Abstract
NKX family members are involved in a variety of developmental processes such as cell fate determination in the central nervous system, gastrointestinal tract, and pancreas. However, whether NKX6.3 contributes to gastric carcinogenesis remains unclear. The objective of this study was to examine roles of NKX6.3 depletion in mutagenesis and gastric carcinogenesis, focusing on its effects on genetic alterations and expression of genes. Our results revealed that NKX6.3 depletion induced multiple genetic mutations in coding regions, including high frequency of point mutations such as cytosine-to-thymine and guanine-to-adenine transitions caused by aberrant expression of AICDA/APOBEC family in human gastric epithelial cells. Interestingly, NKX6.3 downregulated AICDA/APOBEC family, NFκB, and CBFβ genes by acting as a transcription factor while inhibiting deaminase activity in gastric epithelial cells. Functional relevance of NKX6.3 was validated in xenograft mice injected with NKX6.3 depleting cells. NKX6.3 depletion resulted in tumor formation and mutations of tumor-associated genes, including p53 and E-cadherin. Moreover, expression levels of NKX6.3 and its target genes were analyzed in tumors derived from mice implanted with NKX6.3 depleting cells and tissue samples of gastric cancer patients. Our results indicate that NKX6.3 depletion in gastric epithelial cells activates AICDA/APOBEC family, leading to accumulation of genetic mutations and eventually driving the development of gastric cancers.
Similar content being viewed by others
Introduction
Although genetic and epigenetic alterations in oncogenes and tumor suppressor genes are known to be responsible for the development and progression of gastric cancer1, this cancer remains the seventh most common cancer and the third leading cause of cancer-related death worldwide2. Recurrent mutations in gene coding regions and detailed molecular characterization of gastric cancers have been recently identified3,4,5. The majority of gastric cancers are associated with Helicobacter pylori infection. H. pylori seems to cause increased point mutation rates in mouse models6 and elevated expression of activation-induced cytidine deaminase (AICDA)7 belonging to apolipoprotein B mRNA editing catalytic polypeptide (APOBEC) family of proteins8.
NKX family members are involved in a variety of developmental processes such as cell fate determination in the central nervous system, gastrointestinal tract, and pancreas9. NKX6.3 is expressed in the epithelium of the most distal region of the stomach9,10. Previously, we have reported that NKX6.3 functions as a master regulator of gastric differentiation by modulating Sox2 and Cdx2 expression11. It also acts as a gastric tumor suppressor by inhibiting cell proliferation and inducing apoptosis12. In addition, loss of expression and allelic deletion of NKX6.3 have been frequently detected in gastric cancers12. However, whether NKX6.3 contributes to gastric carcinogenesis remains unclear. Therefore, the objective of this study was to examine roles of NKX6.3 depletion in mutagenesis and gastric carcinogenesis, focusing on its effects on genetic alterations and expression of genes.
Here, we found that NKX6.3 depletion enhances genetic mutations by regulating expression of the genes involved in mutagenesis, including the AICDA/APOBEC family, in gastric epithelial cells. This study provides new insight into molecular gastric carcinogenesis, implicating NKX6.3 depletion as a critical event.
Results
Genetic alterations in gastric cancers from TCGA and COSMIC database
To determine whether defects in NKX6.3 gene might lead to genetic alterations in gastric cancers, TCGA and COSMIC data were analyzed. In 395 gastric cancers, cases without NKX6.3 expression showed significantly higher mutation rates than those with NKX6.3 expression (Fig. 1A). Notably, most mutations in hypermutated genes were detected in gastric cancers with reduced or loss of NKX6.3 expression (Fig. 1A). Because TCGA data did not provide NKX6.3 expression level in 363 corresponding non-neoplastic gastric mucosae, we analyzed NKX6.3 expression levels and mutations rates in 32 gastric cancer samples out of 395 TCGA tumors. Compared with HFE-145 immortalized gastric epithelial cells (fold change, mean ± SD: 1.00 ± 0.02) and non-neoplastic gastric mucosae (fold change, 0.91 ± 0.18), nine cancer cell lines (fold change, 0.23 ± 0.04, P < 0.01) and gastric cancer tissues (fold change, 0.14 ± 0.05, P < 0.01) showed reduced NKX6.3 expression (Supplementary Fig. S1A). In addition, NKX6.3 expression was inversely correlated with mutation rates in 32 TCGA gastric cancer tissues (Supplementary Fig. S1B, Spearman rank correlation test; P < 0.0001). As shown in Supplementary Fig. S1C, a high frequency of G to A transition mutations were observed while coding region mutations in genes including TTN, MUC16, and FAT4 were detected mainly in cases with reduced or loss of NKX6.3 expression (Supplementary Fig. S1D). COSMIC data from 22 gastric cancer cell lines demonstrated frequent coding region point mutations, especially in genes such as TTN, MUC16, and TP53 (Supplementary Fig. S1E).
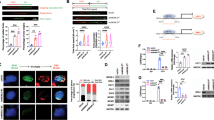
Depletion of NKX6.3 enhances mutations in coding regions. (A) Relationship between NKX6.3 expression and mutations in 395 gastric cancers from TCGA dataset. (B) Number of variants in gene coding regions in HFE-145shNKX6.3#1 and HFE-145shNKX6.3#2 cells compared to HFE-145shCtrl cells. Error bars, SD. P values were derived from t tests. ****P < 0.0001. (C) Types of mutations observed in HFE-145shNKX6.3#1 and HFE-145shNKX6.3#2 cells. Error bars, SD. P values were derived from t tests. **P < 0.01; ***P < 0.001. (D) Left, Venn diagram of NKX6.3 depletion-induced mutant genes with missense/nonsense mutations, mainly G or C to A or T base pair transitions. Error bars, SD. P values were derived from t tests. ***P < 0.001; ****P < 0.0001. Right, NKX6.3 depletion-induced mutant genes with Frame shift/Ins/Del mutations in coding regions. Error bars, SD. P values were derived from t tests. n.s, not significant (P > 0.05). (E) Mutant gene signatures were involved in 15 biological processes in FunrichDB. (F) Mutation sites of PIK3CA, RhoA, EP300, TP53, MDM2, CASP8, PAK2, CDH1, BCL9, and CDX1 induced by NKX6.3 depletion.
Depletion of NKX6.3 leads to genetic mutations in coding regions
Next, we investigated whether depletion of NKX6.3 might induce genetic alterations in gastric epithelial cells. We performed whole genome sequencing for HFE-145shCtrl, HFE-145shNKX6.3#1, and HFE-145shNKX6.3#2 cells with a mean depth of 42.9x, 44.8x, and 39.9x, respectively (98.4%, 98.4%, and 98.3% of the reference human genome were covered by ≥10x, respectively, Supplementary Table S1A,B). Compared to the reference sequence of human genome (hg19), HFE-145shCtrl cells showed 3,544,930 genetic variants, including polymorphism and mutations, possibly caused by serial culture and immortalization. Of these, 386 variants resulted in amino acid changes (data not shown). In HFE-145shNKX6.3#1 and HFE-145shNKX6.3#2 cells, 3,501,785 and 3,492,917 genome-wide genetic variants were observed, respectively, including 386 variants detected in HFE-145shCtrl cells (Supplementary Table S1C). Additionally, 688,704 different genetic variants were found between HFE-145shCtrl and HFE-145shNKX6.3#1 cells and 686,632 different genetic variants were found between HFE-145shCtrl and HFE-145shNKX6.3#2 cells. Of these genetic variants, 674,571 were overlapped between HFE-145shNKX6.3#1 and HFE-145shNKX6.3#2 cells. (data not shown).
In HFE-145shNKX6.3#1 and HFE-145shNKX6.3#2 cells, 1,635.5 ± 39.5 genetic mutations were newly identified in the coding regions on whole chromosomes, including non-synonymous and synonymous, codon insertion/deletion, frame shift, and start/stop codon mutations (Supplementary Fig. S2A) compared to HFE-145shCtrl cells (Fig. 1B and Supplementary Table S2). Among these mutations, a high frequency (93%) of point mutations such as missense, silent, and nonsense mutations rather than frameshift, deletion, and insertion were found. The percentage of missense mutations in point mutations was higher than that of silent and nonsense mutations (Fig. 1C). In addition, substitutions from C to T and from G to A showed high percentages (Supplementary Fig. S2B). In detail, missense and nonsense mutations were found in coding regions of 750 genes in HFE-145shNKX6.3#1 cells and 767 genes in HFE-145shNKX6.3#2 cells. Of them, 633 genes were overlapped. Mutations in these overlapped genes were found to be mainly C to T and G to A transitions (54.7%). In addition, frame shift, insertion, and deletion mutations were observed in coding regions of 95 genes in HFE-145shNKX6.3#1 and 104 genes in HFE-145shNKX6.3#2 cells. Of them, 69 genes were overlapped (Fig. 1D). Next, we analyzed the associated biological pathway using FunRich software. Results of analysis showed that NKX6.3 depletion-induced mutant genes including TP53, RhoA, EP300, and PIK3CA were involved in biological pathways such as intracellular junction assembly and/or maintenance, cellular proliferation, and apoptosis (Fig. 1E, Supplementary Fig. S2C and Supplementary Table S3). Mutation sites of these genes are shown in Fig. 1F. These results strongly indicate that depletion of NKX6.3 can lead to genetic mutations in several genes involved in cell growth, cell proliferation, and apoptosis.
NKX6.3 downregulates APOBEC gene family at transcriptional level
To examine whether NKX6.3 might be involved in regulating AICDA/APOBEC gene family expression, we analyzed mRNA expression of AICDA/APOBEC gene family, AICDA, Ung, and Apex1 genes in AGS, MKN1, and HFE-145 cells. NKX6.3 depletion in HFE-145shNKX6.3 dramatically increased expression levels of APOBEC gene family, AICDA, Ung, and Apex1 genes whereas stable NKX6.3 expression in AGSNKX6.3 and MKN1NKX6.3 cells significantly reduced mRNA expression levels of these genes (Fig. 2A and Supplementary Fig. S3A). To determine binding of NKX6.3 to promoter upstream sequences of APOBEC gene family in HFE-145shCtrl, HFE-145shNKX6.3, AGSMock, MKN1Mock, AGSNKX6.3, and MKN1NKX6.3 cells, chromatin immunoprecipitation (ChIP) assay was performed followed by qPCR. Interestingly, there were 3, 7, 4, 3, 6, and 4 putative NKX6.3 binding motifs (TAAT) in promoter regions from −0.5 kb to −3.5 kb relative to the transcription start site of APOBEC3B, 3C, 3G, 3H, Ung, and Apex1 genes, respectively (Supplementary Fig. S3C). NKX6.3 binding activity to these APOBEC family genes in HFE-145shCtrl, AGSNKX6.3, and MKN1NKX6.3 cells was clearly observed (Fig. 2A and Supplementary Fig. S3D). Results of luciferase activity assay showed that mRNA expression levels of these genes were significantly increased in HFE-145shNKX6.3 cells but significantly decreased in AGSNKX6.3 and MKN1NKX6.3 cells (Fig. 2A and Supplementary Fig. S3E). In addition, NKX6.3 depletion markedly induced expression of APOBEC3A, 3B, 3C, 3G, AICDA, and Ung proteins whereas ectopic expression of NKX6.3 reduced their expression levels (Fig. 2B and Supplementary Fig. S3B).
NKX6.3 regulates AICDA/APOBEC gene family expression. (A) Real-time RT-PCR (left), ChIP-qPCR (middle), and luciferase activity (right) analysis of APOBEC family, AICDA, Ung, and Apex1 in NKX6.3 depleted cells (n = 3). Error bars, SD. P values were derived from t tests. *P < 0.05, **P < 0.01. (B) Immunoblot analysis of APOBEC3A, 3B, 3C, 3G, AICDA, and Ung protein expression in NKX6.3 depleted cells. (C) Deaminase activity in HFE-145shNKX6.3 cells by FRET assay (n = 3). Error bars, SD. P values were derived from t tests. ****P < 0.0001. (D) ChIP-qPCR (upper left), luciferase activity (upper right), real-time RT-PCR (lower left), and immunoblot (lower right) analyses of CBFβ and NFκB p65 genes in NKX6.3 depleted HFE-145shNKX6.3 cells (n = 3). Error bars, SD. P values were derived from t tests. **P < 0.01; ***P < 0.001. (E) Protein expression of AICDA/APOBEC gene family in HFE-145shNKX6.3 cells with CBFβ or NFκB p65 knockdown. (F) In 3D-PCR, somatic mutation in TP53 coding region (n = 11 for shCtrl, n = 9 for shNKX6.3, n = 13 for shNKX6.3 with siCBFβ, n = 11 for shNKX6.3 with siNFκB).
To probe the biological significance of NKX6.3 as a transcriptional regulator of APOBEC gene family, effects of NKX6.3 on deaminase activity in HFE-145, AGS, and MKN1 cells were determined using fluorescence resonance energy transfer (FRET)-based assay. Results showed that deaminase activities in HFE-145shNKX6.3, AGSMock, and MKN1Mock cells that expressed AICDA/APOBEC gene family were increased. However, stable NKX6.3 expression in these cell lines resulted in significant decreases in deaminase activity (Fig. 2C and Supplementary Fig. S3F). Thus, we can conclude that NKX6.3 transcriptionally downregulates expression of AICDA/APOBEC gene family, Ung, and Apex1 in gastric epithelial cells.
NKX6.3 regulates CBFβ and NFκB at transcriptional level
Core-binding factor subunit beta (CBFβ) is required for APOBEC3 gene expression13 while NFκB is required for the AICDA gene expression7. Since a high frequency of mutations were found in gastric cancer tissues and cell lines with loss of NKX6.3 expression, we hypothesized that NKX6.3 could modulate the expression of CBFβ and NFκB. As expected, expression levels of CBFβ and NFκB were inversely associated with levels of NKX6.3 expression but positively associated with mutation rates in TCGA datasets (Supplementary Fig. S4A). Importantly, NKX6.3 binding activities to promoter regions of CBFβ and NFκB p65 genes were observed in HFE-145shCtrl, AGSNKX6.3, and MKN1NKX6.3 cells (Fig. 2D and Supplementary Fig. S4B). Both mRNA and protein expression levels of CBFβ and NFκB p65 were significantly increased in HFE-145shNKX6.3 cells but decreased in NKX6.3-expressing cells based on luciferase activity assay, real-time RT-PCR, and immunoblot analysis (Fig. 2D and Supplementary Fig. S4C–F). In addition, knockdown of CBFβ or NFκB p65 significantly decreased mRNA and protein expression of AICDA/APOBEC gene family and deaminase activity in HFE-145shNKX6.3 cells (Fig. 2E and Supplementary Fig. S5A,B). Next, we determined 3D-PCR error rate using the first PCR amplification corresponding to 139 bp to confirm the effect of CBFβ or NFκB p65 on NKX6.3 depletion-induced somatic mutations in TP53 coding region in HFE-145shNKX6.3 cells. By decreasing the denaturation temperature (Td), it was found that 87.9 °C was the minimal temperature allowing amplification of TP53 in HFE-145shCtrl cells. By contrast, 3D-PCR could amplify TP53 gene in HFE-145shNKX6.3 cells at temperatures as low as 85.6 °C. As expected, knockdown of CBFβ or NFκB p65 in HFE-145shNKX6.3 was able to recover DNA at 86.6 °C, which was a higher temperature than that of NKX6.3-depleted cells (Supplementary Fig. S5C). In addition, 3D-PCR products from amplification at Td of 87.9, 85.6, and 86.6 °C were cloned and up to 11, 9, 13, and 11 clones were sequenced per sample. Knockdown of CBFβ or NFκB p65 reduced NKX6.3 depletion-induced somatic mutations in TP53 coding region of HFE-145shNKX6.3 cells (Fig. 2F and Supplementary Fig. S5D). Thus, NKX6.3 might be able to protect gastric epithelial cells against AICDA/APOBEC gene family-induced somatic mutations by downregulating CBFβ and NFκB expression.
NKX6.3 depletion induces AICDA/APOBEC mutagenesis pattern
Because NKX6.3 depletion could stimulate the entire spectrum of mutations (Fig. 1), we next evaluated AICDA/APOBEC mutation pattern in HFE-145shNKX6.3 cells. The strength of AICDA/APOBEC mutation patterns in C and G from genome-wide analysis of coding regions in HFE-145shNKX6.3 cells is shown in Fig. 3A. C to T and C to G changes in tCw motif of AICDA/APOBEC mutation in HFE-145shNKX6.3 cells showed the highest enrichment (Fig. 3A and Supplementary Table S4). In addition, a strong AICDA mutation pattern with the highest enrichment in wrC motif and a strong preference for C to T and C to A changes in wrC motif of AICDA/APOBEC mutation in NKX6.3-depleted cells were found (Fig. 3A and Supplementary Table S4). Furthermore, APOBEC- and AICDA-induced mutations were mainly detected in coding regions and 3′-UTRs (Supplementary Fig. S6A,B). Of mutations associated with NKX6.3 depletion, APOBEC and AICDA-induced mutations were found in 78 and 150 genes, respectively (Fig. 3B,C). According to related biological pathways, we analyzed gene set enrichment of APOBEC- and AICDA-induced mutant genes. Frequent mutations in genes involved in Nef and signal transduction, adrenoceptors and VEGFR3 signaling in lymphatic endothelium (including TP53, PIK3CA, ELMO1, and SMAD2), and genes involved in proline catabolism, DNA replication initiation, and NAD phosphorylation and dephosphorylation (including YWHAB, POLA1, PRIM2, and FFAR1) were found (Fig. 3B,C, Supplementary Fig. S6C,D, and Supplementary Table S5). Knockdown of APOBEC3B and AICDA partially recovered NKX6.3 depletion-induced mutations of PIK3CA and POLA1 genes in HFE-145shNKX6.3 cells (Fig. 3D,E). These results suggest that NKX6.3 depletion can lead to a variety of mutations, including APOBEC- and AICDA-mutation patterns.
NKX6.3 depletion induces AICDA/APOBEC mutagenesis pattern. (A) An APOBEC mutation pattern with the highest enrichment of C to T and C to G changes on the tCw motif (low left), a strong AICDA mutation pattern with the highest enrichment on the wrC motif, and a strong preference for C to T and C to A changes on the wrC motif in NKX6.3 depleted cells (low right). Error bars, SD. P values were derived from t tests. n.s, not significant (P > 0.05). *P < 0.05; **P < 0.01; ***P < 0.001. (B) Venn diagram of APOBEC-induced mutant genes in NKX6.3 depleted cells (upper) and gene set enrichment of APOBEC-induced mutant genes (lower). (C) Venn diagram of AICDA-induced mutant genes in NKX6.3 depleted cells (upper) and gene set enrichment of AICDA-induced mutant genes (lower). (D,E) Knockdown of APOBEC3B and AICDA in HFE-145 cells with transfected shNKX6.3 (D), knockdown of APOBEC3B and AICDA partially recovered NKX6.3 depletion-induced mutation in PIK3CA (E, left) and POLA1 genes (E, right).
Effects of NKX6.3 depletion on apoptosis and invasion
Since NKX6.3 was associated with mutations in CDH1, TP53, RhoA, PIK3CA, and EP300 genes (Fig. 1), we examined the expression of mutant genes and effects of NKX6.3 depletion on apoptosis and invasion. When we examined the expression and activity of E-cadherin, TP53, RhoA, PIK3CA, and EP300 in HFE-145shNKX6.3 cells, NKX6.3 depletion reduced E-cadherin and p21 expression, increased RhoA expression and Akt phosphorylation, and induced decreased and fragmented expression of EP300 possibly due to p53, RhoA, or PIK3CA mutations (Fig. 4A). In addition, increased expression of cleaved PARP and increased caspase-3/7 activity after anti-Fas treatment were significantly inhibited in NKX6.3-depleted cells possibly due to caspase-8 mutation (Fig. 4B). Furthermore, mutations of these genes in HFE-145shNKX6.3 cells were confirmed by sequencing (Fig. 4C).
Effects of NKX6.3 depletion on apoptosis and invasion. (A) Immunoblot analysis of NKX6.3 depletion-induced mutant gene expressions in HFE-145shCtrl and HFE-145shNKX6.3 cells. (B) Caspase 3/7 activity in HFE-145shCtrl and HFE-145shNKX6.3 cells were treated with anti-Fas (n = 3). Error bars, SD. P values were derived from t tests. *P < 0.05; **P < 0.01. (C) Mutations of CDH1, RhoA, EP300, and CASP8 gene in NKX6.3 depleted cells. (D) Cell migration activity in NKX6.3 depleted cells by transwell chemotaxis assays (n = 3). Error bars, SD. P values were derived from t tests. **P < 0.01. (E) Invasiveness of NKX6.3 depleted cells by Matrigel invasion assay (n = 3). Error bars, SD. P values were derived from t tests. ***P < 0.001. (F) Sphere number (left, n = 3) and size (right, n = 13, 40 spheres per group) in HFE-145shCtrl and HFE-145shNKX6.3 cells based on sphere forming assay. Error bars, SD. P values were derived from t tests. ****P < 0.0001.
Next, to determine the effect of NKX6.3 depletion on cell migration and invasion, we performed transwell chemotaxis and Matrigel invasion assays. Cell migration and invasion activities were significantly increased in HFE-145shNKX6.3 cells (Fig. 4D,E). In tumor sphere assay in vitro, depletion of NKX6.3 dramatically increased sphere number and size in HFE-145 cells (Fig. 4F).
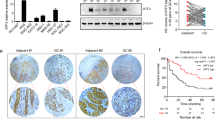
We next investigated whether NKX6.3 regulated gastric tumorigenesis using xenograft mouse model (Fig. 5A). Briefly, 5 × 106 NKX6.3 depleted cells (HFE-145shNKX6.3#1 and HFE-145shNKX6.3#2 cells) or non-target control (HFE-145shCtrl) were inoculated subcutaneously into the lower flank of BALB/C nude mice. Interestingly, mice implanted with HFE-145shNKX6.3#1 and HFE-145shNKX6.3#2 cells developed tumors (n = 4/4, 100%, respectively) whereas no mice subcutaneously injected with HFE-145shCtrl cells developed tumors (n = 0/4, 0%) (Fig. 5A,B). Mice injected with HFE-145shNKX6.3#1 and HFE-145shNKX6.3#2 cells showed significantly reduced body weights compared to HFE-145shCtrl cells injected mice (Fig. 5B). In real-time RT-PCR, immunoblot, and immunofluorescent analyses, tumors derived from mice implanted with HFE-145shNKX6.3#1 and HFE-145shNKX6.3#2 cells exhibited reduced E-cadherin and p21 expression but increased activity of NFκB p65, AICDA, CBFβ, APOBEC3B, RhoA and p-Akt with decreased and fragmented expression of EP300 (Fig. 5C–E). Sequencing HFE-145shNKX6.3#1 and HFE-145shNKX6.3#2 tumors confirmed that NKX6.3 depletion caused mutations in RhoA, PIK3CA, TP53, CDH1, EP300 genes (Fig. 5F). These results suggest that NKX6.3 depletion may inhibit apoptosis and increase invasion and sphere-forming activity due to mutations in genes associated with apoptosis and invasion, subsequently contributing to gastric tumorigenesis.
Depletion of NKX6.3 drives gastric tumorigenesis in xenograft model. (A) All mice implanted with HFE-145shNKX6.3#1 and HFE-145shNKX6.3#2 cells developed tumors whereas all mice subcutaneously injected with HFE-145shCtrl cells did not (each group: n = 4). (B) Tumor volumes and weight in mice implanted with HFE-145shNKX6.3#1 and HFE-145shNKX6.3#2 cells (each group: n = 4). Error bars, SD. P values were derived from t tests. **P < 0.01; ****P < 0.001. (C,D) mRNA (C) and protein (D) expression levels of CDH1, CDKN1A, EP300, NFκB p65, AICDA, CBFβ, APOBEC3B, PIK3CA, p-Akt, and RhoA in HFE-145shNKX6.3#1- and HFE-145shNKX6.3#2-induced xenograft tumors (n = 4). Error bars, SD. P values were derived from t tests. n.s, not significant (P > 0.05). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (E) Immunofluorescent staining for NKX6.3, NFκB p65, CBFβ, AICDA, APOBEC3B, RhoA, E-cadherin, p53, p21, PI3K, p-Akt, and EP300 in HFE-145shCtrl cells or HFE-145shNKX6.3#1- and HFE-145shNKX6.3#2-induced xenograft tumors. (F) Wild and mutant sequences of PIK3CA, CDH1, RhoA, EP300, TP53, and CASP8 gene in HFE-145shCtrl cells or HFE-145shNKX6.3#1- and HFE-145shNKX6.3#2-induced xenograft tumors.
NKX6.3 expression is inversely correlated with AICDA/APOBEC gene family and DNA repair genes in both gastric cancers and non-cancerous gastric mucosae
To further confirm that NKX6.3 expression could regulate the expression of AICDA, NFκB p65, CBFβ, APOBEC3A and APOBEC3B, mRNA expression levels of these genes were compared with NKX6.3 expression in 55 non-neoplastic gastric mucosae. NKX6.3 expression was inversely correlated with expression of these genes (Supplementary Fig. S7A) while expression levels of AICDA, APOBEC3A, and APOBEC3B were positively correlated with expression of NFκB p65 and CBFβ (Supplementary Fig. S7B).
The presence of H. pylori CagA was detected in 26 (47.3%) of 55 non-neoplastic gastric mucosal tissues14. Compared to gastric mucosae without CagA positive H. pylori, gastric mucosae with H. pylori infection showed decreased expression of NKX6.3 but increased expression of AICDA, NFκB p65, CBFβ, APOBEC3A, and APOBEC3B (Fig. 6A). Interestingly, three CagA-positive H. pylori-infected C57BL/6 mice also showed increased mRNA expression of AICDA, NFκB p65, APOBEC3A, and APOBEC3B in gastric mucosal tissues (Fig. 6B). Thus, NKX6.3 might be able to inhibit CagA-induced mutagenesis in gastric mucosal epithelial cells.
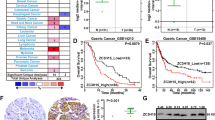
NKX6.3 expression is inversely correlated with AICDA/APOBEC gene family and DNA repair genes in both gastric cancers and non-cancerous gastric mucosae. (A) mRNA expression levels of NKX6.3, AICDA, NFκB p65, CBFβ, APOBEC3A and APOBEC3B in H. pylori CagA positive gastric mucosae compared to those in H. pylori CagA negative gastric mucosae (n = 55, triplicated experiments). Error bars, SD. P values were derived from t tests. **P < 0.01; ****P < 0.0001. (B) Protein levels of NKX6.3 and mRNA expression levels of AICDA, NFκB p65, CBFβ, and APOBEC3 in gastric mucosa tissues from H. pylori-infected mice (Ctrl, n = 2; H.P, n = 3, triplicated experiments). Error bars, SD. P values were derived from t tests. *P < 0.05; **P < 0.01; ***P < 0.001. (C) mRNA expression levels of AICDA, NFκB p65, CBFβ, APOBEC3A, APOBEC3B, CDH1, CDKN1A, EP300, RhoA, ROCK1, ROCK2, PIK3CA, and CCND1 in gastric cancers with negative and positive NKX6.3 expression (upper panel) and in H. pylori CagA positive and negative gastric mucosae (lower panel) (n = 65, triplicated experiments). Error bars, SD. P values were derived from t tests. n.s, not significant (P > 0.05); **P < 0.01; ***P < 0.001; ****P < 0.0001. (D) Expression of NKX6.3, p53, AICDA, and EP300 proteins in gastric cancers (n = 151) by immunohistochemistry. (E) Kaplan-Meier curves for overall survival of patients (n = 151) with gastric cancer with expression levels of NKX6.3, p53, EP300, and AICDA proteins. (F) Schematic model showing that NKX6.3 depletion in gastric epithelial cells induces mutagenesis of various genes by increasing APOBEC/AICDA activity through enhancing NFκB and CBFβ, finally resulting in the development of gastric cancer.
In gastric cancers, mRNA expression levels of AICDA, NFκB p65, CBFβ, APOBEC3A, and APOBEC3B were markedly increased, similar to their expression profiles in gastric mucosae with H, pylori infection. However, NKX6.3 expression was inversely correlated with expression of these genes (Fig. 6C). In addition, NKX6.3 mRNA expression was significantly lower in gastric cancers with higher TNM stage (Supplementary Fig. S7C), consistent with results of our previous study15. As expected, expression levels of AICDA, NFκB p65, CBFβ, APOBEC3A, and APOBEC3B were increased in gastric cancers with higher TNM stage (Supplementary Fig. S7C). We also found that mRNA expression levels of CDH1, CDKN1A, and EP300 were significantly reduced while those of RhoA, ROCK1, ROCK2, PIK3CA, and CCND1 were significantly increased in gastric cancers with reduced or loss of NKX6.3 expression compared to those in NKX6.3 positive cases (Fig. 6C). We also examined the presence of CagA gene in gastric cancer tissues by western blot analysis, as described previously14. Results showed that expression levels of NKX6.3, CDH1, CDKN1A, and EP300 were decreased while expression levels of NFκB p65, AICDA, CBFβ, APOBEC3A, APOBEC3B, RhoA, ROCK1, ROCK2, PIK3CA, and CCND1 were increased in CagA positive gastric cancers compared to those in CagA negative cases (Fig. 6C).
Next, we determined the relationship of NKX6.3 expression with p53, AICDA, and EP300 protein levels based on immunohistochemistry data of 151 human gastric cancer tissue specimens15,16,17. In gastric cancer tissues, NKX6.3, p53, AICDA, and EP300 proteins were expressed in 33 (21.9%), 65 (43%), 54 (35.8%), and 60 (39.7%) of 151 specimens, respectively (Fig. 6D and Table 1). There was a significant relationship between altered expression levels of NKX6.3, p53, AICDA, and EP300 proteins and clinicopathologic parameters, including depth of invasion (Chi-Square test: P = 0.0026, P = 0.2302, P = 0.039, and P = 0.0553, respectively), lymph node metastasis (Chi-Square test: P < 0.0001, P = 0.148, P = 0.4184, and P = 0.0889, respectively), and TNM stage (Chi-Square test: P < 0.0001, P = 0.1373, P = 0.0539, and P = 0.029, respectively). Importantly, NKX6.3 protein expression was positively correlated with expression of EP300 and but inversely correlated with expression of AICDA protein (Spearman’s test: P < 0.0001). EP300 protein expression was inversely correlated with that of AICDA protein (Spearman’s test: P = 0.0058) (Table 1). Kaplan-Meier analysis showed that there was no significant correlation between patient survival and p53 expression, EP300 expression, or AICDA expression alone (data not shown). However, combined analysis revealed that patients with negative NKX6.3 expression and/or negative p53, negative EP300, and positive AICDA expression had shorter overall survival time compared to those with positive NKX6.3 expression and/or positive p53, positive EP300, negative AICDA expression (P < 0.0001) (Fig. 6E). In 32 gastric cancers from TCGA datasets, cases with APOBEC3B and/or AICDA expression without NKX6.3 expression showed significantly higher number of mutations than those with NKX6.3 expression (Supplementary Fig. S7D). Taken together, these results indicate that NKX6.3 can regulate cell proliferation and apoptosis as well as cell motility and invasion processes by preventing accumulation of genetic alterations through downregulation of AICDA, NFκB, CBFβ, and APOBEC3B expression in gastric epithelial cells, thereby contributing to the development and progression of gastric cancer.
Discussion
Recently, several studies have revealed genetic alterations underlying gastric carcinogenesis by multi-omics profiling3,4,5,18. A mutational signature highlights specific carcinogenic and mutation processes in gastric cancer, including microsatellite instability, CpG associated deamination, and activation of cytidine deaminases such as AICDA and APOBEC3B3,4,7,18. Here, we observed that NKX6.3 depletion could lead to somatic mutations, mainly G or C to A or T base pair transitions in coding regions in gastric epithelial cells, consistent with results of TCGA-based analysis (Fig. 1A and Supplementary Fig. 1). For example, NKX6.3 depletion induced some driver mutations in CDH1, TP53, RhoA, PIK3CA, and EP300 genes, consistent with previous findings in gastric cancer3,4,5. CDH1 acts as a tumor suppressor. Decreased expression of CDH1 has been observed in gastric cancer with somatic mutation and methylation in the gene19,20. TP53 is another important tumor suppressor that negatively regulates cell cycle. Aberrant activity of TP53 caused by mutations is required for tumor formation21. Loss of p53 function can lead to impaired DNA replication, malignant transformation, genetic instability, and increased survival of cells with increased mutational load22. RhoA is a key member of the Rho family of small GTP-binding proteins that act as mediators between cell surface receptors and different intracellular signaling proteins23. RhoA overexpression has been observed in various cancers and RhoA activity has been implicated in tumorigenesis and tumor cell invasion24. In addition, highly recurrent mutations of RhoA have been detected in diffuse-type of gastric cancers5,25. Mutations in PIK3CA which encodes p110 catalytic subunit of PI3K have been observed in various human cancers, including gastric cancers. These mutations can lead to activation of PI3K/AKT and its downstream signaling pathways, thereby contributing to carcinogenesis26,27,28. EP300 functions as a histone acetyltransferase. It is an important modulator in cell proliferation and differentiation through transcriptional regulation via chromatin remodeling29,30. Loss of EP300 by inactivating mutations may contribute to tumorigenesis in human cancers, including gastric cancers31. Caspase-8 is activated by its recruitment to Fas-mediated apoptosis32. Somatic mutations of caspase-8 can lead to attenuation of its proapoptotic function and contribute to gastric carcinogenesis33. Here, we found that depletion of NKX6.3 could drive tumorigenesis in xenograft mouse model while tumor associated genes including p53, PI3K, E-cadherin, and EP300 were aberrantly expressed in these tumors. Consistent with results of NKX6.3 depleted cells, xenograft tumor also showed mutations in these genes (Figs 1, 4 and 5). Thus, NKX6.3 depletion might lead to multiple genetic mutations known to be closely associated with gastric tumorigenesis.
APOBECs and AICDA can mutate a host’s DNA. Significant numbers of APOBEC and AICDA-induced mutations have been observed in many types of human cancers34,35,36,37,38. Gastric cancer cells are highly enriched in mutation signatures [TC(A|T) → (T|G)] and [(A|T)(A|G)C → (T|G)] that are characteristic of a subclass of APOBEC and AICDA cytidine deaminases39,40,41. Two members of the APOBEC family, APOBEC3A and APOBEC3B, contribute substantially to mutations in cancers by deaminating cytosines in the TpCpW context34,35,36,37,40,41. It has been found that TpCpW mutations occur more frequently in gastric cancers with APOBEC expression38. APOBEC3B could activate genome-wide mutagenesis in various cancers34,35,36,37,40. In addition, aberrant expression of AICDA via NFκB activation causes accumulation of mutation in gastric epithelium with H. pylori infection7. AICDA-induced mutagenesis can lead to genome-wide alterations in the known preferred AICDA target sequence such as WRC motifs39,42. Moreover, CBFβ is required for APOBEC3 gene expression while NFκB is required for AICDA gene expression7,13. In the present study, we found that expression of CBFβ and NFκB was significantly increased in gastric cancer tissues while knockdown of CBFβ or NFκB dramatically inhibited the expression of APOBEC family and AICDA genes, diminished deaminase activity, and reduced the frequency of NKX6.3 depletion-induced mutations in TP53, PIK3CA, and POLA1 genes. Notably, binding of NKX6.3 to the promoter region of CBFβ and NFκB downregulated their expression while depletion of NKX6.3 dramatically increased expression of APOBEC gene family and AICDA genes at mRNA and protein levels, suggesting that NKX6.3 might function as a transcriptional repressor of these genes and downregulate deaminase activity in gastric epithelial cells (Fig. 2). Thus, NKX6.3 might inhibit the APOBEC- and AICDA-induced mutations by acting as a transcriptional repressor of APOBEC family and AICDA genes, thereby protecting gastric epithelial cells against genome-wide genetic mutations.
In non-neoplastic gastric mucosae and gastric cancer tissues, expression levels of CBFβ, NFκB p65, APOBEC family, and AICDA were increased in the cases with reduced or loss of NKX6.3 expression while NKX6.3 expression was inversely correlated with expression of these genes. In addition, expression levels of CDH1, CDKN1A, and EP300 were reduced while expression levels of RhoA, ROCK1, ROCK2, PIK3CA, and CCND1 were increased in gastric cancer tissues with reduced or loss of NKX6.3 expression (Fig. 6). Previously, we have reported that NKX6.3 expression is involved in gastric cancer progression and patients’ survival15. Concordant with these results, gastric cancer patients with reduced or loss of NKX6.3 expression and/or high mutation rates had shorter overall survival time (Supplementary Fig. S8E). In immunohistochemistry analysis, NKX6.3 expression was inversely associated with AICDA expression but positively associated with EP300 expression in 151 gastric cancer tissues (Fig. 6D). Thus, patients with reduced or loss of NKX6.3 expression and/or negative p53, negative EP300, and positive AICDA expression had shorter overall survival time (Fig. 6E), providing strong support for our hypothesis.
In conclusion, our study provides a new paradigm for the role of NKX6.3 in the pathogenesis of gastric cancer. We demonstrated that NKX6.3 depletion could lead to aberrant expression of AICDA and APOBEC family genes in gastric epithelial cells, thereby generating widespread genetic mutations and eventually driving the development and progression of gastric cancer (Fig. 6F). These findings unambiguously indicate that depletion of NKX6.3 in gastric epithelial cells can prompt accumulation of genetic mutations, providing important genetic evidence for the molecular mechanism involved in the development of gastric cancers.
Materials and Methods
Human Gastric Samples
A total of 65 frozen gastric cancers were obtained from Chonnam National University Hwasun Hospital supported by the Ministry of Health, Welfare and Family Affairs. In addition, a total of 55 patients with sporadic gastric cancer who underwent gastrectomy at Seoul St. Mary’s Hospital were included. Fifty-five non-neoplastic gastric mucosae remote from the tumor (>5 cm) at each corresponding region were enrolled in this study. Informed consent was provided according to the Declaration of Helsinki. Written informed consent was obtained from all subjects. This study was approved by the Institutional Review Board of The Catholic University of Korea, College of Medicine (approval numer: MC16SISI0130). There was no evidence of familial cancer in any of these patients.
For immunohistochemical analysis, tissue microarray recipient blocks were constructed, containing 151 gastric cancer tissues from formalin-fixed paraffin embedded specimens at Seoul St. Mary’s Hospital. Three tissue cores from each cancer (2 mm in diameter) were taken and placed in a new recipient paraffin block using a commercially available microarray instrument (Beecher Instruments, Micro-Array Technologies, Silver Spring, MD, USA) according to established methods15. One cylinder of normal gastric mucosa adjacent to each tumor was also transferred to the recipient block. Sections were cut in thickness of 2 μm the day before use. They were stained according to standard protocols.
Other details regarding cell culture, transfection, whole genome sequencing, gene set enrichment analysis, detecting AICDA/APOBEC mutation pattern, in vitro deaminase assay, 3D-PCR, cloning, sequencing, validation detection, caspase 3/7 activity assay, motility and invasion assays, spheroid cell culture, in vivo xenograft mouse experiment, RT–qPCR and ChIP–qPCR, immunoblotting and immunofluorescence (IF), immunohistochemistry (IHC), construction of plasmids, transient reporter assay, bacterial strain and animal infection, datasets, and statistical analyses are available in Supplementary Materials and Methods.
References
Hu, B. et al. Gastric cancer: Classification, histology and application of molecular pathology. J Gastrointest Oncol 3, 251–261 (2012).
Ferlay, J. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136, E359–386 (2015).
Wang, K. et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet 43, 1219–1223 (2011).
Bass, A. J. et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 (2014).
Wang, K. et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet 46, 573–582 (2014).
Touati, E. et al. Chronic Helicobacter pylori infections induce gastric mutations in mice. Gastroenterology 124, 1408–1419 (2003).
Matsumoto, Y. et al. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat Med 13, 470–476 (2007).
Conticello, S. G. The AICDA/APOBEC family of nucleic acid mutators. Genome Biol 9, 229 (2008).
Alanentalo, T. et al. Cloning and analysis of Nkx6.3 during CNS and gastrointestinal development. Gene Expr Patterns 6, 162–170 (2006).
Choi, M. Y. et al. Requirement of the tissue-restricted homeodomain transcription factor Nkx6.3 in differentiation of gastrin-producing G cells in the stomach antrum. Mol Cell Biol 28, 3208–3218 (2008).
Yoon, J. H. et al. Inactivation of NKX6.3 in stomach leads to abnormal expression of CDX2 and SOX2 required for gastric-to-intestinal transdifferentiation. Mod Pathol 29, 194–208 (2016).
Yoon, J. H. et al. NKX6.3 controls gastric differentiation and tumorigenesis. Oncotarget 6, 28425–28439 (2015).
Anderson, B. D. & Harris, R. S. Transcriptional regulation of APOBEC3 antiviral immunity through the CBF-β/RUNX axis. Sci Adv 1, e1500296 (2015).
Yoon, J. H. et al. Gastrokine 1 inhibits the carcinogenic potentials of Helicobacter pylori CagA. Carcinogenesis 35, 2619–2629 (2014).
Yoon, J. H. et al. NKX6.3 Is a Transcription Factor for Wnt/β-catenin and Rho-GTPase Signaling-Related Genes to Suppress Gastric Cancer Progression. EBioMedicine 9, 97–109 (2016).
Kim, C. J. et al. Activation-induced cytidine deaminase expression in gastric cancer. Tumour Biol 28, 333–339 (2007).
Eun, J. W. et al. MicroRNA-495-3p functions as a tumour suppressor by regulating multiple epigenetic modifiers in gastric carcinogenesis. J Pathol 244, 107–119 (2017).
Wong, S. S. et al. Genomic landscape and genetic heterogeneity in gastric adenocarcinoma revealed by whole-genome sequencing. Nat Commun 5, 5477 (2014).
Guilford, P. et al. E-cadherin germline mutations in familial gastric cancer. Nature 392, 402–405 (1998).
Strathdee, G. Epigenetic versus genetic alterations in the inactivation of E-cadherin. Semin. Cancer Biol 12, 373–379 (2002).
Weisz, L., Oren, M. & Rotter, V. Transcription regulation by mutant p53. Oncogene 26, 2202–2211 (2007).
Yeo, C. Q. et al. p53 Maintains Genomic Stability by Preventing Interference between Transcription and Replication. Cell Rep 15, 132–146 (2016).
Ridley, A. J. et al. Cell migration: integrating signals from front to back. Science 302, 1704–1709 (2003).
Sahai, E. & Marshall, C. J. RHO-GTPases and cancer. Nat Rev Cancer 2, 133–142 (2002).
Zhou, J., Hayakawa, Y., Wang, T. C. & Bass, A. J. RhoA mutations identified in diffuse gastric cancer. Cancer Cell 26, 9–11 (2014).
Samuels, Y. et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 304, 554 (2004).
Samuels, Y. et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell 7, 561–573 (2005).
Shigaki, H. et al. PIK3CA mutation is associated with a favorable prognosis among patients with curatively resected esophageal squamous cell carcinoma. Clin Cancer Res 19, 2451–2459 (2013).
Kawasaki, H. et al. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature 393, 284–289 (1998).
Yao, T. P. et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93, 361–372 (1998).
Gayther, S. A. et al. Mutations truncating the EP300 acetylase in human cancers. Nat Genet 24, 300–303 (2000).
Nagata, S. Apoptosis by death factor. Cell 88, 355–365 (1997).
Soung, Y. H. et al. CASPASE-8 gene is inactivated by somatic mutations in gastric carcinomas. Cancer Res 65, 815–821 (2005).
Burns, M. B., Temiz, N. A. & Harris, R. S. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 45, 977–983 (2013).
Burns, M. B. et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494, 366–370 (2013).
Chan, K. et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet 47, 1067–1072 (2015).
Seplyarskiy, V. B. et al. APOBEC-induced mutations in human cancers are strongly enriched on the lagging DNA strand during replication. Genome Res 26, 174–182 (2016).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Rogozin, I. B. et al. Activation induced deaminase mutational signature overlaps with CpG methylation sites in follicular lymphoma and other cancers. Sci Rep 6, 38133 (2016).
Kuong, K. J. & Loeb, L. A. APOBEC3B mutagenesis in cancer. Nat Genet 45, 964–965 (2013).
Roberts, S. A. et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 45, 970–976 (2013).
Matsumoto, T., Shimizu, T., Takai, A. & Marusawa, H. Exploring the Mechanisms of Gastrointestinal Cancer Development Using Deep Sequencing Analysis. Cancers 7, 1037–1051 (2015).
Acknowledgements
The biospecimens and data used for this study were provided by the Biobank of Chonnam National University Hwasun Hospital, a member of the Korea Biobank Network and Seoul St. Mary’s Hospital, Seoul, Korea. This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2018R1A2A2A14019713 to W.S.P. and 2017R1D1A1A09000781 to J.H.Y).
Author information
Authors and Affiliations
Contributions
J.H.Y. and W.S.P. designed the study; J.H.Y. conducted experiments; J.H.Y., O.K., J.W.E. and S.S.C. performed research and analyzed results; J.H.Y. and J.W.E. analyzed data; O.K., H.A., D.T.S., and S.W.N. writing-review and edited the manuscript; J.H.Y. and W.S.P. draft the manuscript; J.H.Y. and W.S.P. funding acquisition; W.S.P. supervision.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yoon, J.H., Kim, O., Eun, J.W. et al. Multiple genetic mutations caused by NKX6.3 depletion contribute to gastric tumorigenesis. Sci Rep 8, 17609 (2018). https://doi.org/10.1038/s41598-018-35733-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-35733-5
Keywords
This article is cited by
-
NKX6-1 mediates cancer stem-like properties and regulates sonic hedgehog signaling in leiomyosarcoma
Journal of Biomedical Science (2021)
-
Depletion of NK6 Homeobox 3 (NKX6.3) causes gastric carcinogenesis through copy number alterations by inducing impairment of DNA replication and repair regulation
Oncogenesis (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.