Abstract
Micavibrio aeruginosavorus is an obligate Gram-negative predatory bacterial species that feeds on other Gram-negative bacteria by attaching to the surface of its prey and feeding on the prey’s cellular contents. In this study, Serratia marcescens with defined mutations in genes for extracellular cell structural components and secreted factors were used in predation experiments to identify structures that influence predation. No change was measured in the ability of the predator to prey on S. marcescens flagella, fimbria, surface layer, prodigiosin and phospholipase-A mutants. However, higher predation was measured on S. marcescens metalloprotease mutants. Complementation of the metalloprotease gene, prtS, into the protease mutant, as well as exogenous addition of purified serralysin metalloprotease, restored predation to wild type levels. Addition of purified serralysin also reduced the ability of M. aeruginosavorus to prey on Escherichia coli. Incubating M. aeruginosavorus with purified metalloprotease was found to not impact predator viability; however, pre-incubating prey, but not the predator, with purified metalloprotease was able to block predation. Finally, using flow cytometry and fluorescent microscopy, we were able to confirm that the ability of the predator to bind to the metalloprotease mutant was higher than that of the metalloprotease producing wild-type. The work presented in this study shows that metalloproteases from S. marcescens could offer elevated protection from predation.
Similar content being viewed by others
Introduction
Predatory prokaryotes are obligatory predators that prey on other bacteria. The most studied predatory bacteria are those from the genus Bdellovibrio. However, many other predatory bacteria can be found in nature, one of which includes Micavibrio. First isolated from wastewater in 19821, Micavibrio spp. are small (~0.5–1.0 μm) curved, Gram-negative, α-proteobacteria bacteria2 with a single polar flagellum. In order to grow, Micavibrio spp. utilizes an epibiotic life cycle in which free-swimming motile attack cells attach to the cell surface of prey bacteria in a polar or non-polar manner, followed by an extracellular growth phase as it divides via binary fission3,4. In 2011, the complete genome sequence of Micavibrio aeruginosavorus ARL-13 was published in addition to transcriptome analysis of the attack and attach growth phase5. Additional insight regarding the biology of M. aeruginosavorus was presented by Pasternak et al.4 in a study in which the epibiotic predators M. aeruginosavorus ARL-13 and strain EPB were compared to an additional epibiotic predator Bdellovibrio exovorus JSS and two periplasmic predators Bdellovibrio bacteriovorus HD100 and B. marinus SJ. Further studies focused on the prey range of M. aeruginosavorus6,7,8,9, its potential use as a live antibiotic to control infection10, its ability to prey on drug resistant pathogens and biofilms6,11,12, and the predator’s non-toxic attributes when exposed to mammalian cells lines in vitro8,13. Additional studies demonstrated the predators’ non-pathogenic characteristics using several animal infection models including rabbit eye wound infection models14, mice and rat intranasal and intravenous inoculation models15,16,17, and gastrointestinal inoculation models used to measure the impact on gut bacterial microbiota18.
Although our knowledge of the biology, genetics, and potential antimicrobial use of Micavibrio has increased considerably in the last few years, the mechanisms governing predation and prey-predator interactions are not yet well understood. In order to prey, Micavibrio needs to attach reversibly and then irreversibly to its prey as well as withstand any secreted metabolites or ‘virulence/antimicrobial factors’ produced by the prey cell. In this study, we have used Serratia marcescens, an opportunistic human pathogen well studied for its virulence factors19,20,21, as the model prey to examine prey components that may play a role in predation. By using specific mutants defective in key extracellular cell structures and secreted factors, we have identified secreted metalloproteases from S. marcescens as being able to alter predation dynamics by reducing the ability of the predator to attach to the prey without affecting predator viability. Genetic manipulation confirmed the role of the metalloprotease in enhancing predation tolerance. Finally, exogenous addition of purified metalloprotease was able to restore predation tolerance to the S. marcescens metalloprotease mutant, as well as providing Escherichia coli protection from predation.
Results
Effect of prey extracellular cell structures and secreted compounds on predation
Extracellular cell structures are the first-thing that the predator encounters as it reaches the prey, it can potentially serve as an attachment site or an obstacle for binding. In this study we have used S. marcescens K904 as a model prey. An initial experiment confirmed that when co-cultured, M. aeruginosavorus is able to reduce S. marcescens K904 viability by 0.30 ± 0.27 and 1.26 ± 0.07 log10 within 24 and 48 hrs, respectively (from an initial 1.8 ± 0.3 × 109 CFU/ml to 1.08 ± 0.7 × 109 following 24 hrs and 1 ± 0.01 × 108 following 48 hrs of predation). No substantial change was measured in S. marcescens viability following incubation with predator free control (from an initial 1.8 ± 0.3 × 109 CFU/ml to 3.6 ± 1.1 × 109 and 2.3 ± 2.8 × 109 following a 24 and 48 hrs of predation, respectively). Additional confirmation that M. aeruginosavorus is able to attach to K904 was done by SEM imaging (Fig. 1). In order to evaluate if prey extracellular structures play a role in predation by M. aeruginosavorus, predation experiments were conducted in which the predator was co-cultured with S. marcescens mutants deficient in synthesis of candidate extracellular cell structure components that play a role in motility, attachment and protection from environmental challenges (Table 1). No significant difference (p > 0.1) was measured in the ability of M. aeruginosavorus to prey on S. marcescens mutants defective in flagella, fimbriae, and surface layer protein production, when compared to predation measured on the S. marcescens wild-type background strain (Table 1). In addition to cell extracellular structures, microbial secreted compounds might also influence predation. S. marcescens secretes several compounds, which have a role as virulence factors, and having known antimicrobial attributes. As seen in Table 1, no significant difference (p > 0.1) was seen in the ability of M. aeruginosavorus to prey on S. marcescens mutants defective in the production of the prodigiosin and phospholipase-A, compared to the wild-type background. However, the predation on a metalloprotease deficient mutant was significantly higher (p < 0.001) than that measured for the wild type protease producing isolate, with a 2.8 and 1.5 log10 reduction respectively (Table 1). In all experiments, maximum predation reduction was measured at the 48 hr time point (data not show). No reduction was seen in any of the mutants following incubation with predator free control when compared to the initial time point (0.06 ± 0.17 log10 reduction at 48 hrs).
Predation of M. aeruginosavorus on S. marcescens. S. marcescens wild type K904 was incubated in the presence of M. aeruginosavorus for 30 min. The cells were fixed and SEM micrographs were taken. Arrows indicate an attached M. aeruginosavorus prey cell to S. marcescens prey. Small round spheres are believed to be S. marcescens membrane vesicles. Scale bar, 100 nm. Magnification, x50,000.
Predation on S. marcescens metalloprotease mutants
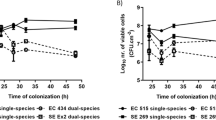
S. marcescens strain K904 produces three serralysin family metalloproteases produced by 3 separate genes prtS, slpB, and slpE22. To determine which of the three genes play a role in providing S. marcescens elevated protection from predation, co-cultures using S. marcescens mutants mutated in each of the genes were conducted. As seen in Fig. 2, a slight increase was measured in the ability of the predator to prey on slpB compared to the wild type (p = 0.01). However, prtS, slpE or a double prtS slpB mutants showed similar enhanced predation characteristics as the triple prtS slpB slpE mutant (Fig. 2), suggesting that the metalloproteases produced by prtS, slpE, and to a lesser extent slpB, were able to provide elevated protection from predation. No differences in cell growth were measured between the wild type and the triple prtS slpB slpE mutant, both reaching similar OD’s following 12 hrs of incubation in LB (OD600 of 1.6 ± 0.01 and 1.5 ± 0.02 for the wild type and mutant, respectively). Furthermore, the ability of the triple prtS slpB slpE mutant to survive in HEPES buffer for 48 hrs was equivalent to that seen in the wild type (from 1.5 × 109 CFU/ ml to 7.0 × 108 CFU/ml for the wild type and from 1.2 × 109 CFU/ ml to 5.8 × 108 CFU/ml for the mutant) (Supplementary material Fig. 1).
Predation on S. marcescens metalloprotease mutants. Co-cultures were prepared by adding M. aeruginosavorus to S. marcescens wild type (WT) prey, single (∆prtS, ∆slpB, and slpE::pMQ118), double (∆prtS ∆slpB) or triple (∆prtS ∆slpB slpE) metalloprotease mutants (n = 9). Values represent the Log10 reduction in prey viability, compared to the predator free control, following 48 hrs of predation. Each value represents the mean and standard deviation of three experiments done in triplicates. Asterisk indicates significant difference from the reduction measured for the WT (p = 0.01). Pound signs indicate significant difference from the reduction measured for the WT (p < 0.001, One Way ANOVA with Tukey’s post-test).
To further validate that the metalloproteases could provide elevated protection from predation, the prtS gene was introduced into the triple metalloprotease deficient mutant under the regulation of an inducible promoter using plasmid pMQ356. As before, the ability of Micavibrio to prey on the wild type metalloprotease producing S. marcescens was reduced compared to that seen on the metalloprotease negative mutant, with 1.3 and 2.3 log10 reduction respectively (Fig. 3). Complementation of the prtS gene into the metalloprotease mutant, under non-inducing conditions, did not alter predation (2.15 log10 reduction, p > 0.1). However, inducing the prtS construct with arabinose complemented the mutant defect and reduced predation to levels that are lower than that measured on the metalloprotease producing wild type (0.78 log10 p < 0.001), and significantly lower (p < 0.001) to that measured on the metalloprotease mutant (Fig. 3, ∆prtS ∆slpB slpE). No change in wild type predation was measured in an arabinose treated co-culture control indicating that the arabinose inducer itself was not responsible for the effect (data not shown).
Genetic complementation. Co-cultures were prepared by adding M. aeruginosavorus to S. marcescens wild- type (WT) prey, triple (∆prtS ∆slpB slpE) metalloprotease mutants (n = 9) or metalloprotease mutants harboring the prtS gene under the regulation of an arabinose inducible promoter (∆prtS ∆slpB slpE::pMQ118 + pMQ356) (n = 9). Co-cultures were incubated with or without arabinose (+ara, −ara). Values represent the Log10 reduction in prey viability, compared to the predator free control, following 48 hrs of predation. Each value represents the mean and standard deviation of three experiments done in triplicates. Asterisks indicate significant differences from the reduction measured for the WT (p < 0.001). Pound signs indicate significant difference from the reduction measured for the arabinose-free triple metalloprotease mutants (p < 0.001, One Way ANOVA with Tukey’s post-test).
Purified serralysin metalloprotease from S. marcescens can protect prey from predation by M. aeruginosavorus
As endogenous production of metalloproteases by S. marcescens was shown to reduce predation, we were interested in examining if exogenous metalloprotease was sufficient to reduce predation. To this end, the PrtS metalloprotease (serralysin) from S. marcescens was purified and added to predator co-cultures. Adding 5 µmol/ml (final concentration) of pure PrtS to the metalloprotease mutant reduced the ability of the predator to prey to levels lower than those measured for the wild type (0.64 and 1.18 log10 predation reduction, respectively) (Fig. 4A). Purified PrtS was also able to render the wild- type strain almost resistant to predation with only a 0.3 ± 0.15 log10 reduction following 48 hrs of incubation (Fig. 3A, WT + PrtS). Similar predation protection was seen when purified exogenous PrtS was added to a non-producing metalloprotease E. coli. Predation was reduced from 2.6 ± 0.09 log10 seen on protease free control (Fig. 4B, E. coli + Mica) to 0.78 ± 0.19 log10 on the E. coli protease treated co-cultures (Fig. 4B, E. coli + Mica + PrtS). Adding purified PrtS to predator-free prey cells did not bring about reduction in E. coli cell viability (from an initial 4.5 × 109 CFU/ml to 5 × 109 following 48 hrs of incubation for S. marcescens and from an initial 2 × 109 CFU/ml to 1.8 × 109 for E. coli). To examine if other proteases have similar predation enhancement effect as metalloproteases, proteinase K and trypsin were added to predator prey co-cultures. No difference in predation was measured when 100 µg/ml (final concentration) proteinase K and 50 µg/ml (final concentration) trypsin was added to a predator co-culture using S. marcescens metalloprotease mutant, with a 2.5 log10, 2.7 log10, and 2.4 log10 predation reduction measured for the metalloprotease mutant alone, with proteinase K or trypsin, respectively.
Exogenous addition of purified metalloprotease. (A) Co-cultures were prepared by adding M. aeruginosavorus to S. marcescens wild-type (WT) prey, or triple (∆prtS ∆slpB slpE) metalloprotease mutants and incubated with or without the addition of purified metalloprotease at 5 µmol/ml final concentration (+PrtS, −PrtS) (n = 9). (B) Cultures were prepared by adding M. aeruginosavorus to E. coli (E.coli + Mica) (n = 9) or E. coli, with predator and purified metalloprotease (E.coli + Mica + PrtS) (n = 9). Values represent the Log10 reduction in prey viability, compared to the predator free control, following 48 hrs of predation. Each value represents the mean and standard deviation of three experiments done in triplicates. Asterisks indicate significant differences from the reduction measured for the WT (A) or E. coli + Mica (B) (p < 0.001). Pound signs indicate significant difference from the reduction measured for the arabinose-free triple metalloprotease mutants (p < 0.001, One Way ANOVA with Tukey’s post-test).
Serralysin metalloprotease does not impact predator viability
One mechanism in which S. marcescens metalloproteases could reduce predation is by toxicity to the predator. To this end, M. aeruginosavorus was incubated for 48 hrs in HEPES buffer or HEPES treated with 5 µmol/ml purified PrtS metalloprotease. No reduction was measured in the viability of the predator incubated with the protease compared to that of the protease free control (from an initial 1.0 × 108 PFU/ml to 1.3 × 108 and 1.2 × 108 following 48 hrs of incubation with protease and protease free control, respectively).
Serralysin metalloprotease impacts attachment to prey cell
A second potential mechanism is that metalloproteases may affect attachment and binding by potentially impacting elements on the surface of the prey or the predator. Protease pre-treatment experiments were conducted in which S. marcescens triple metalloprotease mutant or M. aeruginosavorus were pre-treated for 4 hrs with 5 µmol/ml purified PrtS. The protease was removed, and the cells were used in a predator-prey co-culture. As before, M. aeruginosavorus was able to prey better on the metalloprotease mutant than on the wild type (Fig. 5, ∆prtS ∆slpB slpE). Pre-incubation of the predator (Fig. 5, Mica-pre-treat) with purified metalloprotease did not inhibit the ability of the predator to prey, resulting in similar prey reduction to that measured for the metalloprotease control (1.7 ± 0.2 and 1.8 ± 0.07 log10 predation reduction, respectively). However, pre-incubation of the metalloprotease mutant with PrtS (Fig. 5, ∆prtS ∆slpB slpE-pre-treat) had reduced the ability of the predator to prey, resulting in a 0.27 ± 0.04 log10 reduction after 48 hrs. As before, incubating the predator or prey for 4 hrs with the metalloprotease did not reduce cell viability with the predator numbers remaining at 1 × 108 PFU/ml. Our data suggests that metalloprotease may impact predation by affecting the prey, while transmission electron microscopy (TEM) did not provide additional insight regarding any specific structures that may differ from the wild type and metalloprotease mutant. However, some changes in cell wall surface are evident in the TEM micrographs with what seems as a denser and opaquer cell wall structure in the metalloprotease mutant compared to the wild type (Supplementary material Fig. 2). No difference in yeast agglutination was seen between the wild type and metalloprotease mutant with aggregation occurring 20 seconds after mixing bacteria and yeast cells. Whereas, PBS (no bacteria) control and fimbriae mutant (fimC::pMQ167) failed to agglutinate with yeast (Supplementary material Fig. 4C).
Pre-incubation with purified metalloprotease. Co-cultures were prepared by adding M. aeruginosavorus to S. marcescens wild-type (WT) prey, or triple (∆prtS ∆slpB slpE) metalloprotease mutants. Additional co-cultures included the triple metalloprotease mutants that were pre-incubated for 4 hrs with 5 µmol/ml final concentration of PrtS before being washed and added to the culture (∆prtS ∆slpB slpE-pre-treat) (n = 9) or M. aeruginosavorus that was pre-incubated for 4 hrs with PrtS before being washed and added to the culture (Mica-pre-treat) (n = 9). Values represent the Log10 reduction in prey viability, compared to the predator free control, following 48 hrs of predation. Each value represents the mean and standard deviation of three experiments done in triplicates. Asterisks indicate significant differences from the reduction measured for the WT (p < 0.001). Pound signs indicate significant difference from the reduction measured for the arabinose-free triple metalloprotease mutants (p < 0.001, One Way ANOVA with Tukey’s post-test).
In order to further measure the impact of metalloproteases on predator binding, flow cytometry was used to evaluate initial M. aeruginosavorus - S. marcescens cell-cell attachment. Prey and predator cells were stained, mixed and incubated together for 5 to 90 min. Predator cell association was found to be time dependent with an average of 14%, 34%, 48% and 76% prey attached to the wild type at the 5, 30, 60 and 90 min, respectively. However, the affinity of the predator to bind to the metalloprotease mutant was higher than that measured for the wild type with an average of 17%, 50%, 59% and 84% prey attached at the 5, 30, 60 and 90 min, respectively. Maximum binding difference was seen at the 30 min time point with 47% more cells attached to the mutant than that of the wild type (p = 0.04) (Fig. 6). The enhanced ability of the predator to bind to the metalloprotease mutant was also confirmed by fluorescence microscopy (Fig. 7A). An average of 20.5 ± 4.9% and 21 ± 2.7% of prey cells were found to be attached to the wild type after the 5 and 30 min of co-incubation, respectively. Whereas an average of 40.1 ± 10.5% and 48.3 ± 3.5% of the predator were found to be attached to the metalloprotease mutant at the same time points (p = 0.005) (Fig. 7B).
Attachment dynamics of M. aeruginosavorus. FM 4–64 stained S. marcescens (Wild type) prey or triple (∆prtS ∆slpB slpE) metalloprotease mutant were co-cultured with PKH67 stained M. aeruginosavorus for 5, 30, 60, and 90 min (n = 3). Cells were incubated with the dyes for 10 minutes. Cells were then spun down and resuspended in HEPES to remove excess dye. Immediately after, cells were mixed together for 5, 30, 60, and 90 minutes. The top right number is a representative value of the percent population of M. aeruginosavorus attached to either wild type or triple metalloprotease mutant. The number below represents the average of three independent experiments. Analysis of the data was done on FlowJo v10.4.
Attachment of M. aeruginosavorus. (A) FM 4–64 stained S. marcescens (Wild type) prey or triple metalloprotease mutant (∆prtS ∆slpB slpE) were co-cultured with PKH67 stained M. aeruginosavorus. Cells were incubated for 30 min before being fixed in 1% formaldehyde and examined using a fluorescent microscope with 40x digital zoom and 40x magnification. Predator cells are seen in green, while prey cells seen are in red. The image in the upper right corner represents digital magnification of Micavibrio-Serratia binding. Celena-S and Microsoft PowerPoint software were used for image brightness adjustment and digital magnification. Experiments were conducted 3 times with representative images shown. (B) Attachment of S. marcescens wild type prey or triple metalloprotease mutant following 5 and 30 min of incubation as visualized using Celena-S digital fluorescence microscope with GFP and DAPI filters. Bars represent mean and standard deviation of percent of bound M. aeruginosavorus to S. marcescens calculated from total predator numbers in a field of view. The experiment was performed three times with 6 fields measured for each treatment. Asterisks indicate significant differences from attachment measured for the WT (p = 0.005).
Discussion
In order to feed, survive and proliferate, the epibiotic predator M. aeruginosavorus needs to get in close proximity to its prey before adhering to the prey surface. Thus, we speculated that prey secreted compounds and or surface structures may play a role in predation as inhibiting factors or as facilitators of attachment. In a 2007 study we were unsuccessful in isolating P. aeruginosa PA14 predation-resistant mutants in a screen using a random transposon library6. It was suggested, that prey structures that play a role in predation might be essential, therefore, making them unable to be found in a library of viable mutants. Another explanation for the inability to isolate prey resistant transposon mutants is that genes or pathways required for predator-prey interactions are redundant. In this study, we have used a more direct approach in which specific deletion mutants of S. marcescens K904 were used. S. marcescens is emerging as an important opportunistic pathogen20. Many of the mutants used to study S. marcescens virulence are cell structural components and secreted factors that might also play a role in predation. Although we have previously reported that M. aeruginosavorus ARL-13 is unable to prey on S. marcescens D 217 and PIC3611/3716,7, here we report that M. aeruginosavorus ARL-13 is able to prey on S. marcescens K904. SEM imaging (Fig. 1) and prey enumeration following predation confirmed predator attachment and predation. As previously seen with other prey, maximum prey cell reduction is measured following 48 hrs of predation7,8. Differences in strain specificity and prey susceptibility are well established for both M. aeruginosavorus and Bdellovibrio6,7,8. Thus, it is not uncommon to see predators preying on one strain within a given species and not another.
Deletion of fimbria, which plays a role in cell-cell and cell-surface association23,24,25, did not impact predation. Thus, it could be concluded that fimbria does not serve as an attachment site nor could it interfere in the ability of the predator to reach any putative prey binding sites. Elimination of flagella biosynthesis did not impact predation, as well, suggesting that flagellin does not serve as a predator-binding site. Although the motility of the prey might alter predator-prey encounter, the prey to predator ratio, as well as the small liquid volume used in the study is likely to allow sufficient random collisions of predator and prey, regardless of prey motility26,27. Koval and Hynes suggested that the paracrystalline protein surface layer (S-layer) of Aquaspirillum serpens, A. sinuosum and Aeromonas salmonicida provides protection from B. bacteriovorus predation28. Here we found no significant effect in the ability of Micavibrio to prey on S. marcescens S-layer mutants. Although we confirmed that the slaA mutant does not produce S-layer protein and exhibits a visually different cell wall structure (Supplementary Fig. 3), we could not determine if the S-layer evenly coats the entire cell surface of the wild type. The ability of Micavibrio to easily penetrate the thick extracellular polymeric substances produced by biofilms6,7 may suggest that the S-layer is not sufficient to block predation. Although we have selected several mutants that were deficient in key cell surface structures, future work needs to be conducted in order to evaluate additional structures that may play a role in predation such as bacterial capsule and lipopolysaccharides29. Before encountering the prey, predators can come across a variety of prey secreted proteins, enzymes, lipids, biosurfactants as well as other antimicrobials that might impact predation. Indole, for example, was shown to impact predation of E. coli and Salmonella enterica by B. bacteriovorus30. Medium acidification, in diluted nutrient broth but not HEPES buffer, a byproduct of prey sugar metabolism, was also shown to block predation. The effect was seen in co-cultures using B. bacteriovorus and M. aeruginosavorus, and was attributed to the inability of the predator to survive in acidified environments31. S. marcescens was shown to produce an arsenal of compounds which likely play a role in microbial pathogenesis, among them are phospholipase-A32, proteases33, as well as prodigiosin34,35 which has antimicrobial attributes, as well. In this study, we have found no change in the ability of M. aeruginosavorus to prey on S. marcescens mutants defective in the production of prodigiosin and phospholipase-A. This finding is in accord with the fact that M. aeruginosavorus is able to attack other human pathogens, which are known to secrete a variety of toxins and antimicrobials, such as Burkholderia mallei, P. aeruginosa and Yersinia pestis6,7,8,9,11. An additional virulence factor that is produced by S. marcescens K904 are metalloproteases which are members of the RTX-toxin family36,37 and were shown to be cytotoxic to mammalian cells in vitro36,38,39. Although phospholipase mutant still showed some lipase activity (Supplementary material Fig. 6), the S. marcescens genome harbors numerous predicted lipases and esterase genes that may be active in the lipase assay.
In this study, we confirmed that metalloprotease producing strain S. marcescens K904 was less susceptible to predation than the metalloprotease deficient mutants. Predation protection was found to be mediated by each of the metalloprotease producing genes (Fig. 2). Interestingly, slpB, which showed reduced predation protection capability (Fig. 2), was also shown to produce lower levels of metalloprotease compared to prtS and slpE39. Therefore, metalloprotease predation protection capability may be dose dependent. The enhanced resistance of the wild type following the addition of exogenous purified PrtS also supports the conclusion that metalloprotease predation resistant attributes may be dose dependent (Fig. 4A). Genetic complementation and the ability of purified PrtS to restore predation of the hyper-susceptible triple prtS slpB slpE mutant to wild type levels validated the role of metalloprotease in enhancing resistance to predation. The predation protection effect of metalloprotease was also confirmed by adding purified PrtS to E. coli (Fig. 4B). The resistance conferring activity of the metalloprotease was found to be specific and was not seen when using proteinase K or trypsin that, unlike PrtS, did not alter predation. The ability of proteases to block predation was previously reported in a study that demonstrated that the addition of proteinase K but not trypsin or saliva, which is known to harbor trypsin-like proteases, could block predation of E. coli and Aggregatibacter actinomycetemcomitans by B. bacteriovorus. It suggested that proteinase K may affect specific surface proteins on the prey cell or the B. bacteriovorus that are required for predation40. Although metalloprotease provided enhanced protection from predation by M. aeruginosavorus it did not seem to provide protection from predation by B. bacteriovorus (Supplementary Table 1).
Incubating M. aeruginosavorus with purified PrtS was found not to impact predator viability; however, pre-incubating the prey, but not the predator, with purified metalloprotease was able to block predation (Fig. 5). It could be hypothesized that metalloprotease may affect or modify cell surface components that facilitate predation, causing reduced predation. TEM micrographs showed some changes in the thickness of the cell wall of the wild type compared to the metalloprotease mutant. However, with the limited resolution of TEM and its inability to detect changes that might involve smaller structures such as protein receptors, it is difficult to make any assumptions regarding modified specific structures and their impact on predation. Nonetheless, flow cytometry and microscopy evaluation confirmed that the ability of the predator to bind to the metalloprotease mutant was higher than that of the metalloprotease producing wild type (Figs 6 and 7A,B). No difference in yeast agglutination, a phenotype associated with fimbriae production, was measured between the wild type and the metalloprotease mutant (Supplementary material Fig. 4C). Several species of bacteria such as Erwinia41, which was found not to be preyed upon by M. aeruginosavorus7 and Serratia22 carry up to five highly similar metalloprotease genes in their genomes. Given the functional overlap of the proteins, it is not clear what drives selection to maintain these seemingly redundant proteins. Based on this study, it is tempting to speculate that the one driving force in maintaining these genes is the protection they provide from predatory prokaryotes such as Micavibrio.
In conclusion, the work presented in this study shows that metalloproteases from S. marcescens offer prey elevated protection from Micavibrio predation. This is the first report, to our knowledge, of a mechanism that prey use in order to defend themselves against Micavibrio. This study also offers additional insights on prey factors that may play a role in M. aeruginosavorus prey interaction and prey specificity. Future studies will focus on better understanding the specific prey components that are impacted by metalloprotease and to determine whether metalloproteases from other bacterial prey species provide protection from predation.
Methods
Bacterial strains and growth conditions
The S. marcescens used in the study are listed in Table 2. S. marcescens were grown routinely in 15 ml lysogeny broth (LB) medium in a 25 ml Polystyrene tissue culture flask at 30 °C. When appropriate, gentamicin and kanamycin were used at 10 µg ml−1. Arabinose, which induces expression of PBAD promoter in pMQ356, was added to the culture (0.02% w/v final concentration). Proteolytic activity of S. marcescens was confirmed by a zone of clearance after incubating the bacteria on LB-agar plates containing 2% w/v skim milk. Escherichia coli strain ST-2289, a multi drug resistant KPC-3 isolate42, was kindly provided by Dr. Kreiswirth of Rutgers, New Jersey Medical School, and cultured in LB at 37 °C. M. aeruginosavorus strain ARL-13 was cultured and maintained as described previously7,8,11, using E. coli strain WM3064 as prey. Predator stock-lysates were made by co-culturing the predators with prey cells in HEPES buffer (25 mM) supplemented with 2 mM CaCl2 and 3 mM MgCl2. Co-cultures were incubated at 30 °C for 72 hrs, until the culture cleared (stock-lysates). Stock-lysates were filtered through a 0.45-µm Millex pore-size filter (Millipore) to remove any remaining prey and used in predation experiments (harvested predators). Prey and predator cells were enumerated as colony-forming units (CFU) or plaque forming units (PFU) developing on LB agar plates or lawns of prey cells6.
Cloning and Mutagenesis
To generate the slaA mutant, the 3003 base pair open reading frame was [amplified with primers #3743 5′-aaattctgttttatcagaccgcttctgcgttctgatttaCGCGTAGTGCAGAGTATCCAC-3′ and #3744 5′-aattgtgagcggataacaatttcacacaggaaacagctATGTCATCTCTTGTCTCACAGC-3′ using high fidelity polymerase Phusion (New England Biolabs) and] cloned into allelic replacement vector pMQ46039 using yeast homologous recombination43 and digested with KpnI which removes a slaA internal 1482 base pair fragment. The resulting pMQ460 + ∆slaA plasmid (pMQ566) was used for allelic replacement of the wild type slaA gene as previously described43 and the mutant allele on the chromosome was verified by PCR and sequencing.
Phenotypic analysis was performed to demonstrate that secreted and surface components were made by the wild type bacteria and not made by the mutant bacteria under the conditions used for predation analysis. The presence and lack of surface layer protein in wild type K904 and slaA mutant, respectively, were determined and confirmed by SDS-PAGE analysis as previously described44 (Supplementary material Fig. 3B). Furthermore, transmission electron microscopy (TEM) images demonstrate a change in cell wall structure in the ∆slaA mutant consistent with lack of the S-layer (Supplementary material Fig. 3B,C). TEM was also used to confirm the absence of flagella in the fimC mutant and presence of flagella in the wild type (Supplementary material Fig. 4A,B). Cells were counted and 0 out of 79 fimC mutant cells had flagella, whereas 36.8% (14/38) of the wild type had flagella, p < 0.0001 two-tailed Fisher Exact Test. Yeast agglutination assay was conducted in order to conform the inability of the fimbriae mutant (fimC) to bind to yeast (Supplementary material Fig. 4C), a phenomenon that requires fimbriae45. The absence of prodigiosin (red pigment) in the ∆pigA mutant and production by the wild type under predation experimental conditions was verified visually (Supplementary material Fig. 5). Reduction in phospholipase activity in phospholipase A mutant was confirmed using EnzChek Phospholipase A1 Assay-Kit (ThermoFisher Scientific E1) (Supplementary material Fig. 6).
Predation Experiments
Predation experiments were performed as described previously with some modifications7,8,11. Co-cultures of predator and prey were prepared in 14 ml Falcon™ round-bottom polypropylene tubes by adding 0.4 ml of harvested predators (∼5 × 107 PFU/ml) to 0.4 ml of HEPES washed prey cells (∼2 × 109 CFU/ml) and 1.2 ml HEPES medium. Co-cultures were incubated at 30 °C on a rotary shaker set at 30 r.p.m. Predation was measured by the change in prey population, enumerated by dilution plating, during a 48 hr incubation period. As maximum prey reduction by M. aeruginosavorus occurs following 2 days of incubation (see result section), all predation experiments were conducted and reported for the 48 hr time point. Experiments were done at least three times in triplicates. Experiments requiring the addition of purified metalloprotease were conducted twice in triplicates. Phenotypic characterization of prey cells under predation conditions could be seen in Supplementary Figs 1–6.
Electron microscopy
For scanning electron microscopy (SEM) M. aeruginosavorus ARL-13 was co-cultured with S. marcescens as described above. Samples were removed following 30 min of incubation and placed on a Cell-Tak coated glass coverslip. Samples were fixed using 2.5% glutaraldehyde for 1 hr, washed with PBS and post-fixed in aqueous 1% OsO4. Coverslips were dehydrated through a graded series of 30% to 100% ethanol and washed with Hexamethyldisilazane. Samples were sputter coated with 6 nm of gold/palladium (Cressington Auto 108, Cressington, UK). Imaging was done using a JEOL JSM- 6335 F scanning electron microscope (Peabody, MA) at 3 kV with the SEI detector. For transmission electron microscopy (TEM) aliquots of cultures grown overnight were washed and suspended in HEPES buffer and applied to formvar coated grids, treated with uranyl acetate (1%), and imaged using a JEM-1210 electron microscope as previously described24.
Yeast agglutination assay
To measure fimbriae production by both the wild type and triple metalloprotease mutant (∆prtS ∆slpB slpE), yeast agglutination was measured as previously described25. Bacteria were grown overnight in LB and resuspended in HEPES. The OD was set to 1.0. Twenty-five microliters of S. cerevisiae (Sigma-Aldrich YSC2) in HEPES buffer (0.2 g/10 ml) was mixed with test bacteria (25 µl), or PBS (25 µl) as a negative control on a glass microscope slide and mixed using a rotary rotator (Barnstead Multipurpose Rotator). The rotary platform was set to half of maximum speed and the time to aggregation was measured using a stopwatch.
Purification of PrtS
Serralysin was overexpressed in E. coli and purified under denaturing conditions as previously described46. E. coli PrtS was induced with 0.02% w/v arabinose for 4 hrs and cells were then centrifuged and resuspended in 50 mM tris and 150 mM NaCl. Cells were lysed by sonication and lysates were cleared by centrifugation. Inclusion bodies were then resuspended in 50 mM Tris, 150 mM NaCl and 6 M GuHCl, prior to purification on a Ni-NTA column (GE Life Sciences). Fractions containing the protease were eluted using 400 mM imidazole and further purified using sephacryl S200 Hi-Prep gel filtration column (GE Life Sciences). The denatured protein was then stored at 4 °C. Refolding and protein concentration were done as previously described by rapid dilution while residual GuHCl was removed using size exclusion chromatography47,48. Protease activity was confirmed using a fluorescent casein substrate (Enzchek, ThermoFisher).
Flow Cytometry
Serratia marcescens wild type and triple protease mutant were grown in LB medium at 30 °C overnight. Cells were centrifuged and resuspended in HEPES buffer. Predator stock-lysates were prepared and filtered as described above. Prey and predator cells were stained with red FM 4–64 (Sigma-Aldrich) and green PKH67 (Invitrogen) dyes at 2 nM and 2 mM final concentrations, respectively, and incubated for 10 min. Thereafter, cells were washed by centrifugation and resuspended in HEPES. Prey and predator cells were mixed and incubated for 5, 30, 60 and 90 min, on a rotary shaker at 30 r.p.m. Samples were acquired on the BD LSRFortessa X-20 flow cytometer on low at an approximate rate of 2500ev/sec. Samples were first gated on S. marcescens (FM 4–64 positive) and then measured for binding of M. aeruginosavorus (PKH67 intensity). Data was acquired on the FITC channel (530/30) and the PE-Cy5 channel (670/30). FlowJo v10.4 software was used for data analysis.
Fluorescence microscopy
Predator and prey were prepared and stained with red FM 4–64 green PKH67 as described for flow cytometry. Prey and predator cells were mixed in HEPES buffer, as described for predation assays and incubated for 30 min. Following incubation, the bacteria were fixed with 1% formaldehyde for 10 min and mounted on a poly-L-lysine coated slide. Slides were imaged using a Celena-S Digital Fluorescence Microscope with GFP and DAPI filters, 40x digital zoom and a 40x objective lens. Celena-S and Microsoft PowerPoint software were used for image brightness adjustment and digital magnification.
Statistical analysis
A one-way analysis of variance (ANOVA) with Tukey’ s post-test was performed using Graphpad Prism 6 software. Statistical significance was set to p < 0.001 with standard deviation (SD) values.
References
Lambina, V. A., Afinogenova, A. V., Romai Penabad, S., Konovalova, S. M. & Pushkareva, A. P. Micavibrio admirandus gen. et sp. nov. Mikrobiologiia 51, 114–117 (1982).
Davidov, Y., Huchon, D., Koval, S. F. & Jurkevitch, E. A new alpha-proteobacterial clade of Bdellovibrio-like predators: implications for the mitochondrial endosymbiotic theory. Environ Microbiol 8, 2179–2188, https://doi.org/10.1111/j.1462-2920.2006.01101.x (2006).
Lambina, V. A., Afinogenova, A. V., Romay Penobad, Z., Konovalova, S. M. & Andreev, L. V. New species of exoparasitic bacteria of the genus Micavibrio infecting gram-positive bacteria. Mikrobiologiia 52, 777–780 (1983).
Pasternak, Z. et al. In and out: an analysis of epibiotic vs periplasmic bacterial predators. ISME J 8, 625–635, https://doi.org/10.1038/ismej.2013.164 (2014).
Wang, Z., Kadouri, D. E. & Wu, M. Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13. BMC Genomics 12, 453, https://doi.org/10.1186/1471-2164-12-453 (2011).
Kadouri, D., Venzon, N. C. & O’Toole, G. A. Vulnerability of pathogenic biofilms to Micavibrio aeruginosavorus. Appl Environ Microbiol 73, 605–614, https://doi.org/10.1128/AEM.01893-06 (2007).
Dashiff, A., Junka, R. A., Libera, M. & Kadouri, D. E. Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J Appl Microbiol 110, 431–444, https://doi.org/10.1111/j.1365-2672.2010.04900.x (2011).
Shanks, R. M. et al. An Eye to a Kill: Using Predatory Bacteria to Control Gram-Negative Pathogens Associated with Ocular Infections. PLoS One 8, e66723, https://doi.org/10.1371/journal.pone.0066723 (2013).
Russo, R. et al. Susceptibility of Select Agents to Predation by Predatory Bacteria. Microorganisms 3, 903–912, https://doi.org/10.3390/microorganisms3040903 (2015).
Shatzkes, K., Connell, N. D. & Kadouri, D. E. Predatory bacteria: a new therapeutic approach for a post-antibiotic era. Future Microbiol 12, 469–472, https://doi.org/10.2217/fmb-2017-0021 (2017).
Kadouri, D. E., To, K., Shanks, R. M. & Doi, Y. Predatory bacteria: a potential ally against multidrug-resistant Gram-negative pathogens. PLoS One 8, e63397, https://doi.org/10.1371/journal.pone.0063397 (2013).
Dharani, S., Kim, D. H., Shanks, R. M. Q., Doi, Y. & Kadouri, D. E. Susceptibility of colistin-resistant pathogens to preda tory bacteria. Res Microbiol https://doi.org/10.1016/j.resmic.2017.09.001 (2017).
Gupta, S., Tang, C., Tran, M. & Kadouri, D. E. Effect of Predatory Bacteria on Human Cell Lines. PLoS One 11, e0161242, https://doi.org/10.1371/journal.pone.0161242 (2016).
Romanowski, E. G. et al. Predatory bacteria are nontoxic to the rabbit ocular surface. Sci Rep 6, 30987, https://doi.org/10.1038/srep30987 (2016).
Shatzkes, K. et al. Examining the safety of respiratory and intravenous inoculation of Bdellovibrio bacteriovorus and Micavibrio aeruginosavorus in a mouse model. Sci Rep 5, 12899, https://doi.org/10.1038/srep12899 (2015).
Shatzkes, K. et al. Predatory Bacteria Attenuate Klebsiella pneumoniae Burden in Rat Lungs. MBio 7, https://doi.org/10.1128/mBio.01847-16 (2016).
Shatzkes, K. et al. Examining the efficacy of intravenous administration of predatory bacteria in rats. Sci Rep 7, 1864, https://doi.org/10.1038/s41598-017-02041-3 (2017).
Shatzkes, K. et al. Effect of predatory bacteria on the gut bacterial microbiota in rats. Sci Rep 7, 43483, https://doi.org/10.1038/srep43483 (2017).
Richards, M. J., Edwards, J. R., Culver, D. H. & Gaynes, R. P. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect Control Hosp Epidemiol 21, 510–515, https://doi.org/10.1086/501795 (2000).
Iguchi, A. et al. Genome evolution and plasticity of Serratia marcescens, an important multidrug-resistant nosocomial pathogen. Genome Biol Evol 6, 2096–2110, https://doi.org/10.1093/gbe/evu160 (2014).
Rodrigues, A. P. et al. Virulence factors and resistance mechanisms of Serratia marcescens. A short review. Acta Microbiol Immunol Hung 53, 89–93, https://doi.org/10.1556/AMicr.53.2006.1.6 (2006).
Stella, N. A. et al. SlpE is a calcium-dependent cytotoxic metalloprotease associated with clinical isolates of Serratia marcescens. Res Microbiol 168, 567–574, https://doi.org/10.1016/j.resmic.2017.03.006 (2017).
Labbate, M. et al. Quorum-sensing regulation of adhesion in Serratia marcescens MG1 is surface dependent. J Bacteriol 189, 2702–2711, https://doi.org/10.1128/JB.01582-06 (2007).
Shanks, R. M. et al. A Serratia marcescens OxyR homolog mediates surface attachment and biofilm formation. J Bacteriol 189, 7262–7272, https://doi.org/10.1128/JB.00859-07 (2007).
Shanks, R. M., Stella, N. A., Brothers, K. M. & Polaski, D. M. Exploitation of a “hockey-puck” phenotype to identify pilus and biofilm regulators in Serratia marcescens through genetic analysis. Can J Microbiol 62, 83–93, https://doi.org/10.1139/cjm-2015-0566 (2016).
Baker, M. et al. Measuring and modelling the response of Klebsiella pneumoniae KPC prey to Bdellovibrio bacteriovorus predation, in human serum and defined buffer. Sci Rep 7, 8329, https://doi.org/10.1038/s41598-017-08060-4 (2017).
Dattner, I. et al. Modelling and parameter inference of predator-prey dynamics in heterogeneous environments using the direct integral approach. J R Soc Interface 14 https://doi.org/10.1098/rsif.2016.0525 (2017).
Koval, S. F. & Hynes, S. H. Effect of paracrystalline protein surface layers on predation by Bdellovibrio bacteriovorus. J Bacteriol 173, 2244–2249 (1991).
Negus, D. et al. Predator Versus Pathogen: How Does Predatory Bdellovibrio bacteriovorus Interface with the Challenges of Killing Gram-Negative Pathogens in a Host Setting? Annu Rev Microbiol 71, 441–457, https://doi.org/10.1146/annurev-micro-090816-093618 (2017).
Dwidar, M., Nam, D. & Mitchell, R. J. Indole negatively impacts predation by Bdellovibrio bacteriovorus and its release from the bdelloplast. Environ Microbiol 17, 1009–1022, https://doi.org/10.1111/1462-2920.12463 (2015).
Dashiff, A., Keeling, T. G. & Kadouri, D. E. Inhibition of predation by Bdellovibrio bacteriovorus and Micavibrio aeruginosavorus via host cell metabolic activity in the presence of carbohydrates. Appl Environ Microbiol 77, 2224–2231, https://doi.org/10.1128/AEM.02565-10 (2011).
Shimuta, K. et al. The hemolytic and cytolytic activities of Serratia marcescens phospholipase A (PhlA) depend on lysophospholipid production by PhlA. BMC Microbiol 9, 261, https://doi.org/10.1186/1471-2180-9-261 (2009).
Matsumoto, K. Role of bacterial proteases in pseudomonal and serratial keratitis. Biol Chem 385, 1007–1016, https://doi.org/10.1515/BC.2004.131 (2004).
Williamson, N. R., Fineran, P. C., Leeper, F. J. & Salmond, G. P. The biosynthesis and regulation of bacterial prodiginines. Nat Rev Microbiol 4, 887–899, https://doi.org/10.1038/nrmicro1531 (2006).
Suryawanshi, R. K., Patil, C. D., Koli, S. H., Hallsworth, J. E. & Patil, S. V. Antimicrobial activity of prodigiosin is attributable to plasma-membrane damage. Nat Prod Res 31, 572–577, https://doi.org/10.1080/14786419.2016.1195380 (2017).
Zhang, L., Morrison, A. J. & Thibodeau, P. H. Interdomain Contacts and the Stability of Serralysin Protease from Serratia marcescens. PLoS One 10, e0138419, https://doi.org/10.1371/journal.pone.0138419 (2015).
Wu, D., Ran, T., Wang, W. & Xu, D. Structure of a thermostable serralysin from Serratia sp. FS14 at 1.1 A resolution. Acta Crystallogr F Struct Biol Commun 72, 10–15, https://doi.org/10.1107/S2053230X15023092 (2016).
Marty, K. B., Williams, C. L., Guynn, L. J., Benedik, M. J. & Blanke, S. R. Characterization of a cytotoxic factor in culture filtrates of Serratia marcescens. Infect Immun 70, 1121–1128 (2002).
Shanks, R. M. et al. Identification of SlpB, a Cytotoxic Protease from Serratia marcescens. Infect Immun 83, 2907–2916, https://doi.org/10.1128/IAI.03096-14 (2015).
Dashiff, A. & Kadouri, D. E. Predation of oral pathogens by Bdellovibrio bacteriovorus 109J. Mol Oral Microbiol 26, 19–34, https://doi.org/10.1111/j.2041-1014.2010.00592.x (2011).
Ghigo, J. M. & Wandersman, C. A fourth metalloprotease gene in Erwinia chrysanthemi. Res Microbiol 143, 857–867 (1992).
Chavda, K. D., Chen, L., Jacobs, M. R., Bonomo, R. A. & Kreiswirth, B. N. Molecular Diversity and Plasmid Analysis of KPC-Producing Escherichia coli. Antimicrob Agents Chemother 60, 4073–4081, https://doi.org/10.1128/aac.00452-16 (2016).
Shanks, R. M., Kadouri, D. E., MacEachran, D. P. & O’Toole, G. A. New yeast recombineering tools for bacteria. Plasmid 62, 88–97, https://doi.org/10.1016/j.plasmid.2009.05.002 (2009).
Brothers, K. M., Stella, N. A., Romanowski, E. G., Kowalski, R. P. & Shanks, R. M. EepR Mediates Secreted-Protein Production, Desiccation Survival, and Proliferation in a Corneal Infection Model. Infect Immun 83, 4373–4382, https://doi.org/10.1128/IAI.00466-15 (2015).
Kalivoda, E. J., Stella, N. A., O’Dee, D. M., Nau, G. J. & Shanks, R. M. Q. The Cyclic AMP-Dependent Catabolite Repression System of Serratia marcescens Mediates Biofilm Formation through Regulation of Type 1 Fimbriae. Applied and Environmental Microbiology 74, 3461–3470, https://doi.org/10.1128/AEM.02733-07 (2008).
Butterworth, M. B., Zhang, L., Liu, X., Shanks, R. M. & Thibodeau, P. H. Modulation of the epithelial sodium channel (ENaC) by bacterial metalloproteases and protease inhibitors. PLoS One 9, e100313, https://doi.org/10.1371/journal.pone.0100313 (2014).
Zhang, L., Conway, J. F. & Thibodeau, P. H. Calcium-induced folding and stabilization of the Pseudomonas aeruginosa alkaline protease. J Biol Chem 287, 4311–4322, https://doi.org/10.1074/jbc.M111.310300 (2012).
Butterworth, M. B., Zhang, L., Heidrich, E. M., Myerburg, M. M. & Thibodeau, P. H. Activation of the epithelial sodium channel (ENaC) by the alkaline protease from Pseudomonas aeruginosa. J Biol Chem 287, 32556–32565, https://doi.org/10.1074/jbc.M112.369520 (2012).
Kalivoda, E. J. et al. Cyclic AMP negatively regulates prodigiosin production by Serratia marcescens. Res Microbiol 161, 158–167, https://doi.org/10.1016/j.resmic.2009.12.004 (2010).
Shanks, R. M. et al. Serratamolide is a hemolytic factor produced by Serratia marcescens. PLoS One 7, e36398, https://doi.org/10.1371/journal.pone.0036398 (2012).
Acknowledgements
We would like to thank Sukhwinder Singh and colleagues at Rutgers-NJMS Flow Cytometry Core Facility for his technical assistance and input. The research was sponsored by the Research to Prevent Blindness, Inc., New York, New York; Eye and Ear Foundation of Pittsburgh and NIH grants EY024785, EY08098 to R.M.Q.S and NIH grant DK083284 to P.H.T., as well as the U.S. Army Research Office and the Defense Advanced Research Projects Agency and was accomplished under Cooperative Agreement Number W911NF-15-2-0036 to D.E.K, R.M.Q.S. The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the official policies, either expressed or implied, of the Army Research Office, DARPA, or the U.S. Government. The U.S. Government is authorized to reproduce and distribute reprints for Government purposes notwithstanding any copyright notation hereon.
Author information
Authors and Affiliations
Contributions
D.E.K., R.M.Q.S. and C.J.G. conceived and designed the experiments. C.J.G., M.E., M.T. N.A.S. and A.P. performed the experiments. S.G. assisted in predator preparation and training. C.J.G. and D.E.K. analyzed the data. Electron microscopy imaging was done by J.M.F. and N.A.S. P.H.T. purified and provided the metalloproteases. J.D.C., N.A.S. and R.M.Q.S. provided all S. marcescens mutants. C.J.G. and D.E.K. drafted the manuscript. R.M.Q.S. revised the manuscript. D.E.K. supervised the study. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Garcia, C.J., Pericleous, A., Elsayed, M. et al. Serralysin family metalloproteases protects Serratia marcescens from predation by the predatory bacteria Micavibrio aeruginosavorus. Sci Rep 8, 14025 (2018). https://doi.org/10.1038/s41598-018-32330-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-32330-4
Keywords
This article is cited by
-
A natural symbiotic bacterium drives mosquito refractoriness to Plasmodium infection via secretion of an antimalarial lipase
Nature Microbiology (2021)
-
Biologically active pigment and ShlA cytolysin of Serratia marcescens induce autophagy in a human ocular surface cell line
BMC Ophthalmology (2020)
-
Compounds affecting predation by and viability of predatory bacteria
Applied Microbiology and Biotechnology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.