Abstract
Marine invertebrates associate with diverse microorganisms. Microorganisms even inhabit coelomic fluid (CF), namely, the fluid filling the main body cavity of echinoderms. The CF microbiota potentially impacts host health and disease. Here, we analysed the CF microbiota in two common coastal starfish species, Patiria pectinifera and Asterias amurensis. Although microbial community structures were highly variable among individual starfish, those of P. pectinifera were compositionally similar to those in the surrounding seawater. By contrast, many A. amurensis individuals harboured unique microbes in the CF, which was dominated by the unclassified Thiotrichales or previously unknown Helicobacter-related taxon. In some individuals, the Helicobacter-related taxon was the most abundant genus-level taxon, accounting for up to 97.3% of reads obtained from the CF microbial community. Fluorescence in situ hybridization using a Helicobacter-related-taxon-specific probe suggested that probe-reactive cells in A. amurensis were spiral-shaped, morphologically similar to known Helicobacter species. Electron microscopy revealed that the spiral cells had a prosthecate-like polar appendage that has never been reported in Helicobacter species. Although culture of Helicobacter-related taxon was unsuccessful, this is the first report of the dominance of a Helicobacter-related taxon in invertebrates and non-digestive organs, reshaping our knowledge of the phylogeography of Helicobacter-related taxa.
Similar content being viewed by others
Introduction
Marine animals live with a diverse array of microorganisms. Like the human microbiome, fish gut bacteria have been intensively studied and suggested to play various significant roles in nutrition, immunity, and defence1,2,3. However, relatively little attention has been paid to the marine invertebrates’ association with bacteria, except those of sponge, corals, squid, and animals endemic to deep-sea vents. In deep-sea vents, episymbiotic or endosymbiotic chemolithoautotrophic bacteria that synthesize organics from CO2 provide nutrition to invertebrates, allowing survival in these extreme environments4. In coastal areas, gut microbes of invertebrates may provide their host with growth factors, such as vitamins and amino acids, assist in food digestion and produce antimicrobial agents that protect their host from pathogen infections5. Notably, microbes have been detected not only from the gut and skin but also from haemolymph, namely, the circulatory fluid that is functionally comparable to the blood and lymph of vertebrates6, 7. Playing an analogous role to the endophytes of terrestrial plants, the haemolymph microbiota has important roles in host health and protection8, 9. Although understood in many phyla of marine invertebrates, the natural diversity of host-associated microbiota has little been studied in Echinodermata members such as starfish, sea urchins, and sea cucumbers10, 11.
Coelomic fluid (CF) is the fluid enclosed in the main body cavity of echinoderms, which is equivalent to the haemolymph of invertebrates. The inorganic ion composition of echinoderm CF is slightly different from that of seawater12. Echinoderm CF also contains organic compounds, including amino acids, reduced sugars, proteins, lipids and nitrogenous waste13. In addition, the CF of echinoderms contains circulating cells, called coelomocytes, whose functions range from metabolite transport to immunity14. The coelomocytes phagocytose cell debris and the microorganisms, e.g. microorganisms that invade into the body after injury and autotomy15. Despite this immune system activity, the CF of sea cucumber was reported to harbour unique microbial communities including Sulfurospirillum- and Sulfuricurvum-related taxa within the class Epsilonproteobacteria 16.
Asterias and Patiria are common starfish and keystone species in coastal environments in northern Pacific and Atlantic Oceans. Although these starfish are top predators in some marine ecosystems, they are under attack by viruses and bacteria17. The body wall and CF of starfish exhibits strong antimicrobial activity, probably due to the saponin glycosides, peptidoglycan recognition proteins, and reactive oxygen species18,19,20. As mentioned above, the starfish CF also contains coelomocytes with phagocytic activity21, 22. No commercial benefit of the ability of starfish to synthesize saponins has yet been identified, and starfish are instead increasingly recognized as vermin in fisheries23, 24. Although some microorganisms were isolated in pure cultures from starfish body25, little is known regarding microbial community structures associated with starfish.
In this study, by the combined use of culture-dependent and -independent methods, we analysed microbial communities associated with common starfish species, A. amurensis and P. pectinifera (Echinodermata: Asteroidea). Specifically, we addressed the questions of whether the CF bacterial composition of A. amurensis and P. pectinifera differs from that of the surrounding seawater, and whether the composition shows a geographical pattern.
Results
Coelomic fluid (CF)
CF samples from common coastal starfish, A. amurensis (19 individuals and 1 pooled sample) and P. pectinifera (8 individuals and 1 pooled sample), were analysed. The inorganic ion composition of representative CF samples was similar to that of seawater, although pH and most ion concentrations were slightly higher in the CF (Supplementary Table 1).
Characteristics of sequence data
Microbiota in two common starfish species and their surrounding seawater was studied using 16S rRNA gene amplicon sequencing. A total of 2,473,889 quality-filtered sequence reads were obtained from 34 samples (Supplementary Table 2). Rarefaction curves showed that most starfish samples reached the saturation phase (Supplementary Figure 1). The rarefaction plateaus appeared after fewer reads in A. amurensis CF samples, suggesting a lower microbial richness in A. amurensis than in P. pectinifera. The mean Good’s coverage index of all samples was 0.99, indicating adequate depth of sequencing (Supplementary Table 3).
Microbiota associated with starfish
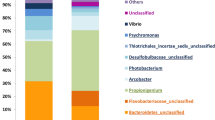
The CF microbiota mostly consisted of bacteria, but markedly varied among individual starfish. Results from the analysis of the alpha diversity metrics on the basis of equal numbers of sequence reads (n = 22,006) confirmed significantly lower microbial richness (Chao1 and observed OTUs) and Shannon’s diversity index for the A. amurensis CF compared with both P. pectinifera CF and seawater samples (p < 0.05 in all cases) (Supplementary Table 3). At the phylum level, the microbial taxonomic composition of CF samples showed a high relative abundance of Proteobacteria (59.9% on average), followed by Bacteroidetes (8.8%), while some CF samples of A. amurensis showed almost complete dominance of Proteobacteria (Supplementary Table 4). At the family level, no phylotype was shared across all CF samples with >3.0% relative abundance (Supplementary Table 5), suggesting great variability in the starfish CF microbiota. Nevertheless, the families Flavobacteriaceae (Bacteroidetes) and Rhodobacteraceae (Alphaproteobacteria) were frequently recovered from many P. pectinifera and several A. amurensis individuals. Members of these families were abundantly detected in seawater samples as well (Supplementary Table 5). When all quality-filtered sequence reads from the 5 seawater samples were pooled, Rhodobacteraceae was the most abundant, accounting for 12.3%, followed by Flavobacteriaceae. The similarity among microbiota within the many P. pectinifera CF, some A. amurensis CF and seawater was generally supported by cluster analysis (Fig. 1) and principal coordinates analysis (Fig. 2). Members of the family Mycoplasmataceae were dominantly recovered from one individual of P. pectinifera (sample ID, 2016U-ICF-2) (Fig. 1). At the genus level, the sequences (a total of 4 OTUs from 2016U-ICF-2) were affiliated with the genus Candidatus Hepatoplasma. This phylotype was not widely distributed, but was dominantly observed in the CF of A. amurensis (2014U-MCF-Bulk) collected from the same sampling site, suggesting the biogeography of starfish CF microbiota. We were generally unable to obtain results for the microbiome on the starfish body surface, probably due to the low microbial biomass there. Nevertheless, the body surface microbiome was successfully studied in one individual of A. amurensis, which was similar to that found in seawater (Fig. 1).
Histogram showing the relative abundance of 16S rRNA gene amplicon reads assigned to family level taxa in the various samples. UPGMA (Unweighted Pair Group Method with Arithmetic mean) clustering based on weighted UniFrac distances is also shown. Each colour on the graph represents a distinct family. Families with >10% relative abundance in any sample are presented, and the rest and unassigned taxa are indicated as ‘others’. The family Helicobacteraceae is divided into Helicobacter-related taxon and Helicobacter-unrelated taxa (mainly Sulfuricurvum, Sulfurimonas, and Sulfurovum relatives). Sample names represent sampling year, sampling site (N or H, Nemuro City; U or M, Hakodate City), starfish species (A. amurensis, M; P. pectinifera, I), body parts (CF, coelomic fluid; S, body surface) or seawater (SW), and individual number. “-Bulk” indicates bulk samples. A. amurensis, P. pectinifera, seawater, and starfish body surface samples are shown in red, blue, green, and black, respectively. Read numbers and ratios of Helicobacter-related taxon against bacteria estimated by quantitative PCR are shown in parentheses on right side of histograms. Detailed taxonomic information is provided in Supplementary Table 5.
As suggested by diversity indices, relatively limited but unique phylotypes were detected in the CF of A. amurensis. Although the A. amurensis CF microbiota greatly varied between starfish individuals (Fig. 1), the non-parametric multivariate analysis of variance (PERMANOVA) suggested that the microbiota variation resulted from differences in sampling site (at city level) and sampling year (p < 0.05). The biogeographical pattern could be due to the small number of A. amurensis samples obtained from Hakodate City. The family Helicobacteraceae and unclassified Thiotrichales were dominantly detected from some A. amurensis individuals collected from the coast of Nemuro City in 2015 and 2016 (Fig. 1, Supplementary Table 5). Although these phylotypes were rarely detected from seawater, P. pectinifera, or even A. amurensis individuals collected from another sampling site, they often showed almost complete dominance in some individuals. Although members of the family Helicobacteraceae were detected from both starfish species, their phylogenetic properties were different according to starfish species. All Helicobacteraceae abundantly (average >0.5%) found in P. pectinifera (3 OTUs) were related to known sulfur-oxidizing bacteria, that is, Sulfuricurvum, Sulfurimonas and Sulfurovum. Additionally, Helicobacteraceae detected in two individuals of A. amurensis from Hakodate (4.2% and 6.5% in 2016M-MCF-A and 2016M-MCF-C, respectively) were closely related to Sulfurovum species. In contrast, Helicobacteraceae in A. amurensis from Nemuro were distantly related to known Helicobacteraceae members, and their phylogeny could not be inferred in detail from the short-read sequence from MiSeq.
To clarify the phylogenetic position of Helicobacteraceae dominantly found in A. amurensis, cloning and sequencing of the 16S rRNA gene were performed on one representative sample, 2014N-MCF-2. Half of the cloned sequences (14 out of 28 sequenced clones; one OTU at the 97% cut-off) were affiliated with the family Helicobacteraceae, and a BLAST search revealed that the representative clone sequence (1,286 bp in length) was most closely related to Helicobacter species, for example, H. marmotae and H. rodentium, but the identity score was low (89%) (Fig. 3). No known environmental clone sequence was more similar to the Helicobacteraceae from A. amurensis. The qPCR results generally confirmed the limited but dominant occurrence of the Helicobacter-related taxon in A. amurensis of Nemuro City (Fig. 1). Likewise, the sequence of almost the full-length (1,348 bp in length) of the 16S rRNA gene of Thiotrichales was determined from A. amurensis (sample ID, 2016N-MCF-K), which was most closely related to some uncultured clone sequences from seawater (up to 97% identity), but distantly related to validly described species, for example, Francisella species (up to 87% identity), sulfur-oxidizing Piscirickettsiaceae species (up to 85% identity), and Candidatus Endoecteinascidia (up to 84% identity) (Supplementary Figure 2). Even with the almost full-length 16S rRNA gene sequence, the Thiotrichales phylotype could not be classified precisely than at the order level.
Phylogenetic tree of representative members of the genera Helicobacter and Wolinella inferred from 16S rRNA gene sequences by the neighbour joining method using 856 homologous sequence positions. The accession numbers are shown in parentheses. Bootstrap values (expressed as percentages of 1,000 replications) higher than 50% are shown at branching points. The sequence found in the CF of A. amurensis is shown in bold. The scale bar represents 0.01 substitutions per nucleotide position.
Microscopy
By FISH analysis with a probe designed for the Helicobacter-related taxon, we successfully observed probe-positive cells in the CF of A. amurensis. All of the probe-positive cells were spiral-shaped, similar to those of some Helicobacter species (Fig. 4). In addition, spiral-shaped cells were observed by transmission electron microscopy (TEM) (Fig. 5A and B) and scanning electron microscopy (SEM) (Fig. 5C and D). Electron microscopy revealed that the spiral-shaped cells had a prosthecate-like polar appendage (Fig. 5B and D), which has never been reported for known Helicobacter species.
Cultivation
We attempted to cultivate the Helicobacter-related taxon found in the CF of A. amurensis by using a modified version of a medium designed for the selective cultivation of Helicobacter species with the addition of basal saline under microaerobic conditions. A total of 34 isolates were obtained from the A. amurensis CF (sample ID, 2016N-MCF-J). Phylogenetic analysis based on the 16S rRNA gene sequence suggested that isolates were closely related to the genera Idiomarina (17 strains), Pseudoalteromonas (11 strains), Vibrio (4 strains), Alcanivorax (1 strain) and Thalassospira (1 strain). The number of colony-forming units (CFUs) was low (≤4.8 × 103 ml−1).
Pathogenicity
To assess the presence of a representative pathogenic gene (cagA) and urease gene of Helicobacter species in the Helicobacter-related taxon that dominated the CF of A. amurensis, we performed PCR using specific primers. No amplicon was obtained from all samples (data not shown), suggesting that the risk to humans from the Helicobacter-related taxon of starfish is not urgent.
Discussion
In this study, microbial diversity was assessed for the CF of common coastal starfish species. Given that the antimicrobial activity is well known15, 26, the occurrence of microbial diversity within the echinoderm CF might be considered unlikely. We found that the CF of echinoderms could be a reservoir for unique microorganisms including potentially pathogenic and/or symbiotic bacteria. Since the CF microbiota greatly differed among individuals, they are probably not vertically inherited but rather horizontally acquired from the surroundings. This is supported by the general similarity in microbial communities between the CF of P. pectinifera and surrounding seawater. Therefore, the bacterial communities in the CF of starfish may be allochthonous, involving the transient passage in and out of surrounding seawater. Many factors potentially influence the composition of starfish CF microbiota, including genetic background, diet, age, stress and environmental factors (e.g. temperature). Nevertheless, A. amurensis harboured specific microbiota, namely, Helicobacter-related taxon and unclassified Thiotrichales members, although this species lives together with P. pectinifera. These microorganisms often exhibited almost complete dominance in some individuals. The host specificity of CF microbiota may result from active selection by the host. The dominance of potentially sulfur-oxidizing microbiota might be associated with the starfish feeding behavior; for example, A. amurensis has been observed digging up and eating bivalves buried (depth < 10 cm) in sediments27, 28. In both starfish species collected in Hakodate City, members of the genus Candidatus Hepatoplasma were dominantly detected, although they were not detected in all starfish individuals collected there. Members of this group of bacteria are related to the genus Mycoplasma, and have been intensively studied as symbionts living within the hepatopancreas of isopods29, 30. Such symbionts are thought to improve the survival of host isopods under low-nutrient conditions31. The isopod Hepatoplasma may be inherited from parents to offspring through horizontal transmission30. Even in the isopod, however, the infection ratio was reported to significantly vary30. Among the sampling sites, the common coastal isopod, Ligia exotica, was found to be abundant only in Hakodate City, which might be associated with the near absence of Candidatus Hepatoplasma in the starfish CF from Nemuro City.
Mainly from the A. amurensis CF collected from Nemuro City in 2016, unclassified members of the order Thiotrichales were dominantly detected. Although it was not possible to classify them precisely, the 16S rRNA gene sequence was distantly related to those of Francisella, sulfur-oxidizing Piscirickettsiaceae and Candidatus Endoecteinascidia. Members of the genus Francisella are strictly aerobic, facultatively intracellular heterotrophs, which include the fatal human pathogen, F. tularensis 32. Francisella members also infect various fish and shellfish species, and are known as the causative agent of severe disease33. In Japan, F. halioticida caused the mass mortality of the abalone Haliotis gigantea 33, 34. In contrast, in sulfidic marine environments, symbiotic sulfur-oxidizing Thiotrichales both participate in the detoxification of sulfide and heavy metals and fix carbon for their host invertebrates4. Candidatus Endoecteinascidia is a commercially important symbiotic bacterium of the Caribbean mangrove tunicate Ecteinascidia turbinata 35. This symbiont produces an anti-tumour compound, ET-743, and potentially protects the host from predators36. Precise characterization of the physiology of the starfish Thiotrichales appears to be necessary, considering its potential positive and negative impacts on fisheries23, 24.
Among members of the class Epsilonproteobacteria, Arcobacter species (family Campylobacteraceae) were previously detected in the gut of sea urchins37, 38. In this study, some P. pectinifera individuals harboured close relatives of known species of the genus Arcobacter (up to 22.4% in relative abundance of sequences). In addition, the Sulfuricurvum-related OTU detected in P. pectinifera (up to 6.7%) was closely related to the phylotype previously detected in the CF of sea cucumber16, suggesting a potential similarity in settings among sea urchin guts, sea cucumber CF and P. pectinifera CF. Although these Epsilonproteobacteria were scarcely detected in A. amurensis CF, previously unknown Helicobacter-related taxon was mainly detected from A. amurensis collected from Nemuro City in 2015.
The 16S rRNA gene sequence of Helicobacter-related taxon was most closely related to Helicobacter rodentium and H. marmotae, which were respectively isolated from laboratory mice39 and livers of woodchucks and intestines of cats40. Members of the genus Helicobacter are diverse in vertebrate hosts, infecting the digestive tracts of a wide array of mammals and birds41. Recently, Helicobacter species were also found in reptiles42. Well-characterized Helicobacter-related taxa include Wolinella species, which are the cattle rumen-associated non-pathogenic members of the Epsilonproteobacteria 43. The HelicobacterWolinella group has never been isolated from invertebrates44. Although some studies detected Helicobacter species in coastal seawater45, 46, these microbes were probably in a dormant state. Although the starfish Helicobacter-related taxon was not successfully cultured in this study, this is the first report to describe the dominance of Helicobacter-related taxon in invertebrates and non-digestive organs. The failure of cultivation of the Helicobacter-related taxon from A. amurensis might suggest that it had lost the ability to grow outside the host or differs physiologically from known Helicobacter species. Considering that many marine Helicobacteraceae species have the ability to oxidize sulfide4, the Helicobacter-related taxon might aid sulfide detoxification, as hypothesized for unclassified Thiotrichales. Although virulence gene cagA and the urease gene were not detected by PCR, primers used in this study were designed for H. pylori. In addition, some Helicobacter species, including H. pylori strains, are known to lack these genes47. More genomic and physiological characterization of the Helicobacter-related taxon found in A. amurensis are necessary in the future. Nevertheless, the successful isolation of diverse bacteria suggested the suitability of the main body cavity of starfish for habitation by microbes. Previous comparative genomic analysis revealed that epsilonproteobacterial autotrophy in deep-sea vents has provided the core of virulence for important human/animal pathogens including Helicobacter 48, 49. The occurrence of Helicobacter-related taxon within the coastal echinoderm CF may fill the evolutionary gap between the deep-sea vents and terrestrial vertebrate intestinal habitats. In this context, Echinodermata are amongst the most abundant animals in the deep-sea, and our findings warrant the assessment of deep-sea echinoderm microbiota.
The CF microbiota potentially have adverse effects on the health of starfish. In contrast, the bacteria may confer some competitive advantages to starfish by allowing starfish survival under nutrient stress, producing bioactive compounds to repel predators, and/or detoxifying sulfide. Considering their resistance to cultivation, further studies using metagenomics or single-cell genomics are required to elucidate the precise nature of the starfish-bacteria association. Such studies should be combined with a functional approach in which controlled in vivo experiments are used to determine the degree of dependence of the starfish on its microbiota, e.g. during regeneration.
Methods
Sampling
Starfish individuals were collected from piers using a net in coastal areas of two different cities in Hokkaido, Japan: Nemuro City (43.2–3′N, 145.58–59′E) and Hakodate City (41.8–9′N, 140.6–9′E). Two sites in each city were studied. At the sampling sites, A. amurensis and P. pectinifera live together, although A. amurensis was rarely found in the area of Nemuro with turbid seawater. All starfish individuals appeared healthy, and no evidence of autotomy was observed. For some individuals, outer surfaces of the body wall were sampled before sampling the CF, using sterile cotton-tipped swabs (Osakimedical, Nagoya, Japan). A single arm tip was cut using sterilized scissors to pour CF into a sterile tube. The volume of CF fluctuated markedly among the individuals, but at least 5 ml was used for the analysis described below. Although over 100 ml of CF was collected from an individual in some A. amurensis, the CF from small individuals was pooled and then analyzed (sample IDs: 2014U-MCF-Bulk and 2015N-ICF-Bulk). For FISH and electron microscopy, the CF microbiota was fixed in 4% paraformaldehyde immediately after the CF collection. Digestive glands were also collected after dissection with sterilized scissors. Seawater samples (100-500 ml) were collected using a VanDorn sampler and immediately filtered through 0.22-µm-pore-size polyethersulfone filters. All samples (CF, filters, and digestive glands) were kept on ice and transported to the laboratory as quickly as possible. Because no laboratory facility was available in Nemuro, the most eastern city of Hokkaido, sample transportation took up to 30 hours after sampling (up to 10 hours for samples from Hakodate). Details of samples are summarized in Supplementary Table 2.
Immediately upon arrival at the laboratory, host cells in the CF were pelleted at 800 g for 5 min50, and the supernatant containing microbial cells was further centrifuged at 21,500 g for 10 min. The obtained supernatants were then subjected to ion chromatography. The microbial cell pellets and filters from seawater samples were kept at −80 °C for use in DNA extraction. For FISH and electron microscopy, fixed cells were washed three times with PBS, and preserved in 50% (v/v) ethanol in PBS at −30 °C.
Ion chromatography
Anion and cation concentrations were determined by ion chromatography using a Shim-pack IC-A3 column (Shimadzu, Kyoto, Japan) and a Shim-pack IC-C3 column (Shimadzu), respectively. The Shimadzu HPLC CBM-20A system controller and CDD-10AVP conductivity detector were used.
DNA analysis
Genomic DNA was extracted from cell pellets of CF samples, seawater filters, and swabs using the Ultraclean Microbial DNA isolation kit (MoBio Laboratories, Carlsbad, CA) following the manufacturer’s instructions. For digestive gland samples, DNeasy blood and tissue kit (Qiagen, Valencia, CA) was used, but no microbial components were detected (data not shown), potentially due to the presence of strong DNase and/or PCR inhibitors. The V4 regions of 16S rRNA genes were amplified using the LA Taq (TaKaRa Bio Inc., Otsu, Japan). Negative controls without template DNA were included in each PCR trial. The first PCR step (20-35 cycles) was performed using primers 515F (5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTGTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGGACTACHVGGGTWTCTAAT-3′). These primers are suitable for the amplification of both bacterial and archaeal DNA. The amplified products from the minimum PCR cycles with visible PCR product were used for a second round of PCR after the purification with AMPure XP (Beckman Coulter, Mississauga, Canada). The second amplification step (10 cycles) used the first PCR product as a template and was performed using primers with a tag sequence. After the second PCR, amplicons were purified again using AMPure XP. After measuring the concentrations of each PCR product using the Qubit fluorometer (Thermo Fisher Scientific Inc., MA, USA), amplicons were sent to FASMAC (Atsugi, Japan) for Illumina sequencing (MiSeq). The raw sequencing data have been submitted at GenBank/EMBL/DDBJ under accession number DRA005566.
Pair-end reads were merged with PEAR51. Mitochondrial sequences were removed from the data set by mapping the reads against mitochondrial genomes of A. amurensis and P. pectinifera using Bowtie 252. After the removal of primers, low-quality (Q score <30 in more than 3% of sequences) and short (<150 bp) reads were removed using a custom perl script. Sequences (253 bp average) were processed using the QIIME software package53. After the removal of chimeric sequences using Usearch6154 in QIIME, OTUs were selected at the 97% similarity level using UCLUST54 and were subsequently assigned to a taxon (at phylum, class, order, family, and genus levels) by comparison with the non-redundant 16S rRNA small subunit SILVA128 database55, using the RDP classifier56 with the default parameters. Alpha and beta diversity analyses were performed on the rarefied OTU table at 22,006 sequences per sample in QIIME. Alpha diversity indices were compared between starfish species with a non-parametric t-test (Monte Carlo, 999 permutations) in QIIME. An unweighted pair group method algorithm (UPGMA) dendrogram was made in QIIME using a weighted UniFrac distance matrix57. Similarly, principal coordinate analysis (PCoA) was performed in QIIME. Differences in CF microbial community by sampling site and year were evaluated using permutational multivariate analysis of variance (PERMANOVA) based on weighted UniFrac distance matrix in QIIME.
To obtain nearly the full-length sequence of the 16S rRNA gene, cloning and sequencing were performed using the 27F and 1492R primers, as previously described58 for two samples. Phylogenetic analysis was performed using ARB software59 as described previously58. Furthermore, real-time PCR was performed on all DNA samples by using the Helicobacter-related-taxon-specific primer Hel398F (5′-GAGGATGACGGCTTTCGAGT-3′) and the universal primer 533R (5′-TTACCGCGGCKGCTGRCAC-3′)60. Targets of these primers were checked using the Probe Match tool of ARB software59. The Hel398F primer had at least three mismatches to non-target sequences. The SYBR Premix Ex Taq kit (TaKaRa) was used with the Thermal Cycler Dice Real Time System Lite (TaKaRa). The qPCR mixtures contained 12.5 µl of SYBR Premix Ex Taq II, 2 µl of each forward and reverse primer (10 pmol ul−1), 6.5 µl of PCR grade water, and 2 µl of the DNA template. Purified PCR products amplified from a CF DNA sample (2015N-MCF-E) with 338F61/533R primers and 398F/533R primers were used as quantification standards of bacteria and Helicobacter-related taxon, respectively. Concentrations of the standards were measured fluorometrically with Qubit (Thermo Fisher Scientific). The PCR conditions were 95 °C for 1 min, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. Dissociation curve analysis and gel electrophoresis were performed for quality assurance. The 16S rRNA gene sequences of Helicobacter-related taxon and unclassified Thiotrichales reported in this study are available under accession numbers LC214978 and LC278460, respectively.
To assess the presence of pathogenic genes in the starfish Helicobacter-related taxon, PCR was performed using primer sets designed for cagA 62 and ureA 63. Genomic DNA of H. pylori strain NCTC 11637 was purchased from American Type Culture Collection (Manassas, VA) and used as a positive control.
Cultivation
To cultivate microorganisms found in the CF of A. amurensis, some CF samples were transferred to the solid medium, modified Columbia horse blood agar HP (CHBHP). The medium (per litre) contains yeast extract 2 g, peptone 0.5 or 0.1 g, agar 12 g, horse blood 100 ml, and the Helicobacter pylori selective supplement 2 vials (Thermo Fisher Scientific). The medium composition was slightly modified from CHBHP medium64. Two-thirds diluted MMJ synthetic seawater65 or the supernatant of CF (after 21,500 g for 10 min) was used as the basal saline. The anaeropack (Mitsubishi Gas Co., Ltd.) was used for microaerobic conditions. Plates were incubated at the in-situ temperature (17 °C).
Microscopy
Fluorescence in-situ hybridization (FISH) was performed on one representative CF sample, 2015N-MCF-D (fixed at the sampling site), as described previously66, using Helicobacter-specific probe Hel395F with Alexa Fluor 488 (5′-CKAAAWCCKTCATCCTCCAC-3′) (designed in this study). Formamide concentration (set to 30%) and specificity was predicted using mathFISH67. After 4′-6-diamidino-2-phenylindole (DAPI) staining, samples were observed under a confocal laser scanning microscope (TCS-SP8; Leica Microsystems, Wetzlar, Germany) equipped with filters for Alexa Fluor 488 (excitation 488 nm; emission 500-550 nm) and DAPI (excitation 405 nm; emission 423-456 nm). Non338 was used as a negative control probe68. For transmission electron microscopy, the cell pellet from a CF sample, 2015N-MCF-D (fixed at the sampling site), was stained with 1% (v/v) phosphotungstic acid containing 0.01% (w/v) sodium azide and 0.4% (w/v) sucrose (pH7.6). Micrographs were captured using a Technai G2 20 electron microscope (FEI, Hillsboro, OR) operated at 200 kV. Cells were also observed with a JEOL JSM-6700F scanning electron microscope.
References
Gajardo, K. et al. A high-resolution map of the gut microbiota in Atlantic salmon (Salmo salar): A basis for comparative gut microbial research. Sci. Rep. 6, 30893 (2016).
Gatesoupe, F. J. et al. The highly variable microbiota associated to intestinal mucosa correlates with growth and hypoxia resistance of sea bass, Dicentrarchus labrax, submitted to different nutritional histories. BMC Microbiol. 16, 266 (2016).
Wong, S. et al. Aquacultured rainbow trout (Oncorhynchus mykiss) possess a large core intestinal microbiota that is resistant to variation in diet and rearing density. Appl. Environ. Microbiol. 79, 4974–4984 (2013).
Nakagawa, S. & Takai, K. Deep-sea vent chemoautotrophs: diversity, biochemistry and ecological significance. FEMS Microbiol. Ecol. 65, 1–14 (2008).
Castro, D., Pujalte, M. J., Lopez-Cortes, L., Garay, E. & Borrego, J. J. Vibrios isolated from the cultured manila clam (Ruditapes philippinarum): numerical taxonomy and antibacterial activities. J. Appl. Microbiol. 93, 438–447 (2002).
Olafsen, J. A., Mikkelsen, H. V., Giaever, H. M. & Hansen, G. H. Indigenous bacteria in hemolymph and tissues of marine bivalves at low temperatures. Appl. Environ. Microbiol. 59, 1848–1854 (1993).
Wang, X. W., Xu, J. D., Zhao, X. F., Vasta, G. R. & Wang, J. X. A shrimp C-type lectin inhibits proliferation of the hemolymph microbiota by maintaining the expression of antimicrobial peptides. J. Biol. Chem. 289, 11779–11790 (2014).
Lokmer, A. et al. Spatial and temporal dynamics of Pacific oyster hemolymph microbiota across multiple scales. Front. Microbiol. 7, 1367 (2016).
Lokmer, A. & Mathias Wegner, K. Hemolymph microbiome of Pacific oysters in response to temperature, temperature stress and infection. ISME J. 9, 670–682 (2015).
Kelly, M. & McKenzie, J. Survey of the occurrence and morphology of sub-cuticular bacteria in shelf echinoderms from the north-east Atlantic Ocean. Mar. Biol. 123, 741–756 (1995).
Yamazaki, Y. et al. Individual Apostichopus japonicus fecal microbiome reveals a link with polyhydroxybutyrate producers in host growth gaps. Sci. Rep. 6, 21631 (2016).
Binyon, J. Physiology of Echinodermata. (Pergamon Press, Oxford, 1972).
Collard, M. et al. Buffer capacity of the coelomic fluid in echinoderms. Comp. Biochem. Physiol. A 166, 199–206 (2013).
Matranga, V. et al. Monitoring chemical and physical stress using sea urchin immune cells in Echinodermata (ed. Matranga, V.) 85–110 (Springer, 2005).
Pinsino, A., Thorndyke, M. C. & Matranga, V. Coelomocytes and post-traumatic response in the common sea star Asterias rubens. Cell Stress Chaperones 12, 331–341 (2007).
Enomoto, M., Nakagawa, S. & Sawabe, T. Microbial communities associated with holothurians: presence of unique bacteria in the coelomic fluid. Microbes Environ. 27, 300–305 (2012).
Hewson, I. et al. Densovirus associated with sea-star wasting disease and mass mortality. Proc. Natl. Acad. Sci. USA 111, 17278–17283 (2014).
Coteur, G. et al. Peptidoglycan recognition proteins with amidase activity in early deuterostomes (Echinodermata). Dev. Comp. Immunol. 31, 790–804 (2007).
Haug, T. et al. Antibacterial activity in Strongylocentrotus droebachiensis (Echinoidea), Cucumaria frondosa (Holothuroidea), and Asterias rubens (Asteroidea). J. Invertebr. Pathol. 81, 94–102 (2002).
Osbourn, A., Goss, R. J. M. & Field, R. A. The saponins – polar isoprenoids with important and diverse biological activities. Nat. Prod. Rep. 28, 1261 (2011).
Coteur, G., DeBecker, G., Warnau, M., Jangoux, M. & Dubois, P. Differentiation of immune cells challenged by bacteria in the common European starfish, Asterias rubens (Echinodermata). Eur. J. Cell. Biol. 81, 413–418 (2002).
Coteur, G., Danis, B., Wantier, P., Pernet, P. & Dubois, P. Increased phagocytic activity in contaminated seastars (Asterias rubens) collected in the Southern Bight of the North Sea. Mar. Pollut. Bull. 50, 1295–1302 (2005).
Burtle, G. J. Invasive aquatic animals in Encyclopedia of Agriculture and Food Systems, Second Edition (ed Van Alfen, N. K.) 58–65 (Elsevier, 2014).
Barkhouse, C., Niles, M. & Davidson, L.-A. A literature review of sea star control methods for bottom and off bottom shellfish cultures. Can. Ind. Rep. Fish. Sci. 279, 1–38 (2007).
Zhang, Y. et al. Pseudoruegeria marinistellae sp. nov., isolated from an unidentified starfish in Sanya, China. Antonie van Leeuwenhoek (2016).
Burnell, D. J. & Apsimon, J. W. Echinoderm saponins in Marine natural products: chemical and biological perspectives, volume 5 (ed Scheuer, P. J.) 287–389 (Academic Press, 1983).
Arima, K., Hamaya, S. & Miyakawa, Y. Feeding behaviour of starfish to bivalves. Scient. Rep. Hokkaido Fish. Exp. Stn. 14, 63–69 (1972).
Fukuyama, A. K. & Oliver, J. S. Sea star and walrus predation on bivalves in Norton Sound, Bering Sea, Alaska. Ophelia 24, 17–36 (1985)
Leclercq, S., Dittmer, J., Bouchon, D. & Cordaux, R. Phylogenomics of “Candidatus Hepatoplasma crinochetorum,” a lineage of mollicutes associated with noninsect arthropods. Genome Biol. Evol. 6, 407–415 (2014).
Wang, Y., Brune, A. & Zimmer, M. Bacterial symbionts in the hepatopancreas of isopods: diversity and environmental transmission. FEMS Microbiol. Ecol. 61, 141–152 (2007).
Fraune, S. & Zimmer, M. Host-specificity of environmentally transmitted Mycoplasma-like isopod symbionts. Environ. Microbiol. 10, 2497–2504 (2008).
Foley, J. E. & Nieto, N. C. Tularemia. Vet. Microbiol. 140, 332–338 (2010).
Birkbeck, T. H., Feist, S. W. & Verner-Jeffreys, D. W. Francisella infections in fish and shellfish. J. Fish Dis. 34, 173–187 (2011).
Brevik, O. J., Ottem, K. F., Kamaishi, T., Watanabe, K. & Nylund, A. Francisella halioticida sp. nov., a pathogen of farmed giant abalone (Haliotis gigantea) in Japan. J. Appl. Microbiol. 111, 1044–1056 (2011).
Perez-Matos, A. E., Rosado, W. & Govind, N. S. Bacterial diversity associated with the Caribbean tunicate Ecteinascidia turbinata. Antonie Van Leeuwenhoek 92, 155–164 (2007).
Schofield, M. M., Jain, S., Porat, D., Dick, G. J. & Sherman, D. H. Identification and analysis of the bacterial endosymbiont specialized for production of the chemotherapeutic natural product ET-743. Environ. Microbiol. 17, 3964–3975 (2015).
Hakim, J. A. et al. An abundance of Epsilonproteobacteria revealed in the gut microbiome of the laboratory cultured sea urchin, Lytechinus variegatus. Front. Microbiol. 6, 1047 (2015).
Nelson, L. et al. Molecular analysis of gut microflora in captive-raised sea urchins (Lytechinus variegatus). J. World Aquac. Soc. 41, 807–815 (2010).
Shen, Z. et al. Helicobacter rodentium sp. nov., a urease-negative Helicobacter species isolated from laboratory mice. Int. J. Syst. Bacteriol. 47, 627–634 (1997).
Fox, J. G. et al. Helicobacter marmotae sp. nov. Isolated from livers of woodchucks and intestines of cats. J. Clin. Microbiol. 40, 2513–2519 (2002).
On, S. L. et al. Genus I. Helicobacter in Bergey’s manual of systematic bacteriology. Vol. 2 Part C (eds Brenner, D. J., Krieg, N. R., Staley, J. T. & Garrity, G. M.) 1169–1189 (Springer, 2005).
Gilbert, M. J. et al. Occurrence, diversity, and host association of intestinal Campylobacter, Arcobacter, and Helicobacter in reptiles. PLoS One 9, e101599 (2014).
Baar, C. et al. Complete genome sequence and analysis of Wolinella succinogenes. Proc. Natl. Acad. Sci. USA 100, 11690–11695 (2003).
Garrity, G. M., Bell, J. A. & Lilburn, T. Family II. Helicobacteraceae fam. nov. in Bergey’s Manual of Systematic Bacteriology. Second ed. Volume 2, Part C (eds Brenner, D. J., Krieg, N. R., Staley, J. T. & Garrity, G. M.) 1168–1194 (Springer, 2005).
Twing, K. I., Kirchman, D. L. & Campbell, B. J. Temporal study of Helicobacter pylori presence in coastal freshwater, estuary and marine waters. Water Res. 45, 1897–1905 (2011).
Carbone, M. et al. Occurrence of Helicobacter pylori DNA in the coastal environment of southern Italy (Straits of Messina). J. Appl. Microbiol. 98, 768–774 (2005).
Solnick, J. V. & Schauer, D. B. Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin. Microbiol. Rev. 14, 59–97 (2001).
Nakagawa, S. et al. Deep-sea vent epsilon-proteobacterial genomes provide insights into emergence of pathogens. Proc. Natl. Acad. Sci. USA 104, 12146–12150 (2007).
Perez-Rodriguez, I., Bolognini, M., Ricci, J., Bini, E. & Vetriani, C. From deep-sea volcanoes to human pathogens: a conserved quorum-sensing signal in Epsilonproteobacteria. ISME J. 9, 1222–1234 (2015).
Nakagawa, S. et al. Allying with armored snails: the complete genome of gammaproteobacterial endosymbiont. ISME J. 8, 40–51 (2014).
Zhang, J., Kobert, K., Flouri, T. & Stamatakis, A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2012).
Wang, Q., Garrity, G. M., Tiedje, J. M. & Cole, J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007).
Lozupone, C. & Knight, R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235 (2005).
Nakagawa, S. et al. Distribution, phylogenetic diversity and physiological characteristics of epsilon-Proteobacteria in a deep-sea hydrothermal field. Environ. Microbiol. 7, 1619–1632 (2005).
Ludwig, W. et al. ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371 (2004).
Watanabe, K., Kodama, Y. & Harayama, S. Design and evaluation of PCR primers to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. J. Microbiol. Methods 44 (2001).
Huse, S. M. et al. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4, e1000255 (2008).
Yamaoka, Y. et al. Relationship of vacA genotypes of Helicobacter pylori to cagA status, cytotoxin production, and clinical outcome. Helicobacter 3, 241–253 (1998).
Clayton, C., Kleanthous, H., Coates, P. J., Morgan, D. D. & Tabaqchali, S. Sensitive detection of Helicobacter pylori by using polymerase chain reaction. J. Clin. Microbiol. 30, 192–200 (1992).
Hasegawa, M. et al. A multicenter study of a new Helicobacter pylori selective medium. Columbia horse blood agar HP. Kansenshogaku Zasshi 76, 341–346 (2002).
Nakagawa, S. & Takai, K. The isolation of thermophiles from deep-sea hydrothermal environments. Methods Microbiol. 35, 55–91 (2006).
Takai, K. et al. Spatial distribution of marine crenarchaeota group I in the vicinity of deep-sea hydrothermal systems. Appl. Environ. Microbiol. 70, 2404–2413 (2004).
Yilmaz, L. S., Parnerkar, S. & Noguera, D. R. mathFISH, a web tool that uses thermodynamics-based mathematical models for in silico evaluation of oligonucleotide probes for fluorescence in situ hybridization. Appl. Environ. Microbiol. 77, 1118–1122 (2011).
Wallner, G., Amann, R. & Beisker, W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14, 136–143 (1993).
Acknowledgements
We thank T. Sawabe for assistance in sample preparation in Hakodate City. This work was partially supported by the Institute for Fermentation, Osaka (IFO), the Noguchi Institute, and the Suhara Memorial Foundation.
Author information
Authors and Affiliations
Contributions
S.N. conceived the experiments, S.N., H.S., A.T., M.H., H.Y., and H.M. conducted the experiments, S.N., S.S., T.S., and Y.T. analysed the results. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nakagawa, S., Saito, H., Tame, A. et al. Microbiota in the coelomic fluid of two common coastal starfish species and characterization of an abundant Helicobacter-related taxon. Sci Rep 7, 8764 (2017). https://doi.org/10.1038/s41598-017-09355-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-09355-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.