Abstract
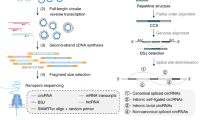
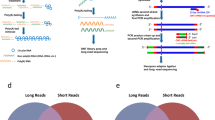
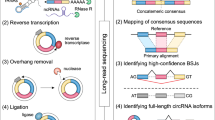
Circular RNAs (circRNAs) have important roles in regulating developmental processes and disease progression. As most circRNA sequences are highly similar to their cognate linear transcripts, the current short-read sequencing-based methods rely on the back-spliced junction signal for distinguishing circular and linear reads, which does not allow circRNAs’ full-length structure to be effectively reconstructed. Here we describe a long-read sequencing-based protocol, CIRI-long, for the detection of full-length circular RNAs. The CIRI-long protocol combines rolling circular reverse transcription and nanopore sequencing to capture full-length circRNA sequences. After poly(A) tailing, RNase R treatment, and size selection of polymerase chain reaction products, CIRI-long achieves an increased percentage (6%) of circular reads in the constructed library, which is 20-fold higher compared with previous Illumina-based strategies. This method can be applied in cell lines or tissue samples, enabling accurate detection of full-length circRNAs in the range of 100–3,000 bp. The entire protocol can be completed in 1 d, and can be scaled up for large-scale analysis using the nanopore barcoding kit and PromethION sequencing device. CIRI-long can serve as an effective and user-friendly protocol for characterizing full-length circRNAs, generating direct and convincing evidence for the existence of detected circRNAs. The analytical pipeline offers convenient functions for identification of full-length circRNA isoforms and integration of multiple datasets. The assembled full-length transcripts and their splicing patterns provide indispensable information to explore the biological function of circRNAs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The CIRI-long dataset used in this manuscript can be downloaded from the National Genomics Data Center46 (China National Center for Bioinformation: https://bigd.big.ac.cn/gsa) with accession number CRR194209.
Code availability
The CIRI-long software can be downloaded from GitHub at https://github.com/bioinfo-biols/CIRI-long47.
References
Kristensen, L. S. et al. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691 (2019).
Liu, C.-X. & Chen, L.-L. Circular RNAs: characterization, cellular roles, and applications. Cell 185, 2016–2034 (2022).
Wu, W., Ji, P. & Zhao, F. CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 21, 101 (2020).
Gao, Y., Wang, J. & Zhao, F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 16, 4 (2015).
Zhang, J., Chen, S., Yang, J. & Zhao, F. Accurate quantification of circular RNAs identifies extensive circular isoform switching events. Nat. Commun. 11, 90 (2020).
Wu, W., Zhang, J., Cao, X., Cai, Z. & Zhao, F. Exploring the cellular landscape of circular RNAs using full-length single-cell RNA sequencing. Nat. Commun. 13, 3242 (2022).
Ji, P. et al. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep. 26, 3444–3460 e3445 (2019).
Dong, R., Ma, X. K., Li, G. W. & Yang, L. CIRCpedia v2: an updated database for comprehensive circular RNA annotation and expression comparison. Genom. Proteom. Bioinform. 16, 226–233 (2018).
Gan, X. et al. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol. Cancer 19, 45 (2020).
Kristensen, L. S. et al. Spatial expression analyses of the putative oncogene ciRS-7 in cancer reshape the microRNA sponge theory. Nat. Commun. 11, 4551 (2020).
Chen, S. et al. circVAMP3 drives CAPRIN1 phase separation and inhibits hepatocellular carcinoma by suppressing c-Myc translation. Adv. Sci. 9, e2103817 (2022).
Liu, C. X. et al. RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol. Cell. 82, 420–434 e426 (2022).
Chen, S., Zhang, J. & Zhao, F. Screening linear and circular RNA transcripts from stress granules. Genom. Proteom. Bioinform. https://doi.org/10.1016/j.gpb.2022.01.003 (2022).
Yang, Y. et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 27, 626–641 (2017).
Li, Y. et al. HNRNPL circularizes ARHGAP35 to produce an oncogenic protein. Adv. Sci. 8, 2001701 (2021).
Zhang, M. et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 9, 4475 (2018).
Dal Molin, A. et al. CRAFT: a bioinformatics software for custom prediction of circular RNA functions. Brief. Bioinform. https://doi.org/10.1093/bib/bbab601 (2022).
Gao, Y., Zhang, J. & Zhao, F. Circular RNA identification based on multiple seed matching. Brief. Bioinform. 19, 803–810 (2018).
Gao, Y. et al. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat. Commun. 7, 12060 (2016).
Zheng, Y., Ji, P., Chen, S., Hou, L. & Zhao, F. Reconstruction of full-length circular RNAs enables isoform-level quantification. Genome Med. 11, 2 (2019).
Zhang, X. O. et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 26, 1277–1287 (2016).
Zhang, J. et al. Comprehensive profiling of circular RNAs with nanopore sequencing and CIRI-long. Nat. Biotechnol. 39, 836–845 (2021).
Zhang, J. et al. Evaluation of circRNA sequence assembly methods using long reads. Front. Genet. 13, 816825 (2022).
Soutschek, M., Gross, F., Schratt, G. & Germain, P.-L. scanMiR: a biochemically based toolkit for versatile and efficient microRNA target prediction. Bioinformatics 38, 2466–2473 (2022).
Wu, J. et al. CircAST: full-length assembly and quantification of alternatively spliced isoforms in circular RNAs. Genom. Proteom. Bioinform. 17, 522–534 (2019).
Ye, C. Y. et al. Full-length sequence assembly reveals circular RNAs with diverse non-GT/AG splicing signals in rice. RNA Biol. 14, 1055–1063 (2017).
Metge, F., Czaja-Hasse, L. F., Reinhardt, R. & Dieterich, C. FUCHS-towards full circular RNA characterization using RNAseq. PeerJ 5, e2934 (2017).
Hossain, M. T., Peng, Y., Feng, S. & Wei, Y. FcircSEC: an R package for full length circRNA sequence extraction and classification. Int. J. Genom. 2020, 9084901 (2020).
Qin, Y. et al. Reference-free and de novo identification of circular RNAs. Preprint at bioRxiv https://doi.org/10.1101/2020.04.21.050617 (2020).
Zheng, Y. & Zhao, F. Visualization of circular RNAs and their internal splicing events from transcriptomic data. Bioinformatics 36, 2934–2935 (2020).
Zhang, J. & Zhao, F. Reconstruction of circular RNAs using Illumina and Nanopore RNA-seq datasets. Methods 196, 17–22 (2021).
Zhang, J. & Zhao, F. Characterizing circular RNAs using Nanopore sequencing. Trends Biochem. Sci. 46, 785–786 (2021).
Jeck, W. R. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157 (2013).
Pandey, P. R., Rout, P. K., Das, A., Gorospe, M. & Panda, A. C. RPAD (RNase R treatment, polyadenylation, and poly(A)+ RNA depletion) method to isolate highly pure circular RNA. Methods 155, 41–48 (2019).
Panda, A. C. et al. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 45, e116 (2017).
Zhang, Z. & Han, L. Circular RNAs sequenced at last. Nat. Biotechnol. 39, 811–812 (2021).
Vo, J. N. et al. The landscape of circular RNA in cancer. Cell 176, 869–881 e813 (2019).
Xin, R. et al. isoCirc catalogs full-length circular RNA isoforms in human transcriptomes. Nat. Commun. 12, 266 (2021).
Liu, Z. et al. circFL-seq reveals full-length circular RNAs with rolling circular reverse transcription and nanopore sequencing. eLife https://doi.org/10.7554/eLife.69457 (2021).
Rahimi, K., Veno, M. T., Dupont, D. M. & Kjems, J. Nanopore sequencing of brain-derived full-length circRNAs reveals circRNA-specific exon usage, intron retention and microexons. Nat. Commun. 12, 4825 (2021).
Jeck, W. R. & Sharpless, N. E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 32, 453–461 (2014).
Vincent, H. A. & Deutscher, M. P. Substrate recognition and catalysis by the exoribonuclease RNase R. J. Biol. Chem. 281, 29769–29775 (2006).
Xiao, M. S. & Wilusz, J. E. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3′ ends. Nucleic Acids Res. 47, 8755–8769 (2019).
Fukasawa, Y., Ermini, L., Wang, H., Carty, K. & Cheung, M. S. LongQC: a quality control tool for third generation sequencing long read data. G3 (Bethesda) 10, 1193–1196 (2020).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://doi.org/10.48550/arXiv.1303.3997 (2013).
Wang, Y. et al. GSA: Genome Sequence Archive. Genom. Proteom. Bioinform. 15, 14–18 (2017).
Zhang, J. Full-length circular RNA profiling by nannopore sequencing with CIRI-long. https://github.com/bioinfo-biols/CIRI-long, https://doi.org/10.5281/zenodo.7399826 (2022).
Thorvaldsdottir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013).
Diesh, C. et al. JBrowse 2: a modular genome browser with views of synteny and structural variation. Preprint at https://doi.org/10.1101/2022.07.28.501447 (2022).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (32130020, 32025009, 91940306, 32200530) and the National Key R&D Project (2021YFA1300500, 2021YFA1302000).
Author information
Authors and Affiliations
Contributions
L.H. and J.Z. developed the experimental protocol for CIRI-long. L.H., J.Z. and F.Z. wrote the manuscript. F.Z. conceived the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Michael Clark and Leng Han for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Zhang, J. et al. Nat. Biotechnol. 39, 836–845 (2021): https://doi.org/10.1038/s41587-021-00842-6
Supplementary information
Supplementary Code 1
Code used in the paper.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hou, L., Zhang, J. & Zhao, F. Full-length circular RNA profiling by nanopore sequencing with CIRI-long. Nat Protoc 18, 1795–1813 (2023). https://doi.org/10.1038/s41596-023-00815-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00815-w
This article is cited by
-
New insight into circRNAs: characterization, strategies, and biomedical applications
Experimental Hematology & Oncology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.