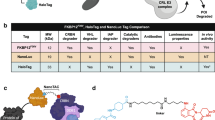
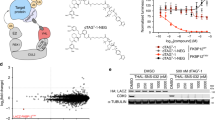
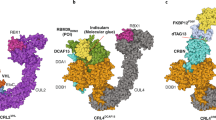
Abstract
The rational development of small-molecule degraders (e.g., proteolysis targeting chimeras) remains a challenge as the rate-limiting steps that determine degrader efficiency are largely unknown. Standard methods in the field of targeted protein degradation mostly rely on classical, low-throughput endpoint assays such as western blots or quantitative proteomics. Here we applied NanoLuciferase- and HaloTag-based screening technologies to determine the kinetics and stability of small-molecule-induced ternary complex formation between a protein of interest and a selected E3 ligase. A collection of live-cell assays were designed to probe the most critical steps of the degradation process while minimizing the number of required expression constructs, making the proposed assay pipeline flexible and adaptable to the requirements of the users. This approach evaluates the underlying mechanism of selective target degraders and reveals the exact characteristics of the developed degrader molecules in living cells. The protocol allows scientists trained in basic cell culture and molecular biology to carry out small-molecule proximity-inducer screening via tracking of the ternary complex formation within 2 weeks of establishment, while degrader screening using the HiBiT system requires a CRISPR–Cas9 engineered cell line whose generation can take up to 3 months. After cell-line generation, degrader screening and validation can be carried out in high-throughput manner within days.
Key points
-
This protocol describes the application of NanoLuciferase- and HaloTag-based screening technologies to measure the whole cascade of targeted protein degradation from binary complexes to live-cell target degradation kinetics.
-
Compared with conventional endpoint assays such as western blots or quantitative proteomics, live-cell assays take dynamic cellular processes into account such as the complete degradation machinery, intracellular competitors or posttranslational modifications during time-resolved measurements.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Any additional information required to reanalyze the data reported in this manuscript is available upon request from the lead contact. Plasmids have been made available on Addgene.
References
Schwalm, M. P. & Knapp, S. BET bromodomain inhibitors. Curr. Opin. Chem. Biol. 68, 102148 (2022).
Hu, Z. & Crews, C. M. Recent developments in PROTAC-mediated protein degradation: From bench to clinic. Chembiochem 23, e202100270 (2022).
Petrylak, D. P. et al. First-in-human phase I study of ARV-110, an androgen receptor (AR) PROTAC degrader in patients (pts) with metastatic castrate-resistant prostate cancer (mCRPC) following enzalutamide (ENZ) and/or abiraterone (ABI). J. Clin. Oncol. 38, 3500–3500 (2020).
Paiva, S. L. & Crews, C. M. Targeted protein degradation: elements of PROTAC design. Curr. Opin. Chem. Biol. 50, 111–119 (2019).
Nemec, V., Schwalm, M. P., Muller, S. & Knapp, S. PROTAC degraders as chemical probes for studying target biology and target validation. Chem. Soc. Rev. 51, 7971–7993 (2022).
Roy, M. J. et al. SPR-measured dissociation kinetics of PROTAC ternary complexes influence target degradation rate. ACS Chem. Biol. 14, 361–368 (2019).
Casement, R., Bond, A., Craigon, C. & Ciulli, A. Mechanistic and structural features of PROTAC ternary complexes. Methods Mol. Biol. 2365, 79–113 (2021).
Li, K. & Crews, C. M. PROTACs: past, present and future. Chem. Soc. Rev. 51, 5214–5236 (2022).
Gadd, M. S. et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 13, 514–521 (2017).
Yamamoto, J., Ito, T., Yamaguchi, Y. & Handa, H. Discovery of CRBN as a target of thalidomide: a breakthrough for progress in the development of protein degraders. Chem. Soc. Rev. 51, 6234–6250 (2022).
Moehler, T. M., Hillengass, J., Glasmacher, A. & Goldschmidt, H. Thalidomide in multiple myeloma. Curr. Pharm. Biotechnol. 7, 431–440 (2006).
Teo, S. et al. Thalidomide in the treatment of leprosy. Microbes Infect. 4, 1193–1202 (2002).
Li, D. et al. Discovery of a dual WDR5 and Ikaros PROTAC degrader as an anti-cancer therapeutic. Oncogene 41, 3328–3340 (2022).
Domostegui, A., Nieto-Barrado, L., Perez-Lopez, C. & Mayor-Ruiz, C. Chasing molecular glue degraders: screening approaches. Chem. Soc. Rev. 51, 5498–5517 (2022).
Che, Y., Gilbert, A. M., Shanmugasundaram, V. & Noe, M. C. Inducing protein–protein interactions with molecular glues. Bioorg. Med. Chem. Lett. 28, 2585–2592 (2018).
Schwalm, M. P. et al. Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization. Cell Chem. Biol. 30, 753–765.e8 (2023).
Mo, X. L. & Fu, H. BRET: NanoLuc-based bioluminescence resonance energy transfer platform to monitor protein–protein interactions in live cells. Methods Mol. Biol. 1439, 263–271 (2016).
Dixon, A. S. et al. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 11, 400–408 (2016).
Schwalm, M. P. et al. A toolbox for the generation of chemical probes for Baculovirus IAP repeat containing proteins. Front Cell. Dev. Biol 10, 886537 (2022).
Vasta, J. D. et al. Quantitative, wide-spectrum kinase profiling in live cells for assessing the effect of cellular ATP on target engagement. Cell Chem. Biol. 25, 206–214 e211 (2018).
Machleidt, T. et al. NanoBRET—a novel BRET platform for the analysis of protein–protein interactions. ACS Chem. Biol. 10, 1797–1804 (2015).
Riching, K. M. et al. Quantitative live-cell kinetic degradation and mechanistic profiling of PROTAC mode of action. ACS Chem. Biol. 13, 2758–2770 (2018).
Owens, D. D. G. et al. A chemical probe to modulate human GID4 Pro/N-degron interactions. Preprint at bioRxiv https://doi.org/10.1101/2023.01.17.524225 (2023).
Mortison, J. D. et al. Rapid evaluation of small molecule cellular target engagement with a luminescent thermal shift assay. ACS Med. Chem. Lett. 12, 1288–1294 (2021).
RA, M. S. et al. Development of the first covalent monopolar spindle kinase 1 (MPS1/TTK) inhibitor. J. Med. Chem. 65, 3173–3192 (2022).
Moon, S. B., Kim, D. Y., Ko, J. H. & Kim, Y. S. Recent advances in the CRISPR genome editing tool set. Exp. Mol. Med. 51, 1–11 (2019).
Degorce, F. et al. HTRF: a technology tailored for drug discovery - a review of theoretical aspects and recent applications. Curr. Chem. Genomics 3, 22–32 (2009).
Tolvanen, T. A. Current advances in CETSA. Front. Mol. Biosci. 9, 866764 (2022).
Shyu, Y. J., Suarez, C. D. & Hu, C. D. Visualization of ternary complexes in living cells by using a BiFC-based FRET assay. Nat. Protoc. 3, 1693–1702 (2008).
Bielefeld-Sevigny, M. AlphaLISA immunoassay platform—the ‘no-wash’ high-throughput alternative to ELISA. Assay Drug Dev. Technol. 7, 90–92 (2009).
Kowarz, E., Loscher, D. & Marschalek, R. Optimized sleeping beauty transposons rapidly generate stable transgenic cell lines. Biotechnol J 10, 647–653 (2015).
Spitzer, J., Landthaler, M. & Tuschl, T. Rapid creation of stable mammalian cell lines for regulated expression of proteins using the Gateway(R) recombination cloning technology and Flp-In T-REx(R) lines. Methods Enzymol. 529, 99–124 (2013).
Chen, Y. H. et al. Rapid lentiviral vector producer cell line generation using a single DNA construct. Mol. Ther. Methods Clin. Dev. 19, 47–57 (2020).
Khan, S. H. Genome-editing technologies: concept, pros, and cons of various genome-editing techniques and bioethical concerns for clinical application. Mol. Ther. Nucleic Acids 16, 326–334 (2019).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Smith, J. D. et al. Quantitative CRISPR interference screens in yeast identify chemical–genetic interactions and new rules for guide RNA design. Genome Biol. 17, 45 (2016).
Doudna, J. A. & Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR–Cas9. Science 346, 1258096 (2014).
Desjardins, P. & Conklin, D. NanoDrop microvolume quantitation of nucleic acids. J. Vis. Exp. https://doi.org/10.3791/2565 (2010).
Cheng, Y. & Prusoff, W. H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 (1973).
Acknowledgements
M.P.S., K.S., S.M. and S.K. are grateful for support by the Structural Genomics Consortium, a registered charity (no. 1097737) that receives funds from Bayer AG, Boehringer Ingelheim, Bristol Myers Squibb, Genentech, Genome Canada through Ontario Genomics Institute, EU/EFPIA/OICR/McGill/KTH/Diamond Innovative Medicines Initiative 2 Joint Undertaking (EUbOPEN grant 875510), Janssen, Pfizer and Takeda and by the German Cancer Research Center DKTK and the Frankfurt Cancer Institute. M.P.S. is funded by the Deutsche Forschungsgemeinschaft (German Research Foundation), CRC1430 (project ID 424228829). M.P.S. is thankful for the help of B.-T. Berger during assay establishment.
Author information
Authors and Affiliations
Contributions
M.P.S. conceived the study and designed experiments. The manuscript and figures were prepared by M.P.S. and edited by K.S., S.M. and S.K.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Schwalm, M. P. et al. Front. Cell Dev. Biol. 10, 886537 (2022): https://doi.org/10.3389/fcell.2022.886537
Schwalm, M. P. et al. Cell Chem. Biol. 30, 753–765.e8 (2023): https://doi.org/10.1016/j.chembiol.2023.06.002
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Schwalm, M.P., Saxena, K., Müller, S. et al. Luciferase- and HaloTag-based reporter assays to measure small-molecule-induced degradation pathway in living cells. Nat Protoc (2024). https://doi.org/10.1038/s41596-024-00979-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41596-024-00979-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.