Abstract
Tissue-specific transcriptional programs control most biological phenotypes, including disease states such as cancer. However, the molecular details underlying transcriptional specificity is largely unknown, hindering the development of therapeutic approaches. Here, we describe novel experimental reporter systems that allow interrogation of the endogenous expression of HIF2A, a critical driver of renal oncogenesis. Using a focused CRISPR-Cas9 library targeting chromatin regulators, we provide evidence that these reporter systems are compatible with high-throughput screening. Our data also suggests redundancy in the control of cancer type-specific transcriptional traits. Reporter systems such as those described here could facilitate large-scale mechanistic dissection of transcriptional programmes underlying cancer phenotypes, thus paving the way for novel therapeutic approaches.
Similar content being viewed by others
Introduction
Genetic loss-of-function screening is a powerful approach for mechanistic dissection of biological phenomena. Functional screens can also be useful for the identification of molecular mediators in various disease states. Given the recent methodological developments, such as improved RNAi and CRISPR-Cas91,2, performing genetic screens in vitro has become relatively easy, with the limiting factor now being suitable functional assays that model relevant biology accurately. For example, in the field of cancer biology, in vitro assays that measure cell proliferation or survival under various conditions can be powerful3,4. However, many relevant cancer phenotypes cannot be easily modelled in vitro. On the other hand, even though some in vivo genetic screens have been successful5,6, large-scale genetic screening in vivo remains a challenge. Thus, novel functional assays are needed for high-throughput genetic dissection of clinically relevant cancer dependencies.
An example of a cancer phenotype that displays experimental context-specificity is the role of hypoxia-inducible factor 2 (HIF2A), encoded by the EPAS1 gene, as a driver of renal tumorigenesis. HIF2A is activated in clear cell renal cell carcinoma (ccRCC) through mutations in the von Hippel-Lindau tumour suppressor gene (VHL), which under normal conditions targets HIF2A for proteosomal degradation7. Under hypoxia and in cancers that have lost VHL, a stereotypical genetic event in ccRCC8, HIF2A accumulates and drives the expression of multiple target genes, some which mediate renal cancer growth and metastasis7,9,10. Despite abundant evidence demonstrating the relevance of the VHL-HIF2A pathway for ccRCC growth in vivo, HIF2A inhibition directly or via VHL restoration does not have an obvious growth phenotype in tissue culture systems7.
We set out to develop experimental systems that would allow genetic interrogation of the molecular mechanisms underlying tissue-specific HIF2A expression in ccRCC. We established and validated fluorescence-based reporters of endogenous HIF2A expression in ccRCC cell lines. As a proof-of-principle, we conducted a pooled CRISPR-Cas9 screen focusing on chromatin factors, demonstrating the feasibility of using these systems in high-throughput genetic screens. Our screening data suggest that HIF2A expression in ccRCC may be supported by a robust regulatory network, which is insensitive to perturbations in individual chromatin factors. We conclude that genetically engineered systems such as those described here could facilitate large-scale dissection of essential transcriptional programmes underlying cancer phenotypes, with potential implications for the identification of novel targets for cancer therapy development.
Results
CRISPR-Cas9-based knock-in of a fluorescent reporter into the endogenous EPAS1 locus
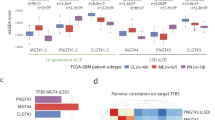
Analysis of RNA-seq data from 26 different tumour types from the TCGA cohort11 revealed that HIF2A mRNA was highly expressed in ccRCC when compared to other tumour types (Fig. 1a). HIF2A mRNA expression was also elevated in ccRCC when compared to normal kidney (Fig. 1a). Given that VHL mutations are remarkably specific for ccRCC12,13, this suggested the possibility that high HIF2A mRNA expression levels in ccRCC may modulate the potential at which VHL loss promotes tumorigenesis. To analyse the molecular basis of this transcriptional specificity, we developed a CRISPR-Cas9-based knock-in strategy for the integration of an mCherry fluorescence reporter into the endogenous EPAS1 locus of ccRCC cells using homology-directed repair (Fig. 1b), an approach that has previously been successfully applied in other biological contexts14,15. A T2A self-cleaving peptide was placed between endogenous HIF2A and mCherry, ensuring that HIF2A and mCherry proteins were produced at a stoichiometric ratio16,17. After hygromycin selection, surviving cells were sorted into single cell clones based on mCherry fluorescence, followed by genetic analysis of the integration site. Of the cell lines tested, we identified three clones (henceforth referred to as HIF2A-mCherry-single-cell-clone 1–3 or H2AmC1–3), all derived from UOK101 cells, a ccRCC-derived VHL-mutant cell line that transcriptionally resembles ccRCCs with poor prognosis18,19, with the correct integration pattern both at the genomic and cDNA level (Fig. 1c,d). No wild-type product was observed by genomic PCR indicating homozygous integration at both EPAS1 alleles (Fig. 1c). Sanger sequencing verified the integration site (Fig. 1e and Supplementary Fig. 1). mCherry positivity of the clones was confirmed by flow cytometry (Fig. 1f) and confocal microscopy (Fig. 1g).
Generation of endogenous HIF2A reporter systems. (a) TCGA data analysis of HIF2A mRNA expression in ccRCC compared to different tumour types and normal kidney. P-value by Wilcoxon rank sum test. (b) Schematic of CRISPR-Cas9-based knock-in strategy of an mCherry fluorescent gene into the exon 16 of EPAS1. Plasmid template consists of an mCherry fluorescent gene with a hygromycin selection marker that are flanked by homology arms (HA). Sequencing primers used for genomic and cDNA amplifications in (c) and (d), respectively, are shown. (c) Genomic amplification of the HIF2A-mCherry integration site in single-cell derived clones. WC, water control. GC, genomic control. (d) cDNA amplification of the HIF2A-mCherry integration site. WC, water control. GC, genomic control. (e) Sanger sequencing of the HIF2A-mCherry integration sites in H2AmC2 cells. (f) FACS analysis of mCherry fluorescence in the three H2AmC clones compared to the parental UOK101 cells. (g) mCherry fluorescence in the three H2AmC clones compared to the parental control. DAPI in blue, mCherry in red.
Functional validation of endogenous HIF2A reporter systems
To functionally validate the H2AmC1-3 reporter systems we evaluated mCherry expression alongside HIF2A mRNA and protein expression. We used lentivirally delivered CRISPR interference (CRISPRi)20, i.e. sgRNA-mediated recruitment of a repressive dCas9-KRAB protein to a specific genomic locus, to inhibit endogenous HIF2A mRNA expression using three sgRNA constructs (iHIF2A-1, iHIF2A-2, iHIF2A-3) targeting the EPAS1 transcription start site (Fig. 2a). In comparison to the non-targeting sgRNA control, iHIF2A-1 and iHIF2A-2 caused a significant reduction in HIF2A mRNA level while no reduction was observed in iHIF2A-3 transduced cells (Fig. 2b). These effects translated into similar changes at the protein level (Fig. 2c). The reporter lines expressed a strong HIF2A protein band of the expected size, and the HIF2A antibody did not detect any larger bands of ~150 kDa, the expected size of a HIF2A-mCherry fusion protein, indicating that the T2A peptide was efficiently cleaved in our systems (Supplementary Fig. 2a). Importantly, flow cytometry on the H2AmC2 cells showed that mCherry fluorescence in the non-targeting sgRNA control cells stayed close to the level seen in untransduced cells and cells transduced with iHIF2A-3, while iHIF2A-1 and iHIF2A-2 transduced cells showed a clear reduction in mCherry fluorescence, closely mimicking the parental UOK101 cells (Fig. 2d). These results were replicated in the H2AmC3 cells (Fig. 2e). HIF2A targeting in the H2AmC1 cells resulted in only modest changes in mCherry fluorescence, suggesting the possibility that in this clone mCherry was expressed at least partially independently of HIF2A (data not shown). H2AmC1 was therefore excluded from further analysis. To further test the interdependence of HIF2A mRNA, protein and mCherry fluorescence in our systems we transduced both H2AmC2 and H2AmC3 cells with wildtype VHL. As expected, this resulted in a robust reduction in HIF2A protein expression (Fig. 2f). However, no change was observed in HIF2A mRNA expression or mCherry fluorescence (Fig. 2g and Supplementary Fig. 2b,c). These results confirmed that mCherry activity in the H2AmC2 and H2AmC3 systems reflected HIF2A mRNA expression, and they also suggested that HIF2A does not significantly regulate its own expression.
Validation of endogenous HIF2A reporter systems. (a) Schematic of CRISPRi-based inhibition of reporter activity. RNAPol, RNA Polymerase II. KRAB, Kruppel associated box. (b) Relative HIF2A mRNA levels in H2AmC2 and H2AmC3 dCas9 cells transduced with HIF2A-targeting sgRNAs or a non-targeting control. *P < 0.05, **P < 0.01, ***P < 0.005, n.s., non-significant. P-value by Student’s t-test. (c) Western blot analysis of H2AmC2 and H2AmC3 dCas9 cells transduced with HIF2A-targeting sgRNAs or a non-targeting control. B-actin acts as a loading control. Full-length blots are presented in Supplementary Fig. 5. (d) FACS analysis of mCherry fluorescence in H2AmC2 cells transduced with different HIF2A-targeting sgRNA constructs. Untransduced H2AmC2 and UOK101 cells serve as a positive and negative control, respectively. (e) FACS analysis of mCherry fluorescence in H2AmC3 cells transduced with different HIF2A tandem sgRNAs compared to controls. (f) Western blot analysis of VHL-reintroduced H2AmC2 and H2AmC3 cells compared to the empty vector (EV) control. B-actin acts as a loading control. Full-length blots are presented in Supplementary Fig. 5. (g) FACS analysis of mCherry fluorescence in H2AmC2 and H2AmC3 cells transduced with HA-VHL and empty vector.
FACS-based evaluation of reporter activity at the single cell level
To test the possibility of using the H2AmC2 and H2AmC3 cells in pooled CRISPR-Cas9-based screening, we transduced them with the HIF2A-targeting sgRNAs under conditions of low multiplicity of infection (MOI) and evaluated the effects on mCherry fluorescence. For these experiments, the sgRNAs were cloned into vectors that also expressed EGFP, which allowed the evaluation of the relationship between the presence of HIF2A-targeting constructs and mCherry activity at the single cell level in a mixed population. Using a population in which ~10% of the cells were EGFP positive (Fig. 3a), leading to an estimated MOI of 0.11 with ~95% of the EGFP positive cells being infected by a single virus, we evaluated the percentage of EGFP positive cells within the population of lowest 10% of mCherry fluorescence (Fig. 3b). As expected, the non-targeting sgRNA or iHIF2A-3 demonstrated no enrichment of EGFP positive cells within the mCherry low population, however, there was a marked increase in EGFP positive cells within the mCherry low fraction in the population that contained 10% of iHIF2A-1 or iHIF2A-2 (Fig. 3c,d). Next, we evaluated the effects of different mCherry population cut-offs on the level of enrichment for EGFP positive cells. This confirmed that decreasing levels of mCherry fluorescence selected for an increasing fraction of EGFP positive cells in iHIF2A-1 and iHIF2A-2 transduced cells whereas no changes were observed in cells transduced with the non-targeting sgRNA or iHIF2A-3 (Fig. 3e and Supplementary Fig. 3a). However, even the lowest 1% of mCherry expressing cells had a significant fraction of EGFP negative cells. Finally, we wanted to test the reporter systems in conjunction with CRISPR-Cas9-based mutational gene targeting. In order to achieve uniform Cas9 expression, we first isolated single cell-derived H2AmC2 and H2AmC3 subclones transduced with a constitutively expressed Cas9-EGFP (Fig. 3f). We then tested the effects of single HIF2A-targeting sgRNAs on mCherry fluorescence at low MOI. As expected, a combined analysis of both control and HIF2A-targeted cells revealed an enrichment of HIF2A-targeted cells in the population of lowest 10% of mCherry fluorescence (Fig. 3g and Supplementary Fig. 3b). In sum, these experiments gave proof-of-principle evidence that a pooled CRISPR-Cas9 genetic screen could be coupled to FACS-based isolation of low mCherry cells in order to identify genes that support HIF2A expression in ccRCC.
HIF2A sgRNA enrichment as a function of mCherry depletion. (a) EGFP fluorescence in sgRNA-transduced H2AmC2 CRISPRi cells. (b) mCherry fluorescence of a H2AmC2 population that contains 10% of EGFP+ sgRNA-expressing cells. The bottom 10% of mCherry population was selected for further analysis of EGFP fluorescence. (c,d) EGFP fluorescence in H2AmC2 (c) and H2AmC3 (d) cells with the lowest 10% mCherry fluorescence from analysis similar to that shown in panel (b). In iHIF2A-1 and iHIF2A-2 cells the fraction of EGFP positive cells is increased compared to the unselected population (panel (a)) or the control or iHIF2A-3 cells, indicating enrichment of HIF2A-targeting sgRNAs in the mCherry low population. (e) Relative abundance of EGFP positive (EGFP+) and negative (EGFP-) H2AmC2 cells in populations with different levels of mCherry fluorescence. (f) Western blot analysis of H2AmC2 and H2AmC3 cells transduced with lenti-Cas9-EGFP compared to untransduced control cells. B-actin act as a loading control. Full-length blots are presented in Supplementary Fig. 5. Accompanying EGFP fluorescence analysis of single cell derived clones of the respective H2AmC2 and H2AmC3 lenti-Cas9-EGFP pools on the right. (g) Left, mCherry fluorescence in H2AmC3-Cas9 cells transduced with non-targeting control sgRNA (CTRL), sgEPAS1-1 or sgEPAS1-5. Right, normalized cell counts for each construct in the combined population with the lowest 10% of mCherry fluorescence.
A pooled CRISPR-Cas9 screen for chromatin regulators supporting HIF2A expression
Chromatin factors as supporters of cancer-specific transcriptional programmes are emerging as potential therapeutic target molecules21. In order to interrogate the chromatin factor dependencies of HIF2A expression in ccRCC, we generated a focused library of 7617 sgRNAs targeting a total of 836 known and potential chromatin regulators as well as HIF2A as a positive control (Supplementary Tables 1 and 2). The library contained on average nine sgRNAs per gene and 100 non-targeting negative controls. A total of 120 million H2AmC2 and H2AmC3 cells were transduced with the chromatin library at a low MOI, resulting in ~12 million infected cells and thus ensuring >1000X representation of sgRNAs. We then used FACS to isolate the population with the lowest mCherry expression and assessed the representation of sgRNAs by high-throughput sequencing (Fig. 4a). Unsorted cells propagated for an equal time were used as controls. We observed a good correlation between the experimental systems (Fig. 4b and Supplementary Table 2). Only <1.5% of sgRNAs had <5 counts/million reads in the initial population and excluding such sgRNAs retained 100% of the target genes (Fig. 4c) with most genes retaining all sgRNAs (Fig. 4d). The excluded sgRNAs were either lost during library preparation due to technical reasons, or they targeted genes that affected cell viability, such as SF3B3 and PCNA, genes included as control genes in the library. A formal analysis comparing the unsorted control samples to the plasmid that was used for virus production confirmed that known essential genes22 were highly represented among the most significantly depleted genes (Supplementary Fig. 4). Looking specifically at HIF2A as a positive control, we found that 8/9 sgRNAs showed evidence of enrichment in the mCherry low population with good correlation between the two reporter systems (Fig. 4e), one sgRNA was excluded due to low representation. In a global analysis, HIF2A sgRNAs stood out as being the most enriched, with 6/10 top scoring sgRNAs targeting HIF2A (Fig. 4f). In a gene level analysis combining data from all sgRNAs for each gene, HIF2A was by far the most strongly enriched gene in the mCherry low population (Fig. 4g). To test the statistical significance of the result we used a permutation-based approach to calculate empirical P-values for the gene level enrichment scores. Comparing the observed data to an expected background clearly highlighted HIF2A as the most significant hit with an adjusted P-value of <0.001 (nominal P-value < 1.195e-06) (Fig. 4h,i). However, no other factor reached statistical significance, a result confirmed by manual inspection of the top-scoring sgRNAs: no other gene contained more than one sgRNA within the top 50 of the list whereas non-targeting control sgRNAs appeared twice. In sum, the only significant hit from our screen was the positive control HIF2A, confirming the technical validity of our approach, but we found no evidence for individual chromatin factors that would be required for the strong HIF2A expression in ccRCC.
A CRISPR-Cas9 screen targeting chromatin regulators. (a) Schematic showing the screening process. A pool of sgRNAs targeting chromatin factors was cloned into a lentiviral vector. H2AmC2 and H2AmC3 clones expressing Cas9 were transduced with the virus, followed by isolation of the cells with the lowest 10% of mCherry fluorescence. sgRNA enrichment was assessed by high-throughput sequencing. (b) Correlation plot of the normalized sgRNA counts in unsorted H2AmC2 and H2AmC3 cells. (c) Breakdown of total sgRNAs, control sgRNAs and total genes analysed in the screen after the removal of sgRNAs with less than 5 normalized counts in the unsorted population. (d) Number of sgRNAs analysed per gene. (e) Fold enrichment of HIF2A sgRNAs in H2AmC2 and H2AmC3 cells. (f) Average enrichment of all sgRNAs analysed in the screen with a snapshot of the top 10 scoring sgRNAs. (g) Z-scores of the median enrichment for all genes in the H2AmC2 and H2AmC3 cells. (h) Permutation analysis (1000x) of gene enrichment scores. Observed data in red, simulated data in blue. (i) Distribution of FDR-corrected P-values for each gene. PCC, Pearson’s correlation coefficient.
Discussion
We report the development of endogenous reporter systems for HIF2A expression in ccRCC cells and their application in high-throughput functional CRISPR-Cas9-based loss-of-function screening. HIF2A is a critical mediator of ccRCC development and a validated therapeutic target23,24,25. Additionally, varying sensitivity of ccRCC cells to HIF2A inhibition has been attributed to different levels of HIF2A expression as well as acquired mutations in the HIF2A complex23,25. Yet, the mechanisms that regulate the high and specific HIF2A expression in ccRCC are incompletely understood. Given the recent evidence that the expression of oncogenic drivers, such as MYC, can be dependent on single chromatin modifiers3, we explored the chromatin factor dependencies of HIF2A expression using our systems. Despite strong signal for HIF2A, the positive control in the screen, we found little evidence for the existence of individual chromatin factors that would be essential for HIF2A expression in ccRCC. This suggests redundancy within the HIF2A gene regulatory machinery, possibly complicating efforts that aim at targeting HIF2A expression in the therapeutic setting26.
CRISPR-Cas9-based genome editing has opened up unprecedented possibilities for biological research. The general efficiency of CRISPR-Cas9 gene editing across the many possible biological contexts remains to be established, however. For example, the degree to which an intact homology-directed repair (HDR) pathway is required for HDR-targeted gene editing in cancer cell lines remains unclear. ccRCCs often carry mutations in the histone methyltransferase SETD2, which promotes homologous repair27,28,29. This suggests that defects in some DNA repair pathways may be a requirement for ccRCC development, possibly hindering the efficiency of HDR-mediated gene editing in ccRCC cells. Our results show that at least some ccRCC cell lines are amenable for HDR-based gene editing, but the efficiency appears to be lower than what has been reported in some other systems14. Several ccRCC cell lines tested also failed to integrate the fluorescent reporter (data not shown). Alternative strategies that depend on smaller template plasmids and NHEJ-based DNA repair could make gene editing-based development of experimental ccRCC systems more efficient30.
While the strong signal for HIF2A in our genetic screen gives technical validation to both the reporter systems and the screening approach, it remains possible that the systems are not sensitive enough to identify factors that only subtly affect HIF2A expression. It is also possible that some chromatin factors were not efficiently targeted due to technical reasons. An alternative interpretation suggests, however, that simultaneous targeting of multiple chromatin factors may be required for efficient inhibition of HIF2A expression. Combinatorial CRISPR-Cas9 screens could thus represent an interesting avenue forward. Targeting alternative gene sets or performing genome-wide screens could also give new insight into HIF2A regulation. The endogenous transcriptional reporters developed herein could also be suitable for unbiased functional analysis of gene regulatory element function31,32.
In conclusion, we have developed and validated experimental systems for the identification of factors that support HIF2A expression in ccRCC cells. We demonstrate that these systems are compatible with pooled genetic screens. Combined with microscopy they may also be compatible with small molecule screens in an arrayed format. More generally, similar systems could be useful for the identification of transcriptional dependencies in other cancer contexts as well.
Methods
Cells and Reagents
UOK101 cell were obtained from the National Cancer Institute (NCI). The identity of the UOK101 cells has been confirmed by Sanger sequencing-based detection of a previously identified homozygous VHL mutation19,33 in June 2018, and STR analysis confirmed that the UOK101 cells had not been cross-contaminated with other commonly used cell lines. The cancer cell lines have been confirmed negative for mycoplasma by biannual tests using the MycoAlert™ Mycoplasma Detection Kit (Lonza, LT07-318). UOK101 cells were cultured in RPMI-1640. HEK293T cells, used for lentivirus production, were cultured in DMEM. Both media were supplemented with 10% fetal bovine serum (FBS), penicillin (100 U ml−1) and streptomycin (μg ml−1). Cell lines were used in experiments within 10 passages from thawing.
Plasmids and cloning
Px330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene #42230)34, pKLV-U6gRNA(BbsI)-PGKPuro2ABFP (Addgene #50946)35 and pKLV-U6gRNA(BbsI)-PGKHyg2AEGFP (adapted from Addgene #50946)35 were used for sgRNA delivery. The HDR template for targeted gene knock-in was cloned into a truncated pcDNA3.1(−) (Thermo #V79520). The template was generated by step-wise insertion of T2A-mCherry, PCR amplified from pU6-sgCXCR4-2 (Addgene #46917)20, PGK-HygR, PCR amplified from pRetroX-Tight-Hygro (Clontech #631034) and the two homology arms that were PCR amplified from genomic DNA by PCR. pHR-SFFV-dCas9-BFP-KRAB (Addgene #46911)20 and tandem HIF2A-targeting sgRNA constructs, cloned as previously described10, were used for HIF2A CRISPRi. Lenti-Cas9-EGFP (adapted from Addgene #52962)36 was used for HIF2A mutagenesis. pLVX-puro (Clontech #632164) was used to express HA-VHL (Addgene #19234)37. Primer and sgRNA sequences used in this study are listed in Supplementary Table 3.
Quantitative RT-PCR
Total RNA from cells was purified using RNAzol RT (Sigma) according to the manufacturer’s protocol. cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Thermo). qRT-PCR was performed using the StepOnePlus instrument (Thermo) with pre-designed TaqMan gene expression assays (Thermo): EPAS1 (Hs01026149_m1), CXCR4 (Hs00607978_s1), VHL (Hs03046964_s1) and TBP (Hs00427620_m1). Signal was quantified using the double delta Ct method and normalized to TBP as the housekeeping control.
Western Blotting
Whole-cell extract was used for Western blotting by lysing cells in RIPA buffer (Sigma-Aldrich, Life Science) supplemented with protease inhibitors. Protein was quantified using the PierceTM BCA Protein Assay Kit (ThermoScientific). Primary antibodies for HIF2A (Novus Biologicals NB100-122), FLAG (Sigma #F3165), VHL (BD Biosciences #564183) and β-actin (Sigma #A1978) were used. Secondary antibodies were polyclonal goat anti-mouse immunoglobulins/HRP (Dako) and polyclonal goat anti-rabbit immunoglobulins/HRP (Dako).
Lentiviral Transduction
Lentivirus was produced by transfecting HEK293T cells with the psPAX2 and pMD2.G viral packaging plasmids using FuGENE 6 transfection reagent (Promega E269A). Viral supernatant was harvested 48 hours after transfection and filtered through a 0.45 mm PVDF syringe filter. Cells were transduced with the virus along with 8 ug/ml of polybrene (Milipore). The number of positive cells was assessed through FACS analysis.
Fluorescence-Activated Cell Sorting (FACS)
Cells were analysed for fluorescence on an LSR Fortessa (BD Biosciences). Single cells were detected on the basis of FSC-A, FSC-H and SSC-A. mCherry (561 nm/610 nm), EGFP (488 nm/509 nm), and BFP (383 nm/445 nm) fluorescence was measured. Cell sorting was performed on an Influx cell sorter (BD Biosciences) using the same settings as described above. Single cells were sorted onto individual wells of a 96-well plate containing cancer cell-conditioned RPMI-1640 supplemented with 10% fetal bovine serum (FBS), penicillin (100 U ml−1) and streptomycin (100 μg ml−1). FlowJo software was used to analyse data obtained.
Immunofluorescence
Cells grown on coverslips in 6-well plates were washed with PBS and incubated with 4% PFA in PBS for 10 min at room temperature (RT). Cells were permeabilized with 0.5% Triton X-100 (Sigma). A drop of ProLong Diamond Antifade Mountant with DAPI (Life Technologies) was placed on microscope slides and coverslips with cells were mounted on them. After 5 min incubation at RT, imaging was done on a Leica TCS SP5 confocal microscope.
Chromatin Factor sgRNA Library Production
Genes encoding known or putative chromatin factors were identified from previously curated lists38,39,40,41 (Supplementary Table 1). The library also contained additional control genes, such as known essential genes, as well as 100 non-targeting control sgRNAs22. The sequences for the sgRNAs were taken from Wang et al.22 with nine sequences selected for each gene. Oligos were ordered from Custom Array Inc. Oligos were amplified and cloned into pKLV2-U6gRNA5(BbsI)-PGKpuro2ABFP-W (Addgene #67974)2 by Gibson assembly42.
Pooled CRISPR-Cas9 Screening
The lentiviral sgRNA library was produced using HEK293T cells as described above. H2AmC2 and H2AmC3 cells were first transduced with the lentiviral library at varying concentrations to determine the amount needed for a MOI of ~0.11. A total of twelve 15 cm dishes were seeded at 5 million cells each. Twenty-four hours later, i.e. after one doubling time, ~120 million cells were transduced with the lentiviral library at a low MOI, leading to ~10% positive cells and ensuring a >1000x representation of the library. Virus was removed the following day. Puromycin treatment, starting 2 days after transduction, was then applied for a total of 4 days. Three weeks post-transduction, the lowest 10% of the mCherry population from both H2AmC2 and H2AmC3 cells were isolated by FACS and the cells were pelleted along with their respective unsorted controls. DNA was extracted and amplified for the region containing the sgRNAs. Samples were then purified using an Agencourt AMPure XP beads purification protocol. Purified samples were pooled and quantified with Qubit before sending for Illumina sequencing an a HiSeq4000 instrument.
Sequencing results were aligned using Bowtie and normalised to one million reads. Normalised counts of less than five were removed from the analysis. The fold change enrichment was calculated for each sgRNA in both H2AmC2 and H2AmC3 by calculating the fold change of the sorted samples compared to their respective unsorted control sample. Next, the median of all the remaining sgRNAs per gene in each system was calculated. This data was normalised to obtain a z-score for each gene. The average of the normalised median enrichment for each gene was then calculated between the two systems. For each gene, empirical P-values were calculated using a resampling-based method with 1000 permutations and multiple testing correction using FDR.
Statistical Analysis
Statistical analyses were conducted in R and Microsoft Excel. P-values lower than 0.05 were considered statistically significant. For qRT-PCR three independent experiments are shown, each of which is the average of three technical replicates, unless stated otherwise in the figure legends.
Data availability
The TCGA data set was downloaded from the TCGA data portal at https://tcga-data.nci.nih.gov/. The datasets and reagents generated during the current study are available from the corresponding author on reasonable request.
References
Fellmann, C. et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 5, 1704–1713 (2013).
Tzelepis, K. et al. A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Rep. 17, 1193–1205 (2016).
Zuber, J. et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528 (2011).
Prahallad, A. et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483, 100–103 (2012).
Knott, S. R. V. et al. Asparagine bioavailability governs metastasis in a model of breast cancer. Nature 554, 378–381 (2018).
Possik, P. A. et al. Parallel In Vivo and In Vitro Melanoma RNAi Dropout Screens Reveal Synthetic Lethality between Hypoxia and DNA Damage Response Inhibition. Cell Rep. 9, 1375–1386 (2014).
Kaelin, W. G. Von Hippel-Lindau disease. Annu. Rev. Pathol. 2, 145–173 (2007).
Network, C. G. A. R. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 (2013).
Vanharanta, S. et al. Epigenetic expansion of VHL-HIF signal output drives multiorgan metastasis in renal cancer. Nat. Med. 19, 50–56 (2013).
Rodrigues, P. et al. NF-kappaB-dependent lymphoid enhancer co-option promotes renal carcinoma metastasis. Cancer Discov. 8, 850–865 (2018).
Grossman, R. L. et al. Toward a Shared Vision for Cancer Genomic Data. N. Engl. J. Med. 375, 1109–1112 (2016).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Cerami, E. et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Yang, H. et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/cas-mediated genome engineering. Cell 154, 1370–1379 (2013).
Rojas-Fernandez, A. et al. Rapid generation of endogenously driven transcriptional reporters in cells through CRISPR/Cas9. Sci. Rep. 5, 9811, https://doi.org/10.1038/srep09811 (2015).
Szymczak, A. L. & Vignali, D. A. A. Development of 2A peptide-based strategies in the design of multicistronic vectors. Expert Opin. Biol. Ther. 5, 627–638 (2005).
Donnelly, M. L. L. et al. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: A putative ribosomal ‘skip’. J. Gen. Virol. 82, 1013–1025 (2001).
Sinha, R. et al. Analysis of renal cancer cell lines from two major resources enables genomics-guided cell line selection. Nat. Commun. 8, 15165, https://doi.org/10.1038/ncomms15165 (2017).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Gilbert, L. A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Bradner, J. E., Hnisz, D. & Young, R. A. Transcriptional Addiction in. Cancer. Cell 168, 629–643 (2017).
Wang, T., Wei, J. J., Sabatini, D. M. & Lander, E. S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014).
Wallace, E. M. et al. A small-molecule antagonist of HIF2a is efficacious in preclinical models of renal cell carcinoma. Cancer Res. 76, 5491–5500 (2016).
Cho, H. et al. On-target efficacy of a HIF-2α antagonist in preclinical kidney cancer models. Nature 539, 107–111 (2016).
Chen, W. et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 539, 112–117 (2016).
Welsh, S. J. et al. Inhibition of the hypoxia-inducible factor pathway by a G-quadruplex binding small molecule. Sci. Rep. 3, 2799, https://doi.org/10.1038/srep02799 (2013).
Aymard, F. et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 21, 366–374 (2014).
Carvalho, S. et al. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. Elife 3, e02482, https://doi.org/10.7554/eLife.02482 (2014).
Kanu, N. et al. SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene 34, 5699–5708 (2015).
Suzuki, K. et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149 (2016).
Korkmaz, G. et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat. Biotechnol. 34, 192–198 (2016).
Rajagopal, N. et al. High-throughput mapping of regulatory DNA. Nat. Biotechnol. 34, 167–174 (2016).
Consortium, C. C. L. E. & Consortium, G. D. S. C. Pharmacogenomic agreement between two cancer cell line data sets. Nature 528, 84–87 (2015).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Koike-Yusa, H., Li, Y., Tan, E. P., Velasco-Herrera, M. D. C. & Yusa, K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 32, 267–273 (2014).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nature Methods 11, 783–784 (2014).
Li, L. et al. Hypoxia-inducible factor linked to differential kidney cancer risk seen with type 2A and type 2B VHL mutations. Mol. Cell. Biol. 27, 5381–5392 (2007).
Cheloufi, S. et al. The histone chaperone CAF-1 safeguards somatic cell identity. Nature 528, 218–224 (2015).
Gurard-Levin, Z. A. et al. Chromatin Regulators as a Guide for Cancer Treatment Choice. Mol. Cancer Ther. 15, 1768–1777 (2016).
Consortium, T. U. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46, 2699 (2018).
Gonzalez-Perez, A., Jene-Sanz, A. & Lopez-Bigas, N. The mutational landscape of chromatin regulatory factors across 4,623 tumor samples. Genome Biol. 14, r106 (2013).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Acknowledgements
This work was supported by the Medical Research Council (MC_UU_12022/7). We thank M. Linehan for the UOK101 cells.
Author information
Authors and Affiliations
Contributions
S.V. and M.N.Z. designed the experiments, analysed the data and prepared the manuscript. M.N.Z., S.A.P., S.E.S. and P.R. performed the experiments. S.V. supervised the study.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zaini, M.N., Patel, S.A., Syafruddin, S.E. et al. Endogenous HIF2A reporter systems for high-throughput functional screening. Sci Rep 8, 12063 (2018). https://doi.org/10.1038/s41598-018-30499-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-30499-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.