Abstract
The EZH2 histone methyltransferase is a member of the polycomb repressive complex 2 (PRC2) that is highly expressed in diverse human cancers and is associated with a poor prognosis. MUC1-C is an oncoprotein that is similarly overexpressed in carcinomas and has been linked to epigenetic regulation. A role for MUC1-C in regulating EZH2 and histone methylation is not known. Here, we demonstrate that targeting MUC1-C in diverse human carcinoma cells downregulates EZH2 and other PRC2 components. MUC1-C activates (i) the EZH2 promoter through induction of the pRB→E2F pathway, and (ii) an NF-κB p65 driven enhancer in exon 1. We also show that MUC1-C binds directly to the EZH2 CXC region adjacent to the catalytic SET domain and associates with EZH2 on the CDH1 and BRCA1 promoters. In concert with these results, targeting MUC1-C downregulates EZH2 function as evidenced by (i) global and promoter-specific decreases in H3K27 trimethylation (H3K27me3), and (ii) activation of tumor suppressor genes, including BRCA1. These findings highlight a previously unreported role for MUC1-C in activating EZH2 expression and function in cancer cells.
Similar content being viewed by others
Introduction
Histone methylation plays an essential role in the epigenetic control of gene expression in cancer1, 2. The polycomb group (PcG) proteins repress gene expression by maintaining chromatin in a transcriptionally suppressed state and thereby contribute to cell fate, development and cancer1, 3, 4. The PcG proteins form the (i) polycomb repressive complex 2 (PRC2), which predominantly catalyzes trimethylation of histone H3 at lysine 27 (H3K27me3), and (ii) polycomb repressive complex 1 (PRC1), which recognizes H3K27me3 and stabilizes the inactive epigenetic state1, 4. The PRC2 complex includes in part the enhancer of zeste homolog 2 (EZH2), suppressor of zeste 12 homolog (SUZ12) and embryonic ectoderm development (EED). EZH2 is a histone methyltransferase (HMT), which is dependent on the presence of SUZ12 and EED, and mediates H3K27 trimethylation with the downregulation of target genes5, 6. Overexpression of EZH2 in invasive and metastatic breast cancers is associated with a poor prognosis7, 8. EZH2 overexpression promotes tumorigenesis in mouse models of lung cancer9 and has been linked to poor clinical outcomes in patients with non-small cell lung cancer (NSCLC), as well as other types of carcinomas10,11,12,13,14,15,16. In concert with these findings, EZH2 confers a proliferative advantage, induces transformation and drives the epithelial-mesenchymal transition (EMT) program11, 17,18,19. The EZH2-containing PRC2 complex also recruits DNA methyltransferases (DNMTs) and thereby promotes the repression of tumor suppressor genes (TSGs), such as CDH1, by methylation of their promoters2, 20,21,22,23. Overexpression of EZH2 is associated with amplification of the EZH2 locus in certain cancers11. In addition, activation of the E2F pathway contributes to EZH2 transcription11. MYC has also been linked to activation of EZH2 transcription and the regulation of EZH2 mRNA levels by a miR-26a-dependent mechanism24,25,26.
Mucin 1 (MUC1) is a heterodimeric protein that is aberrantly overexpressed in breast, non-small cell lung (NSCL) and other cancers27. Notably, MUC1 consists of two subunits27. The MUC1 N-terminal subunit (MUC1-N) is the mucin component of the heterodimer, which is positioned extracellularly in a complex with the transmembrane C-terminal subunit (MUC1-C)27. The MUC1-N/MUC1-C complex evolved to protect epithelia from stress by (i) a MUC1-N-associated physical barrier and (ii) a MUC1-C-activated signaling cascade that confers self-renewal, repair and survival27, 28. In this capacity and with overexpression in cancer, MUC1-C functions as an oncoprotein that interacts with (i) receptor tyrosine kinases (RTKs) at the cell surface and (ii) certain transcription factors, such as β-catenin/TCF4 and NF-κB p65, in the nucleus29,30,31. For example, MUC1-C activates the MYC gene by a β-catenin/TCF4-mediated mechanism32, 33. In turn, the MUC1-C→MYC pathway drives BMI1 gene transcription and the ubiquitylation of H2A34. MUC1-C also activates the inflammatory TAK1→IKK→NF-κB pathway29, 35,36,37. The MUC1-C cytoplasmic domain binds directly to NF-κB p65 and promotes NF-κB p65 occupancy on the promoters of its target genes29. In this way, MUC1-C drives NF-κB-mediated activation of the ZEB1 gene, suppresses miR-200c expression and promotes EMT37. The interaction between MUC1-C and NF-κB also promotes self-renewal capacity of carcinoma cells, activation of the LIN28B→let-7 pathway, downregulation of E-cadherin and expression of other markers of stemness38, 39. These findings and the demonstration that MUC1-C drives DNMT expression have supported the notion that MUC1-C links the inflammatory NF-κB pathway to epigenetic regulatory mechanisms associated with EMT and a malignant phenotype40, 41.
The present studies demonstrate that targeting MUC1-C in carcinoma cells is associated with downregulation of EZH2, SUZ12 and EED expression, indicating that MUC1-C activates major components of the PRC2 complex. We have focused here on MUC1-C-mediated regulation of EZH2 and demonstrate that MUC1-C drives EZH2 transcription by retinoblastoma protein (pRB)→E2F- and NF-κB p65-mediated mechanisms. We further demonstrate that MUC1-C interacts directly with EZH2 and forms a complex with EZH2 on the CDH1 and BRCA1 promoters. In concert with these results, we show that targeting MUC1-C decreases global and gene promoter-specific H3K27me3 levels. These findings uncover a previously unrecognized role for MUC1-C in driving EZH2-mediated epigenetic regulation in cancer cells.
Results
MUC1-C drives EZH2 expression
EZH2, a member of the PRC2 complex, has been linked to breast and NSCL cancers, among others. We found that stable silencing of MUC1-C in BT-549 triple-negative breast cancer (TNBC) cells is associated with downregulation of EZH2 mRNA levels (Fig. 1A). The PRC2 complex also includes SUZ12 and EED1 and, interestingly, silencing MUC1-C was associated with downregulation of SUZ12 and EED mRNA (Fig. 1A). Similar results were obtained in MDA-MB-231 (Supplemental Fig. S1A) and H460 (Fig. 1B) cells, indicating that MUC1-C drives EZH2, SUZ12 and EED expression in TNBC and NSCLC cells. EZH2 possesses HMT activity, whereas SUZ12 and EED are necessary for EZH2 function42. Accordingly, we focused our studies here on the regulation of EZH2. In concert with the mRNA results, targeting MUC1-C resulted in suppression of EZH2 protein (Fig. 1C, left and right). To extend these observations, we established BT-549 cells stably expressing a tetracycline-inducible MUC1 shRNA (tet-MUC1shRNA) or a control shRNA (tet-CshRNA). Treatment of BT-549/tet-MUC1shRNA cells with doxycycline (DOX) for 7 days resulted in suppression of MUC1-C and EZH2 expression (Fig. 1D, left and right). By contrast, treatment of BT-549/tet-CshRNA cells with DOX had no effect on MUC1-C or EZH2 mRNA levels (Supplemental Fig. S1B). Similar results were obtained with DOX-treated MDA-MB-468/tet-MUC1shRNA and MDA-MB-468/tet-CshRNA breast cancer cells (Fig. 1E, left and right; Supplemental Fig. S1C). We also found that silencing MUC1-C in KRAS mutant A549 NSCLC cells decreases EZH2 mRNA levels (Supplemental Fig. S1D). Moreover, MUC1-C was necessary for EZH2 expression in DU145 prostate cancer cells (Supplemental Fig. S1E), supporting the notion that MUC1-C drives the upregulation of EZH2 in diverse types of cancer cells.
Silencing MUC1-C suppresses EZH2, SUZ12 and EED expression. A and B. BT-549 (A) and H460 (B) cells stably expressing a control scrambled shRNA (CshRNA) or a MUC1shRNA were analyzed for MUC1, EZH2, SUZ12 and EED mRNA levels by qRT-PCR using primers listed in Table S1. The results (mean ± SD) are expressed as relative mRNA levels compared to that obtained for the CshRNA cells (assigned a value of 1). (C) The respective BT-549 (left) and H460 (right) cells expressing a CshRNA or MUC1shRNA were immunoblotted with the indicated antibodies. (D and E) BT-549 (D) and MDA-MB-468 (E) cells were stably transduced to express a tetracycline-inducible MUC1 shRNA (tet-MUC1shRNA). Cells treated with 200 ng/ml DOX for 4 d were analyzed for MUC1 and EZH2 mRNA levels by qRT-PCR. The results (mean ± SD) are expressed as relative mRNA levels compared to that obtained for control DOX-untreated cells (assigned a value of 1) (left). Lysates from cells treated with 200 ng/ml DOX for 7 d were immunoblotted with the indicated antibodies (right). See also Fig. S1.
Targeting the MUC1-C cytoplasmic domain suppresses EZH2 expression
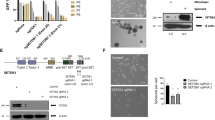
In concert with the above findings, enforced overexpression of MUC1-C resulted in upregulation of EZH2 mRNA and protein (Fig. 2A,B), demonstrating that MUC1-C, and not the MUC1 N-terminal subunit (MUC1-N), is necessary for this response. The MUC1-C subunit consists of a 72-amino acid (aa) intrinsically disordered cytoplasmic domain that is sufficient for promoting self-renewal and tumorigenicity (Fig. 2C)30, 43. Noteworthy is the presence of a CQC motif in the MUC1-C cytoplasmic domain that is required for the formation of MUC1-C homodimers and for MUC1-C-mediated transformation (Fig. 2C)44, 45. Moreover, expression of MUC1-C in which the CQC motif is mutated to AQA suppresses tumorigenicity, consistent with a dominant-negative effect for transformation44, 45. In support of a role for MUC1-C in driving EZH2, expression of the MUC1-C(AQA) mutant resulted in downregulation of EZH2 levels (Fig. 2D). The MUC1-C inhibitor GO-203 (Fig. 2C) binds to the MUC1-C CQC motif and blocks MUC1-C homodimerization and its oncogenic function46, 47. Treatment of BT-549 cells with GO-203, but not with the control peptide CP-2, was associated with downregulation of EZH2 expression (Fig. 2E, left and right). GO-203 has been formulated in polymeric nanoparticles (NPs) for delivery to tumors in mouse models48. Treatment of H460 tumor xenografts with GO-203/NPs32 resulted in marked downregulation of EZH2 (Fig. 2F, left and right), supporting the premise that targeting the MUC1-C cytoplasmic domain in vitro and in vivo is sufficient for suppression of EZH2 expression.
Targeting the MUC1-C cytoplasmic domain downregulates EZH2 expression. (A) H460 cells stably expressing a control or MUC1-C vector were analyzed for MUC1 (left) and EZH2 (right) mRNA levels by qRT-PCR. The results (mean ± SD) are expressed as relative mRNA levels compared to that obtained for vector cells (assigned a value of 1). (B) Lysates from H460 (left) and BT-549 (right) stably expressing a control or MUC1-C vector were immunoblotted with the indicated antibodies. (C) Schema of the MUC1-C subunit with the extracellular domain (ED), transmembrane domain (TM), and the sequence of the 72 aa cytoplasmic domain (CD). MUC1-CD contains a CQC motif that is necessary for MUC1-C homodimerization and oncogenic function. GO-203 is a cell-penetrating peptide that binds the CQC motif and blocks the formation of MUC1-C homodimers. Highlighted are MUC1-C-induced pathways that confer the activation of NF-κB p65 and MYC. (D) BT-549 cells were transfected with a control or MUC1-C(AQA) vector in which the CQC motif was mutated to AQA. Lysates were immunoblotted with the indicated antibodies. (E) BT-549 cells treated with 5 μM CP-2 or 5 μM GO-203 for 12 h were analyzed for EZH2 mRNA levels by qRT-PCR. The results (mean ± SD) are expressed as relative EZH2 mRNA levels compared to that obtained for CP-2 (assigned a value of 1) (left). Lysates from cells treated with 5 μM CP-2 or 5 μM GO-203 for 48 h were immunoblotted with the indicated antibodies (right). (F) H460 tumors obtained on day 14 of treatment with empty NPs or GO-203/NPs32 were analyzed for EZH2 mRNA levels by qRT-PCR. The results (mean ± SD) are expressed as relative EZH2 mRNA levels compared to that obtained for empty NP-treated cells (assigned a value of 1) (left). Lysates were immunoblotted with the indicated antibodies (right).
MUC1-C activates EZH2 transcription by an E2F-dependent mechanism
Studies with a pEZH2-luciferase promoter-reporter (pEZH2-Luc) (Fig. 3A) demonstrated that targeting MUC1-C in BT-549 cells is associated with a decrease in promoter activity (Fig. 3B). Silencing MUC1-C in H460 cells also decreased pEZH2-Luc activation (Fig. 3C), indicating that MUC1-C drives EZH2 transcription. The EZH2 promoter includes a consensus E2F binding site (TTTGGCGC) (Fig. 3A)11, 49, 50; however, there is no known association between MUC1-C and E2F-mediated gene transcription. Nonetheless, MUC1-C has been linked to the induction of CDK4 and phosphorylation of pRB (phospho-pRB)(Fig. 2C, schema)32. In this respect, we found that silencing MUC1-C decreases phospho-pRB levels (Fig. 3D, left and right) and, accordingly, we asked if MUC1-C induces EZH2 transcription by a pRB→E2F-mediated mechanism. Indeed, silencing E2F in H460/MUC1-C cells was associated with downregulation of pEZH2-Luc activity (Fig. 3E) and EZH2 expression (Fig. 3F). Moreover, ChIP studies demonstrated that silencing MUC1-C decreases occupancy of E2F on the EZH2 promoter (Fig. 3G), supporting the notion that MUC1-C→E2F signaling activates the EZH2 promoter.
Targeting MUC1-C suppresses the EZH2 promoter. (A) Schema of the pEZH2-Luc vector with positioning of the E2F binding site at −12 to −5 bp upstream of the transcription start site. (B and C) The respective BT-549 (B) and H460 (C) cells expressing a CshRNA or MUC1shRNA were transfected with the pGL3-Basic Luc or pEZH2-Luc reporter for 48 h and then analyzed for luciferase activity. The results (mean ± SD of 3 determinations) are expressed as the relative luciferase activity compared to that obtained with pGL3-Basic Luc (assigned a value of 1). (D) Lysates from the respective BT-549 (left) and H460 (right) cells expressing a CshRNA or MUC1shRNA were immunoblotted with the indicated antibodies. (E) H460/MUC1-C cells were transduced with lentiviral vectors to express a control shRNA (CshRNA) or a E2F shRNA. The cells were transfected with the pGL3-Basic Luc or pEZH2-Luc reporter for 48 h and then analyzed for luciferase activity. The results (mean ± SD of 3 determinations) are expressed as the relative luciferase activity compared to that obtained with pGL3-Basic Luc (assigned a value of 1). (F) The respective H460/MUC1-C cells were analyzed for EZH2 mRNA levels by qRT-PCR. The results (mean ± SD) are expressed as relative mRNA levels compared to that obtained for the CshRNA cells (assigned a value of 1). (G) Soluble chromatin from H460/CshRNA and H460/MUC1shRNA cells was precipitated with anti-E2F or a control IgG. The final DNA samples were amplified by qPCR with primers for the EZH2 promoter. The results (mean ± SD of three determinations) are expressed as the relative fold enrichment compared with that for the control IgG (assigned a value of 1).
MUC1-C enhances EZH2 activation by an NF-κB-mediated mechanism
MUC1-C activates the inflammatory TAK1→IKK→NF-κB pathway36 and thereby induces DNMT1 and DNMT3b expression40. Based on these findings and the known integration of EZH2 with DNMTs and DNA methylation2, 20,21,22,23, we asked if MUC1-C also regulates EZH2 expression by an NF-κB-mediated mechanism. Intriguingly, we found that (i) silencing NF-κB p65 decreases EZH2 mRNA (Figs 4A,B, left and right), and (ii) treatment with the NF-κB inhibitor BAY-11-7085 decreases EZH2 protein (Fig. 4C). We therefore searched for putative NF-κB binding sites and identified two consensus sequences downstream to the transcription start site at positions + 388 to + 397 and + 439 to + 449 in the first EZH2 intron (Fig. 4D). In support of a potential enhancer function, incorporation of the intron 1 fragment containing two putative NF-κB binding sites in a luciferase reporter (eEZH2-Luc) demonstrated activation in BT-549 and H460 cells by a MUC1-C-dependent mechanism (Fig. 4E,F). Additionally, we found that NF-κB p65 occupies the EZH2 intron 1 region and that silencing MUC1-C suppresses NF-κB occupancy (Fig. 4G). These and the above findings indicate that MUC1-C (i) activates the EZH2 promoter by E2F-dependent signaling, and (ii) enhances EZH2 transcription by an NF-κB p65-mediated mechanism.
MUC1-C activates an enhancer in the EZH2 intron 1 by an NF-κB-medicated mechanism. A and B. BT-549 (A) and H460 (B) cells were transduced with lentiviral vectors to stably express a control shRNA (CshRNA) or a NF-κB p65 shRNA. The indicated cells were analyzed for NF-κB p65 (left) and EZH2 (right) mRNA levels by qRT-PCR using primers listed in Table S1. The results (mean ± SD) are expressed as relative EZH2 mRNA levels compared to that obtained for CshRNA cells (assigned a value of 1). (C) Lysates from BT-549 cells treated with 5 μM BAY-11-7085 or vehicle control for 48 h were immunoblotted with the indicated antibodies. (D) Schema of the EZH2 intron 1 region with positioning of the putative NF-κB binding sites downstream of the transcription start site. (E and F) The respective BT-549 (E) and H460 (F) cells expressing a CshRNA or MUC1shRNA were transfected with the pGL3-Basic Luc or eEZH2-Luc reporter for 48 h and then analyzed for luciferase activity. The results (mean ± SD of 3 determinations) are expressed as the relative luciferase activity compared to that obtained with pGL3-Basic Luc (assigned a value of 1). (G) Soluble chromatin from BT-549/CshRNA and BT-549/MUC1shRNA cells was precipitated with anti-NF-κB p65 or a control IgG. The final DNA samples were amplified by qPCR with primers for the EZH2 intron 1 region (Table S2). The results (mean ± SD of three determinations) are expressed as the relative fold enrichment compared with that obtained with the IgG control (assigned a value of 1).
MUC1-C binds directly to EZH2
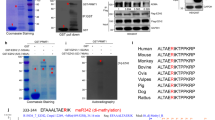
MUC1-C interacts with certain transcriptional complexes28 and contributes to the recruitment of epigenetic regulators, such as the histone acetyltransferase p30031, 32. To determine if MUC1-C interacts with EZH2, we performed ChIP studies on the CDH1 promoter, which is a target for EZH2-mediated repression18, 19 and found occupancy of both EZH2 and MUC1-C (Fig. 5A). Re-ChIP studies further showed that EZH2 and MUC1-C form a complex on the CDH1 promoter (Fig. 5B). Similar results were obtained in studies of the CDH1 promoter in H460 cells; that is, (i) occupancy by both MUC1-C and EZH2 (Supplemental Fig. S2A), and (ii) detection of MUC1-C/EZH2 complexes (Supplemental Fig. S2B). EZH2 consists of 751 aa, which include a WD-repeat binding domain, two adjacent SANT/Myb domains, a CXC domain and a SET domain that catalyzes methylation of H3K27 (NCBI Accession NM_004456; Fig. 5C)51. To further assess the nature of the association between EZH2 and MUC1-C, we first generated GST-EZH2 fragments that included aa 1–500 and 501–751 (Fig. 5C). Incubation of these fragments with the MUC1-C cytoplasmic domain (MUC1-CD) demonstrated binding to EZH2(501–751), and not EZH2(1–500) (Fig. 5D), supporting a direct interaction. Based on these results, we incubated GST-EZH2 with MUC1-CD fragments and found that MUC1-CD(1–45), and not MUC1-CD(46–72), confers the interaction (Fig. 5E, left and right). MUC1-CD contains a CQC motif at residues 1–3 that is necessary for interactions with certain binding partners (Fig. 2C)31, 47. Mutation of both Cys residues to Ala (AQA) blocked the interaction between MUC1-CD and EZH2 (Fig. 5F). The EZH2(501–751) fragment includes a CXC domain (aa 508 to 610) and the SET catalytic domain (aa 617 to 738). Accordingly, we generated GST-EZH2(501–614) and GST-EZH2(615–751). Incubation of these fragments with MUC1-CD(1–45) demonstrated substantially higher binding with EZH2(501–614) as compared with EZH2(615–751) (Fig. 5G). These findings demonstrate that the MUC1-C cytoplasmic domain interacts predominantly with EZH2 at the CXC region adjacent to the catalytic SET domain.
MUC1-C forms complexes with EZH2 by binding directly to the EZH2 CXC domain. (A) Soluble chromatin from BT-549 cells was precipitated with anti-EZH2, anti-MUC1-C or a control IgG. The final DNA samples were amplified by qPCR with primers for the CDH1 promoter (Table S2). The results (mean ± SD of three determinations) are expressed as the relative fold enrichment compared with that obtained with the IgG control (assigned a value of 1). (B) In the re-ChIP analysis, anti-EZH2 precipitates were released and re-immunoprecipitated with anti-MUC1-C or a control IgG. The final DNA samples were amplified by qPCR with primers for the CDH1 promoter. The results (mean ± SD of three determinations) are expressed as the relative fold enrichment compared with that obtained with the IgG control (assigned a value of 1). See also Fig. S2. (C) Schema of the 751 aa EZH2 protein highlighting the WD-repeat binding domain, two SANT/Myb domains, a CXC domain and a SET domain that catalyzes methylation of H3K27. Also highlighted are the GST-EZH2 fragments used for direct binding studies. (D) GST-EZH2(1-500) and GST-EZH2(501–751) were incubated with purified MUC1-C cytoplasmic domain (MUC1-CD). The adsorbates were immunoblotted with anti-MUC1-CD. Input of the GST proteins was assessed by Coomassie blue staining. (E) GST and GST-EZH2 were incubated with purified MUC1-CD(1–45) (left) or MUC1-CD(46–72) (right). The adsorbates were immunoblotted with the appropriate anti-MUC1-CD antibody (CD1, left; CT2, right). Input of the GST proteins was assessed by Coomassie blue staining. (F) GST and GST-EZH2 were incubated with purified MUC1-CD and MUC1-CD(AQA). The adsorbates were immunoblotted with anti-MUC1-CD. Input of the GST proteins was assessed by Coomassie blue staining. (G) GST-EZH2(501–614) and GST-EZH2(615–751) were incubated with purified MUC1-CD(1–45). The adsorbates were immunoblotted with anti-MUC1-CD (MAb CD1). Input of the GST proteins was assessed by Coomassie blue staining
Targeting MUC1-C decreases global and CDH1 promoter-specific H3K27 trimethylation
The demonstration that MUC1-C induces EZH2 expression and binds directly to EZH2 prompted studies to assess the effects of targeting MUC1-C on global H3K27 trimethylation. We found that silencing MUC1-C in BT-549 cells is associated with decreases in global H3K27me3 levels (Fig. 6A). Similar results were obtained in H460 cells (Fig. 6B). Treatment of BT-549/tet-MUC1shRNA (Fig. 6C) and MDA-MB-231/tet-MUC1shRNA (Fig. 6D) cells with DOX was also associated with downregulation of H3K27me3. In addition, overexpression of MUC1-C increased H3K27me3 levels (Supplemental Fig. S3A). In concert with the findings that MUC1-C/NF-κB p65 complexes activate EZH2 transcription, we also found that targeting NF-κB p65 with silencing or BAY-11-7085 decreases H3K27me3 levels (Supplemental Fig. S3B–D). ChIP studies on the CDH1 promoter further demonstrated that silencing MUC1-C decreases H3K27me3 levels in association with increases in E-cadherin expression (Fig. 6E,F, left and right), providing further support for the notion that MUC1-C drives EZH2-mediated H3K27 trimethylation.
Targeting MUC1-C represses global and CDH1 promoter-specific H3K27me3 levels. (A and B) Lysates from the respective BT-549 (A) and H460 (B) cells expressing a CshRNA or MUC1shRNA were immunoblotted with anti-H3K27me3 and anti-histone H3. (C and D) Lysates from BT-549/tet-MUC1shRNA (C) and MDA-MB-231/tetMUC1shRNA (D) cells treated with 200 ng/ml DOX for 4 d were immunoblotted with the indicated antibodies. E and F. Soluble chromatin from the respective BT-549 (E) and H460 (F) cells expressing a CshRNA or MUC1shRNA was precipitated with anti-H3K27me3 or a control IgG. The final DNA samples were amplified by qPCR with primers for the CDH1 promoter. The results (mean ± SD of three determinations) are expressed as the relative fold enrichment compared with that obtained with the IgG control (assigned a value of 1) (left). Cells were also analyzed for E-cadherin mRNA levels by qRT-PCR using primers listed in Table S1. The results (mean ± SD) are expressed as relative mRNA levels compared to that obtained for the CshRNA cells (assigned a value of 1) (right). See also Fig. S3.
MUC1-C→EZH2 signaling represses expression of the BRCA1 tumor suppressor
To extend this investigation of the MUC1-C→EZH2 pathway, we performed RNA-seq analysis of cells without and with MUC1 silencing. An unanticipated outcome was the finding of a highly significant (p < 1 × 10−12) relationship with up- and down-regulated genes encoding effectors of the DNA damage response, including BRCA1, CHK2 and RAD51, among many others (Supplemental Fig. S4A). In keeping with the focus of the present work, we confirmed that MUC1 expression negatively correlates with that of BRCA1 in datasets from breast cancers (Supplemental Fig. S4B) and NSCLCs (Supplemental Fig. S4C). In addition, silencing MUC1 was associated with upregulation of BRCA1 in BT-549 (Fig. 7A, left and right) and H460 (Supplemental Fig. S5A, left and right) cells. MUC1-C has been linked to the repression of TSGs by DNMT- and PRC1-mediated epigenetic mechanisms34, 40. However, to our knowledge there is no reported association between MUC1 or EZH2 and BRCA1 gene repression. We therefore treated cells with the EZH2 inhibitor GSK343 and found upregulation of BRCA1 mRNA and protein levels (Fig. 7B, left and right; Supplemental Fig. S5B, left and right), indicating that, like MUC1-C, targeting EZH2 induces BRCA1 expression. ChIP studies further demonstrated that both MUC1-C and EZH2 occupy the BRCA1 promoter (Fig. 7C, left and right; Supplemental Fig. S5C, left and right). Re-ChIP experiments also showed that MUC1-C and EZH2 form a complex on the BRCA1 promoter (Fig. 7D; Supplemental Fig. S5D). Moreover, silencing MUC1-C was associated with suppression of H3K27 trimethylation of the BRCA1 promoter (Fig. 7E; Supplemental Fig. S5E), supporting a model in which the MUC1-C→EZH2→H3K27me3 pathway promotes repression of the BRCA1 gene.
Targeting MUC1-C activates the BRCA1 promoter by suppressing EZH2 occupancy and H3K27me3 levels. (A) BT-549/CshRNA and BT-549/MUC1shRNA cells were analyzed for BRCA1 mRNA levels by qRT-PCR. The results (mean ± SD) are expressed as relative mRNA levels compared to that obtained for the CshRNA cells (assigned a value of 1) (left). Lysates were immunoblotted with the indicated antibodies (right). (B) BT-549 cells treated with vehicle control or 10 μM GSK343 for 72 h were analyzed for BRCA1 mRNA levels by qRT-PCR. The results (mean ± SD) are expressed as relative mRNA levels compared to that obtained for the Control cells (assigned a value of 1) (left). Lysates were immunoblotted with the indicated antibodies (right). (C) Soluble chromatin from BT-549 cells was precipitated with anti-EZH2 (left), anti-MUC1-C (right) or a control IgG. (D) In the re-ChIP analysis, EZH2 precipitates were released and re-immunoprecipitated with anti-MUC1-C and a control IgG. (E) Soluble chromatin from BT-549/CshRNA and BT-549/MUC1shRNA cells was precipitated with anti-H3K27me3 or a control IgG. The final DNA samples were amplified by qPCR with primers for the BRCA1 promoter. The results (mean ± SD of three determinations) are expressed as the relative fold enrichment compared with that obtained with the IgG control (assigned a value of 1).
Discussion
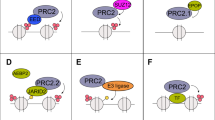
EZH2 has emerged as a highly attractive target based on its elevated expression in human carcinomas and association with poor clinical outcomes52. Gain- and loss-of-function mutations in EZH2 have also been identified in certain hematologic malignancies53,54,55. In addition, CML stem cells are dependent on EZH2 for survival56, 57, further supporting the need for agents that target EZH2 and the PRC2 complex. Indeed, EZH2 has been proposed as a master regulator of gene transcription in the promotion of cancer6, 52. The present studies demonstrate that MUC1-C drives EZH2 expression in TNBC, NSCLC and other types of carcinoma cells. Additionally, we found that MUC1-C promotes the expression of SUZ12 and EED. Therefore, targeting MUC1-C can inactivate the PRC2 complex in multiple ways, including downregulation of EZH2, as well as suppression of SUZ12 and EED, which are required for EZH2 HMT activity1. We focused here on how MUC1-C activates EZH2 based largely on its dysregulation in cancer. Accordingly, subsequent work will be needed to address the role of MUC1-C in driving SUZ12 and EED expression. MUC1-C induces MYC transcription by activation of the β-catenin/TCF4 pathway32, 33. Thus, targeting MUC1-C decreases expression of MYC and its downstream target genes, such as CDK4 32. In turn, targeting MUC1-C indirectly suppresses pRB activity32. The present results uncover a previously unrecognized role for MUC1-C in activation of the pRB→E2F pathway and thereby the EZH2 promoter (Fig. 8). Interestingly, pRB→E2F signaling has also been shown to activate EED gene transcription11. Indeed, in the course of these experiments, we found that MUC1-C also activates EED expression by a pRB→E2F-mediated mechanism.
Schemas depicting the proposed MUC1-C-induced regulation of EZH2 expression and function in epigenetic repression. (A) MUC1-C drives EZH2 expression by inducing (i) the pRB→E2F pathway and in turn E2F-mediated activation of the EZH2 promoter, and (ii) NF-κB p65 occupancy of the EZH2 intron 1 and enhancing EZH2 transcription. (B) MUC1-C binds directly to EZH2, increases EZH2 occupancy on TSG promoters and enhances EZH2-mediated H3K27 trimethylation with repression of gene expression.
The MUC1-C cytoplasmic domain activates the β-catenin/TCF4 pathway by binding directly to β-catenin and promoting β-catenin occupancy on promoters of WNT target genes, such as CCND1 and MYC 30,31,32,33. The MUC1-C cytoplasmic domain also promotes activation of the TAK1→IKK→NF-κB inflammatory pathway, binds directly to NF-κB p65 and promotes occupancy of NF-κB p65 on its target genes, including ZEB1 and LIN28B, among others29, 35,36,37, 39. Overexpression of MUC1-C in carcinomas thereby subverts the NF-κB pathway in driving the induction of EMT37, 39. The effects of MUC1-C on NF-κB p65 activation have also been linked to induction of self-renewal capacity and stemness of cancer cells38, 39. Such characteristics of EMT, self-renewal and stemness depend, at least in part, on epigenetic regulatory mechanisms involving PRC2 to achieve the associated changes in gene expression patterns2. However, to our knowledge, there had been no known link between MUC1-C→NF-κB signaling and the induction of EZH2 expression. In searching for such evidence, we found that MUC1-C/NF-κB p65 complexes occupy consensus NF-κB binding sites in the EZH2 first intron and activate EZH2 transcription. These results and those obtained with E2F support a model in which MUC1-C induces EZH2 expression by the β-catenin/TCF4→MYC and the NF-κB pathways (Fig. 8). Of note, our findings do not exclude the possibility that MUC1-C regulates EZH2 expression by additional mechanisms. For instance, MYC suppresses miR-26a, which targets EZH2 mRNA25, 26, 58. In addition, MUC1-C activates LIN28B and thereby suppresses let-7, another miRNA that targets EZH2 expression39, 59.
MUC1-C induces the expression of DNMT1 and DNMT3b, but not DNMT3a, in carcinoma cells40. As a result, MUC1-C controls global and TSG promoter-specific DNA methylation40. Interestingly in this regard, EZH2 functions as a recruitment platform for DNMTs, linking H3K27 methylation and DNA methylation in gene repression20, 21, 23. An unexpected finding was that, in addition to inducing EZH2 expression in cancer cells, MUC1-C was detectable in complexes with EZH2 on the CDH1 and BRCA1 promoters, invoking the notion that MUC1-C associates with EZH2. EZH2 contains a WD repeat domain that is necessary for binding to EED and thereby activation of the catalytic HMT SET domain. EZH2 also includes SANT DNA binding domains and a highly conserved CXC domain that may contribute to an inactive configuration of the SET domain60. Our results demonstrate that the MUC1-C cytoplasmic domain CQC motif binds directly with the EZH2 CXC domain. The MUC1-C CQC motif is necessary and sufficient for the formation of MUC1-C homodimers and their import into the nucleus44. The MUC1-C CQC motif has also been shown to confer interactions with certain transcription factors, including TCF4 and others31, 37, 61, supporting the premise that this motif is also of importance for binding to nuclear proteins. MUC1-C may thus play dual roles in regulating EZH2; namely, (i) induction of EZH2 expression and (ii) direct binding to the EZH2 CXC motif and thereby affecting the SET domain HMT activity. In this regard, our results further demonstrate that MUC1-C forms a complex with EZH2 on the CDH1 and BRCA1 promoters and enhances H3K27 trimethylation of those regions (Fig. 8).
EZH2-mediated H3K27 trimethylation acts as a site for recruitment of (i) the PRC1 complex, and (ii) DNMTs, and thereby links these epigenetic mechanisms of gene silencing1, 20. MUC1-C is necessary for expression of PRC1 complex members, B cell-specific Moloney murine leukemia virus integration site 1 (BMI1), RING1 and RING234. MUC1-C also binds directly to BMI1 and promotes occupancy of BMI1 on target promoters34. Given the diversity by which MUC1-C drives the functions of PRC2, PRC1 and DNMTs in epigenetic gene silencing, we performed RNA-seq on cells without and with MUC1-C silencing. The findings demonstrated that MUC1-C regulates diverse genes involved in DNA repair pathways. For instance, in the homologous recombination DNA repair pathway, we found that, like BRCA1, MUC1-C represses CHK2 and RAD51 expression by an EZH2-mediated mechanism (unpublished data). Targeting MUC1-C also activates genes in the mismatch repair, base-excision repair and DNA interstrand cross-link repair pathways, suggesting that the overexpression of MUC1-C as found in human carcinomas could contribute to genomic instability. One task at hand is to now investigate which of the potential MUC1-C-induced epigenetic changes involving PRC2, PRC1 and/or DNA methylation contribute to the downregulation of these additional DNA repair genes. The present findings and the involvement of MUC1-C in driving EMT and immune evasion thereby support the integration of multiple phenotypic characteristics of the cancer stem-like cell (CSC) and a mechanistic basis for the development of anti-cancer drug resistance62. Another task at hand is to target MUC1-C and thereby suppress this integrated CSC program in human tumors. For that purpose, the MUC1-C inhibitor, GO-203, has been evaluated in Phase I clinical trials and, based on a favorable safety profile, has been formulated in polymeric nanoparticles for sustained delivery to patients with MUC1-C-expressing cancers48.
Experimental Procedures
Cell culture
Human BT-549 breast cancer, H460 NSCLC, A549 NSCLC and DU145 prostate cancer cells were grown in RPMI1640 medium (ATCC, Manassas, VA, USA). MDA-MB-231 and MDA-MB-468 breast cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Corning, Manassas, VA, USA). BT-20 cells were cultured in Eagle’s Minimum Essential Medium (EMEM) (ATCC). Media were supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS), 100 U/ml penicillin and 100 μg/ml streptomycin. Cells were transduced to stably express a control scrambled CshRNA or a MUC1 shRNA40. Cells stably expressing an empty vector or MUC1-C were generated as described63. Cells were treated with the MUC1-C inhibitor GO-203 or the control CP-2 peptide38. Cells were also treated with the NF-κB inhibitor BAY-11-7085 (Santa Cruz Biotechnology, Dallas, TX, USA) and the EZH2 inhibitor GSK343 (SelleckChem, Houston, TX, USA). Authentication of cells was performed by short tandem repeat (STR) analysis. Cell were monitored for mycoplasma contamination using the MycoAlert® Mycoplasma Detection Kit (Lonza, Rockland, ME, USA).
Tetracycline-inducible MUC1 silencing
MUC1shRNA (MISSION shRNA; Sigma, TRCN0000122938) or a control scrambled CshRNA (Sigma) was inserted into the pLKO-tet-puro vector (Addgene, Cambridge, MA, USA; Plasmid #21915). The viral vectors were produced in HEK293T cells as previously described63, 64. BT-549 and MDA-MB-468 cells expressing tet-CshRNA or tet-MUC1shRNA were selected for growth in 1–3 μg/ml puromycin. Cells were treated with doxycycline (DOX; Sigma).
Real-time quantitative reverse-transcription PCR (qRT-PCR)
Total RNA was isolated using with Trizol reagent (Invitrogen, Carlsbad, CA, USA). Complementary DNA was synthesized from 2.0 μg total RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Grand Island, NY, USA)65. The Power SYBR Green PCR Master Mix (Applied Biosystems) was used with 1 μl of diluted cDNA for each sample. The samples were amplified using the 7300 Realtime PCR System (Applied Biosystems). Primers used for qRT–PCR analysis are listed in Supplemental Table S1.
EZH2 promoter and enhancer luciferase reporter assays
Cells growing in 24-well plates were transfected with (i) an empty pGL3 vector, (ii) a pEZH2-Luc containing EZH2 promoter sequences −703 to + 320 relative to the TSS (Active Motif, Carlsbad, CA, USA), or (ii) eEZH2-Luc containing EZH2 intron 1 sequences +115 to +615 bp downstream to the TSS, and SV-40-Renilla-Luc in the presence of LipofectamineTM 3000 Reagent (Invitrogen). At 48 h after transfection, cell extracts were prepared with passive lysis buffer using the Luciferase® Assay System (Promega, Madison, WI, USA). Luminescence was measured with the Dual-Luciferase® Reporter Assay System (Promega).
Chromatin immunoprecipitation (ChIP) assay
Soluble chromatin was precipitated with anti-MUC1-C (NeoMarkers, Fremont, CA, USA), anti-E2F (Cell Signaling Technology, Danvers, MA, USA), anti-NF-κB p65 (Santa Cruz Biotechnology), anti-H3K27me3 (Abcam, Cambridge, MA, USA), anti-H3K27 (Abcam), or a control non-immune IgG (Santa Cruz Biotechnology). For re-ChIP analysis, complexes from the primary ChIP were eluted and re-immunoprecipitated with a secondary antibody. For real-time ChIP qPCR, the SYBR green system was used with the ABI Prism 7300 sequence detector (Applied Biosystems). Data are reported as relative-fold enrichment40. Primers used for ChIP qPCR are listed in the Supplementary Table S2.
Immunoblot analysis
Whole cells were lysed in NP-40 buffer, containing protease inhibitor cocktail (ThermoScientific, Waltham, MA, USA). Immunoblotting was performed with anti-MUC1-C (NeoMarkers), anti-EZH2 (Cell Signaling Technology), anti-phospho-pRB, anti-pRB (BD Biosciences, San Jose, CA, USA), anti-H3K27me3 (Abcam), anti-E-cadherin (Cell Signaling Technology), anti-NF-κB p65 (Santa Cruz Biotechnology) and anti-β-actin (Sigma).
Protein binding assays
GST-EZH2 was purified from pGEX-EZH2 (Addgene; Plasmid #28060). GST-EZH2(1–500), GST-EZH2(501–751), GST-EZH2(501–614) and GST-EZH2(615–751) fragments were generated by PCR amplification of the pGEX-EZH2 plasmid and cloning into the pGEX-5X-1 bacterial expression plasmid backbone (GE Healthcare, Piscataway, NJ, USA). MUC1-CD, MUC1-CD(AQA) and the MUC1-CD(1–45) and MUC1-CD(46–72) fragments were prepared by expressing the relevant GST-fusion proteins and cleaving the GST tag with thrombin as described37. GST and GST fusion proteins bound to glutathione beads were incubated with purified proteins, washed and the adsorbates were analyzed by immunoblotting with anti-MUC1-C cytoplasmic domain antibodies CD166 and CT2 (NeoMarker).
Statistical analysis
Each experiment was repeated at least three times. Data are expressed as mean ± SD. The unpaired Student’s t-test was used to examine differences between means of two groups. A p-value < 0.05 was considered a statistically significant difference.
References
Mills, A. A. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer 10, 669–682, doi:10.1038/nrc2931 (2010).
Tam, W. L. & Weinberg, R. A. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 19, 1438–1449, doi:10.1038/nm.3336 (2013).
Sparmann, A. & van Lohuizen, M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 6, 846–856, doi:10.1038/nrc1991 (2006).
Bracken, A. P. & Helin, K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer 9, 773–784, doi:10.1038/nrc2736 (2009).
Cao, R. & Zhang, Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell 15, 57–67, doi:10.1016/j.molcel.2004.06.020 (2004).
Comet, I., Riising, E. M., Leblanc, B. & Helin, K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer, doi:10.1038/nrc.2016.83 (2016).
Gong, Y. et al. Polycomb group protein EZH2 is frequently expressed in inflammatory breast cancer and is predictive of worse clinical outcome. Cancer 117, 5476–5484, doi:10.1002/cncr.26179 (2011).
Collett, K. et al. Expression of enhancer of zeste homologue 2 is significantly associated with increased tumor cell proliferation and is a marker of aggressive breast cancer. Clin Cancer Res 12, 1168–1174, doi:10.1158/1078-0432.CCR-05-1533 (2006).
Zhang, X. et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet 48, 176–182, doi:10.1038/ng.3470 (2016).
Varambally, S. et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419, 624–629, doi:10.1038/nature01075 (2002).
Bracken, A. P. et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 22, 5323–5335, doi:10.1093/emboj/cdg542 (2003).
Sauvageau, M. & Sauvageau, G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 7, 299–313, doi:10.1016/j.stem.2010.08.002 (2010).
Behrens, C. et al. EZH2 protein expression associates with the early pathogenesis, tumor progression, and prognosis of non-small cell lung carcinoma. Clin Cancer Res 19, 6556–6565, doi:10.1158/1078-0432.CCR-12-3946 (2013).
Sato, T. et al. PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer. Sci Rep 3, 1911, doi:10.1038/srep01911 (2013).
Chen, S. et al. Enhancer of zeste homolog 2 as an independent prognostic marker for cancer: a meta-analysis. PLoS One 10, e0125480, doi:10.1371/journal.pone.0125480 (2015).
Wang, X. et al. Prognostic significance of EZH2 expression in non-small cell lung cancer: A meta-analysis. Sci Rep 6, 19239, doi:10.1038/srep19239 (2016).
Kleer, C. G. et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA 100, 11606–11611, doi:10.1073/pnas.1933744100 (2003).
Cao, Q. et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 27, 7274–7284, doi:10.1038/onc.2008.333 (2008).
Herranz, N. et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol 28, 4772–4781, doi:10.1128/MCB.00323-08 (2008).
Vire, E. et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439, 871–874, doi:10.1038/nature04431 (2006).
Schlesinger, Y. et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39, 232–236, doi:10.1038/ng1950 (2007).
Cedar, H. & Bergman, Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature reviews. Genetics 10, 295–304, doi:10.1038/nrg2540 (2009).
Rose, N. R. & Klose, R. J. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta 1839, 1362–1372, doi:10.1016/j.bbagrm.2014.02.007 (2014).
Koh, C. M. et al. MYC and prostate cancer. Genes Cancer 1, 617–628, doi:10.1177/1947601910379132 (2010).
Sander, S. et al. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood 112, 4202–4212, doi:10.1182/blood-2008-03-147645 (2008).
Lu, J. et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res 71, 225–233, doi:10.1158/0008-5472.CAN-10-1850 (2011).
Kufe, D. Mucins in cancer: function, prognosis and therapy. Nature Reviews Cancer 9, 874–885, doi:10.1038/nrc2761 (2009).
Kufe, D. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene 32, 1073–1081, doi:10.1038/onc.2012.158 (2013).
Ahmad, R. et al. MUC1-C oncoprotein functions as a direct activator of the NF-κB p65 transcription factor. Cancer Res 69, 7013–7021 (2009).
Huang, L. et al. MUC1 oncoprotein blocks GSK3β-mediated phosphorylation and degradation of β-catenin. Cancer Res. 65, 10413–10422 (2005).
Rajabi, H. et al. MUC1-C oncoprotein induces TCF7L2 transcription factor activation and promotes cyclin D1 expression in human breast cancer cells. J Biol Chem 287, 10703–10713 (2012).
Bouillez, A. et al. Inhibition of MUC1-C suppresses MYC expression and attenuates malignant growth in KRAS mutant lung adenocarcinomas. Cancer Res 76, 1538–1548, doi:10.1158/0008-5472.CAN-15-1804 (2016).
Tagde, A. et al. MUC1-C drives MYC in multiple myeloma. Blood 127, 2587–2597, doi:10.1182/blood-2015-07-659151 (2016).
Hiraki, M. et al. MUC1-C activates BMI1 in human cancer cells. Oncogene, doi:10.1038/onc.2016.439 (2016 Nov 28 [Epub ahead of print]).
Ahmad, R. et al. MUC1 oncoprotein activates the IκB kinase β complex and constitutive NF-κB signaling. Nat Cell Biol 9, 1419–1427 (2007).
Takahashi, H. et al. MUC1-C activates the TAK1 inflammatory pathway in colon cancer. Oncogene 34, 5187–5197, doi:10.1038/onc.2014.442 (2015).
Rajabi, H. et al. MUC1-C oncoprotein activates the ZEB1/miR-200c regulatory loop and epithelial-mesenchymal transition. Oncogene 33, 1680–1689, doi:10.1038/onc.2013.114 (2014).
Alam, M., Rajabi, H., Ahmad, R., Jin, C. & Kufe, D. Targeting the MUC1-C oncoprotein inhibits self-renewal capacity of breast cancer cells. Oncotarget 5, 2622–2634 (2014).
Alam, M., Ahmad, R., Rajabi, H. & Kufe, D. MUC1-C induces the LIN28B → LET-7 → HMGA2 axis and self-renewal in NSCLC cells. Mol Cancer Res 13, 449–460 (2015).
Rajabi, H. et al. DNA methylation by DNMT1 and DNMT3b methyltransferases is driven by the MUC1-C oncoprotein in human carcinoma cells. Oncogene 35, 6439–6445, doi:10.1038/onc.2016.180 (2016).
Tagde, A. et al. MUC1-C induces DNA methyltransferase 1 and represses tumor suppressor genes in acute myeloid leukemia. Oncotarget 7, 38974–38987, doi:10.18632/oncotarget.9777 (2016).
Margueron, R. & Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349, doi:10.1038/nature09784 (2011).
Li, Y., Liu, D., Chen, D., Kharbanda, S. & Kufe, D. Human DF3/MUC1 carcinoma-associated protein functions as an oncogene. Oncogene 22, 6107–6110 (2003).
Leng, Y. et al. Nuclear import of the MUC1-C oncoprotein is mediated by nucleoporin Nup62. J Biol Chem 282, 19321–19330 (2007).
Kufe, D. Functional targeting of the MUC1 oncogene in human cancers. Cancer biology & therapy 8, 1201–1207 (2009).
Raina, D. et al. Targeting cysteine-mediated dimerization of the MUC1-C oncoprotein in human cancer cells. Int J Oncol 40, 1643–1649, doi:10.3892/ijo.2011.1308 (2012).
Raina, D. et al. Characterization of the MUC1-C cytoplasmic domain as a cancer target. PLoS One 10, e0135156, doi:10.1371/journal.pone.0135156 (2015).
Hasegawa, M. et al. Intracellular targeting of the oncogenic MUC1-C protein with a novel GO-203 nanoparticle formulation. Clin Cancer Res 21, 2338–2347, doi:10.1158/1078-0432.CCR-14-3000 (2015).
Muller, H. et al. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev 15, 267–285, doi:10.1101/gad.864201 (2001).
Rabinovich, A., Jin, V. X., Rabinovich, R., Xu, X. & Farnham, P. J. E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Res 18, 1763–1777, doi:10.1101/gr.080622.108 (2008).
Fu, H. L. et al. TET1 exerts its tumor suppressor function by interacting with p53-EZH2 pathway in gastric cancer. J Biomed Nanotechnol 10, 1217–1230 (2014).
Kim, K. H. & Roberts, C. W. Targeting EZH2 in cancer. Nat Med 22, 128–134, doi:10.1038/nm.4036 (2016).
Morin, R. D. et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 42, 181–185, doi:10.1038/ng.518 (2010).
Yap, D. B. et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117, 2451–2459, doi:10.1182/blood-2010-11-321208 (2011).
McCabe, M. T. et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc Natl Acad Sci USA 109, 2989–2994, doi:10.1073/pnas.1116418109 (2012).
Xie, H. et al. Chronic myelogenous leukemia-initiating cells require polycomb group protein EZH2. Cancer Discov 6, 1237–1247, doi:10.1158/2159-8290.CD-15-1439 (2016).
Scott, M. T. et al. Epigenetic reprogramming sensitizes CML stem cells to combined EZH2 and tyrosine kinase inhibition. Cancer Discov 6, 1248–1257, doi:10.1158/2159-8290.CD-16-0263 (2016).
Koh, C. M. et al. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget 2, 669–683, doi:10.18632/oncotarget.327 (2011).
Zhan, M. et al. JMJD1A promotes tumorigenesis and forms a feedback loop with EZH2/let-7c in NSCLC cells. Tumour Biol 37, 11237–11247, doi:10.1007/s13277-016-4999-9 (2016).
Wu, H. et al. Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations. PLoS One 8, e83737, doi:10.1371/journal.pone.0083737 (2013).
Alam, M., Ahmad, R., Rajabi, H., Kharbanda, A. & Kufe, D. MUC1-C oncoprotein activates ERK → C/EBPβ-mediated induction of aldehyde dehydrogenase activity in breast cancer cells. J Biol Chem 288, 30829–30903 (2013).
Rajabi, H. & Kufe, D. MUC1-C oncoprotein integrates a program of EMT, epigenetic reprogramming and immune evasion in human carcinomas. BBA Reviews on Cancer 1868, 117–122, doi:10.1016/j.bbcan.2017.03.003 (2017).
Hiraki, M. et al. MUC1-C stabilizes MCL-1 in the oxidative stress response of triple-negative breast cancer cells to BCL-2 inhibitors. Sci Rep 6, 26643, doi:10.1038/srep26643 (2016).
Hasegawa, M. et al. Functional interactions of the cystine/glutamate antiporter, CD44v and MUC1-C oncoprotein in triple-negative breast cancer cells. Oncotarget 7, 11756–11769, doi:10.18632/oncotarget.7598 (2016).
Hiraki, M. et al. Concurrent targeting of KRAS and AKT by MiR-4689 is a novel treatment against mutant KRAS colorectal cancer. Mol Ther Nucleic Acids 4, e231, doi:10.1038/mtna.2015.5 (2015).
Panchamoorthy, G., Rehan, H., Kharbanda, A., Ahmad, R. & Kufe, D. A monoclonal antibody against the oncogenic mucin 1 cytoplasmic domain. Hybridoma 30, 531–535 (2011).
Acknowledgements
This work was supported by Grants from the National Cancer Institute of the National Institutes of Health under award numbers CA97098 and CA166480.
Author information
Authors and Affiliations
Contributions
H.R., M.H., K.K.W. and D.K. designed the research. H.R., M.H., A.T., M.A., A.B., and C.L.C. performed the research and data analysis. M.S. performed bioinformatics analysis. H.R., K.K.W. and D.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare competing financial interests, D.K. holds equity in Genus Oncology and is a consultant to the company. The other authors disclosed no potential conflicts of interest.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rajabi, H., Hiraki, M., Tagde, A. et al. MUC1-C activates EZH2 expression and function in human cancer cells. Sci Rep 7, 7481 (2017). https://doi.org/10.1038/s41598-017-07850-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-07850-0
This article is cited by
-
MUC1-C intersects chronic inflammation with epigenetic reprogramming by regulating the set1a compass complex in cancer progression
Communications Biology (2023)
-
The multifaceted role of MUC1 in tumor therapy resistance
Clinical and Experimental Medicine (2022)
-
MUC1-C activates the PBAF chromatin remodeling complex in integrating redox balance with progression of human prostate cancer stem cells
Oncogene (2021)
-
High level of EZH2 expression is linked to high density of CD8-positive T-lymphocytes and an aggressive phenotype in renal cell carcinoma
World Journal of Urology (2021)
-
MUC1 oncoprotein mitigates ER stress via CDA-mediated reprogramming of pyrimidine metabolism
Oncogene (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.