extracted using the EndoFree Plasmid Giga Kit (Tiangen, China), and constructs were confirmed by sequencing as described above.
Construction of truncated mutants for the identification of predicted transcription factor (TF) binding sites in the Elovl4a promoter
To determine the potential function of the PPARαb binding sites on the core Elovl4a promoter, four truncated mutations of recombinant plasmids were established. The transcription factor binding site prediction (TFBS)-JASPAR database (http://jaspar.genereg.net/), TRANSFAC®, and MatInspector® were used to search for potential binding sites in the Elovl4a promoter sequence with PPARαb. According to the manufacturer’s protocol, truncated mutants were designed and produced with a Muta-directTM site-directed mutagenesis kit (SBS Genetech, Shanghai, China) from the deletion mutant pGL3-basic-Elovl4a-5, which was wild-type. The prediction of four binding sites (M1, M2, M3, and M4) were directly deleted, and the corresponding TF binding site sequences are shown in Fig. 7A. Furthermore, to acquire the TF binding site mutations, we used the method of PCR augmentation referred to a previous study49. The influence of TF binding site mutations on the promoter activity of ToElovl4a were determined by a dual luciferase assay as described below.
Cell culturing, transfection and luciferase assay
HEK293T cells were cultured in DMEM (Gibco, USA) and supplemented with 10% foetal bovine serum (FBS) (Invitrogen, USA), 100 U mL−1 penicillin, and 100 μg mL−1 streptomycin at 37 °C in a humidified incubator under 5% CO2. Transfection and dual luciferase reporter assays were described by Li et al.15. Relative luciferase activities (firefly and renilla luciferase activities) were measured by a VICTORTM X2 Multi-label Plate Reader (PerkinElmer, Inc., Waltham, MA, USA).
TOCF cells were cultured in L15 media (Gibco, USA) supplemented with 10% FBS, 100 U mL−1 penicillin, and 100 μg mL−1 streptomycin at 28 °C. Before DNA transfection, cells were seeded in 24-well plates until they were 90–100% confluent. Then, small interfering RNA (siRNA) or plasmids were transfected using Lipofectamine RNAiMAX or Lipofectamine 2000 transfection reagent (Invitrogen, USA) according to the manufacturer’s instructions.
Expression analysis of ToElovl4a with ToPPARαb
The ORF of T. ovatus PPARαb (ToPPARαb) (GenBank accession number: MH321826) was amplified with primers incorporating restriction sites for Nhe I and Hind III at the 5′ and 3′ ends, respectively (Supplementary Table 2). The DNA fragment was digested with the same restriction endonucleases (Nhe I and Hind III; Takara, Japan) and ligated into a correspondingly restricted pCDNA3.1-Flag vector (Invitrogen, USA). Transcription factors ToPPARαb and pGL3-basic-Elovl4a-5 of the promoter segment were chosen to determine the regulatory relationship between ToPPARαb and ToElovl4a. Detection of promoter activities were at specific time points (0 h, 3 h, 6 h, 12 h, 24 h, 48 h and 72 h). The siRNA for PPARαb (PPARαb-si) and the negative control (si-NC) were purchased from Genecreate (Wuhan, China). The PPARαb siRNA sequence is listed in Supplementary Table 2. After transfection with TOCF cells, the total protein was isolated at specific time points (0 h, 6 h, 12 h, and 24 h) as described above.
Statistical analysis
SPSS 19.0 software (IBM, USA) was used to conduct the statistical analyses. The data were analysed by the Duncan test using one-way ANOVA. All data from the relative expression represented at least three replications along with means ± standard error of the mean (SE). Differences were considered significant at the p < 0.05 level.
References
Castro, C. L. F., Tocher, D. R. & Monroig, Ó. Long-chain polyunsaturated fatty acid biosynthesis in chordates: Insights into the evolution of Fads and Elovl gene repertoire. Prog. Lipid Res. 62, 25–40 (2016).
Park, W. J., Kothapalli, K. S., Lawrence, P., Tyburczy, C. & Brenna, J. T. An alternate pathway to long-chain polyunsaturates: the FADS2 gene product Delta8-desaturates 20:2n-6 and 20:3n-3. J. Lipid Res. 50, 1195–1202 (2009).
Tocher, D. R. Fatty acid requirements in ontogeny of marine and freshwater fish. Aquac Res. 41, 717–732 (2010).
Monroig, Ó., Li, Y. Y. & Tocher, D. R. Delta-8 desaturation activity varies among fatty acyl desaturases of teleost fish: high activity in delta-6 desaturases of marine species. Comp. Biochem. Physiol. B 159, 206–213 (2011).
Vagner, M. & Santigosa, E. Characterization and modulation of gene expression and enzymatic activity of delta-6 desaturase in teleosts: A review. Aquaculture 315, 131–143 (2011).
Miyazaki, M. & Ntambi, J. M. Fatty acid desaturation and chain elongation in mammals. In: Vance, D. E., Vance, J. E. (Eds), Biochemistry of Lipids, Lipoproteins and Membranes, fifth ed. Elsevier, Amsterdam, pp. 191–212 (2008).
Jakobsson, A., Westerberg, R. & Jacobsson, A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Progress in Lipid Research 45, 237–249 (2006).
Li, Y. Y., Monroig, Ó., Zhang, L., Wang, S. Q. & Zheng, X. Vertebrate fatty acyl desaturase with D4 activity. Proc. Natl. Acad. Sci. USA 107, 16840–16845 (2010).
Kiron, V., Satoh, S., Takeuchi, T. & Yoshizaki, G. Cloning and over-expression of a masu salmon (Oncorhynchus masou) fatty acid elongase-like gene in zebrafish. Aquaculture 282, 13–18 (2008).
Kabeya, N. et al. Modification of the n-3 HUFA biosynthetic pathway by transgenesis in a marine teleost, nibe croaker. J. Biotechnol. 172, 46–54 (2014).
Carmona-Antoñanzas, G., Monroig, Ó., Dick, J. R., Davie, A. & Tocher, D. R. Biosynthesis of very long-chain fatty acids (CN24) in Atlantic salmon: Cloning, functional characterisation, and tissue distribution of an Elovl4 elongase. Comp. Biochem. Physiol. B 159, 122–129 (2011).
Monroig, Ó. et al. Elongation of long-chain fatty acids in rabbitfish Siganus canaliculatus: Cloning, functional characterisation and tissue distribution of Elovl5- and Elovl4-like elongases. Aquaculture 350–353, 63–70 (2012).
Xie, D. Z. et al. Long-chain polyunsaturated fatty acid biosynthesis in the euryhaline herbivorous teleost Scatophagus argus: Functional characterization, tissue expression and nutritional regulation of two fatty acyl elongases. Comp. Biochem. Physiol. B 198, 37–45 (2016).
Jin, M., Monroig, Ó., Navarrod, J. C., Tocher, D. R. & Zhou, Q. C. Molecular and functional characterisation of two elovl4 elongases involved in the biosynthesis of very long-chain (>C24) polyunsaturated fatty acids in black seabream Acanthopagrus schlegelii. Comp. Biochem. Physiol. B 212, 41–50 (2017).
Li, S. L. et al. Functional characterization and differential nutritional regulation of putative Elovl5 and Elovl4 elongases in large yellow croaker (Larimichthys crocea). Sci. Rep. 7, 2303 (2017).
Monroig, Ó. et al. Expression and role of Elovl4 elongases in biosynthesis of very long-chain fatty acids during zebrafish Danio rerio early embryonic development. BBA-Mol. Cell Biol. L. 1801, 1145–1154 (2010).
Kota, B. P., Huang, T. H. & Roufogalis, B. D. An overview on biological mechanisms of PPARs. Pharmacol. Res. 51, 85–94 (2005).
Sampath, H. & Ntambi, J. M. Polyunsaturated fatty acid regulation of genes of lipid metabolism. Annu. Rev. Nutr. 25, 317–340 (2005).
Lefebvre, P., Chinetti, G., Fruchart, J. C. & Staels, B. Sorting out the roles of PPARa in energy metabolism and vascular homeostasis. J. Clin. Invest. 116(3), 571–580 (2006).
Chakravarthy, M. V. et al. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell 138(3), 476–488 (2009).
Navidshad, B. & Royan, M. Peroxisome Proliferator-Activated Receptor Alpha (PPAR alpha), a Key Regulator of Lipid Metabolism in Avians. Critical Reviews in Eukaryotic Gene Expression 26, 303–308 (2016).
Dong, X. J. et al. Regulation of FADS2 transcription by SREBP-1 and PPAR-α influences LC-PUFA biosynthesis in fish. Sci. Rep. 7, 40024 (2017).
Sun, L. Y. et al. Genetic polymorphism of breeding populations of golden pompano (Trachinotus ovatus). South China Fish. Sci. 10, 67–71 (2014).
Zhen, P. L., Ma, Z. H., Guo, H. Y., Jiang, S. G. & Zhang, D. C. Ontogenetic development of caudal skeletons in Trachinotus ovatus larvae. South China Fish. Sci. 10, 45–50 (2014).
Sun, X. X. et al. Feed type regulates the fatty acid profiles of golden pompano Trachinotus ovatus (Linnaeus 1758). J Appl. Anim. Res. 46(1), 60–63 (2018).
Cook, H. W. & McMaster, R. C. R. Fatty acid desaturationand chain elongation in eukaryotes. In (Eds) Vance, D. E., Vance, J. E., Biochemistry of Lipids, Lipoproteins and Membranes, Elsevier, Amsterdam, pp. 181–204 (2004).
Zhu, K. C. et al. The Transcriptional factor PPARαb positively regulates Elovl5 elongase in golden pompano Trachinotus ovatus (Linnaeus 1758). Front. Physiol. 9, 1340 (2018).
Zhu, K. C. et al. Identification of fatty acid desaturase 6 in golden pompano Trachinotus ovatus (Linnaeus 1758) and its regulation by the PPARαb transcription factor. Int. J. Mol. Sci. 20, 23 (2019).
Oboh, A. et al. Two alternative pathways for docosahexaenoic acid (DHA, 22:6n-3) biosynthesis are widespread among teleost fish. Sci. Rep. 7, 3889 (2017).
Tanomman, S., Ketudat-Cairns, M., Jangprai, A. & Boonanuntanasarn, S. Characterization of fatty acid delta-6 desaturase gene in Nile tilapia and heterogenous expression in Saccharomyces cerevisiae. Comp. Biochem. Physiol. B 166, 148–156 (2013).
Bell, J. G. et al. Replacement of fish oil with rapeseed oil in diets of Atlantic salmon (Salmo salar) affects tissue lipid compositions and hepatocyte fatty acid metabolism. J Nutr. 131, 1535–1543 (2001).
Xie, D. Z. et al. Cloning, Functional Characterization and Nutritional Regulation of Delta 6 Fatty Acyl Desaturase in the Herbivorous Euryhaline Teleost Scatophagus Argus. PLOS ONE 9(3), e90200 (2014).
Senadheera, S. D., Turchini, G. M., Thanuthong, T. & Francis, D. S. Effects of dietary α-linolenic acid (18:3n-3)/linoleic acid (18: 2n-6) ratio on fatty acid metabolism in Murray cod (Maccullochella peelii peelii). Journal of agricultural and food chemistry 59(3), 1020–1030 (2011).
Thanuthong, T., Francis, D. S., Senadheera, S. P. S. D., Jones, P. L. & Turchini, G. M. LC-PUFA biosynthesis in rainbow trout is substrate limited: use of the whole body fatty acid balance method and different 18:3n-3/18:2n-6 ratios. Lipids 46, 1111–1127 (2011).
Li, Y. Y. et al. The effects of dietary fatty acids on liver fatty acid composition and Δ6–desaturase expression differ with ambient salinities in Siganus canaliculatus. Comp. Biochem. Physiol. B 151, 183–190 (2008).
O’Malley, B. W., Towle, H. C. & Schwartz, R. J. Regulation of gene expression in eucaryotes. Annu. Rev. Genet. 11, 239–275 (1977).
Zhu, K. C. et al. Molecular characterization and expression patterns of myogenin in compensatory growth of Megalobrama amblycephala. Comp. Biochem. Physiol. B 170, 10–17 (2014).
Larkin, M. A., Blackshields, G., Brown, N. P. & Higgins, D. G. ClustalW and ClustalX version 2. Bioinformatics 23, 2947–2948 (2007).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. 3, 2725–2729 (2013).
Agaba, M. K. et al. Cloning and functional characterisation of polyunsaturated fatty acid elongases from marine and freshwater teleost fish. Comp. Biochem. Physiol. B 142, 342–352 (2005).
Li, M. et al. Characterization of two Δ5 fatty acyl desaturases in abalone (Haliotis discus hannai Ino). Aquaculture 416, 48–56 (2013).
Hastings, N. et al. A vertebrate fatty acid desaturase with Δ5 and Δ6 activities. Proc. Natl. Acad. Sci. USA 98, 14304–14309 (2001).
Zhu, K. C. et al. Genomic structure, expression pattern and polymorphisms of GILT in golden pompano Trachinotus ovatus (Linnaeus 1758). Gene 665, 18–25 (2018).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Chen, N. et al. Study on the immune response to recombinant Hsp70 protein from Megalobrama amblycephala. Immunobiology 219, 850–858 (2014).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular cloning. (Vol. 2). New York: Cold spring harbor laboratory press (1989).
Ding, Y., Ao, J. Q., Huang, X. H. & Chen, X. H. Identification of Two Subgroups of Type I IFNs in Perciforme Fish Large Yellow Croaker Larimichthys crocea Provides Novel Insights into Function and Regulation of Fish Type I IFNs. Front. Immunol. 7, 343 (2016).
Sun, L. Y., Zhang, D. C., Jiang, S. G., Guo, H. Y. & Zhu, C. Y. Isolation and characterization of 21 polymorphic microstatellites in golden pompano Trachinotus ovatus. Conserv. genet. resour. 5, 1107–1109 (2013).
Dong, Y. et al. Hepatocyte Nuclear factor 4α (HNF4α) is a transcription factor of vertebrate fatty acyl desaturase gene as identified in marine teleost Siganus canaliculatus. PLoS one 11, e0160361 (2016).
Acknowledgements
The study was supported by the China Agriculture Research System (CARS-47), Central Public-interest Scientific Institution Basal Research Fund, South China Sea Fisheries Research Institute, CAFS (NO. 2019TS10), the Fishing Port Construction and Fishery Development Special Funds for Guangdong Province (Sci-tech Popularization, 2017A0008), and the National Infrastructure of Fishery Germplasm Resources Project (2018DKA30407).
Author information
Authors and Affiliations
Contributions
K.C.Z., S.G.J. and D.C.Z. designed the research and wrote the paper. S.L and K.C.Z. performed the research. H.Y.G. and N.Z. analyzed the data. B.S.L. and L.G. contributed reagents/materials/analysis tools.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhu, KC., Song, L., Guo, HY. et al. Elovl4a participates in LC-PUFA biosynthesis and is regulated by PPARαβ in golden pompano Trachinotus ovatus (Linnaeus 1758). Sci Rep 9, 4684 (2019). https://doi.org/10.1038/s41598-019-41288-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-41288-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.