Abstract
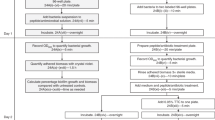
The minimal inhibitory concentration (MIC) assay uses agar or broth dilution methods to measure, under defined test conditions, the lowest effective concentration of an antimicrobial agent that inhibits visible growth of a bacterium of interest. This assay is used to test the susceptibilities of bacterial isolates and of novel antimicrobial drugs, and is typically done in nutrient-rich laboratory media that have little relevance to in vivo conditions. As an extension to our original protocol on MIC assays (also published in Nature Protocols), here we describe the application of the MIC broth microdilution assay to test antimicrobial susceptibility in conditions that are more physiologically relevant to infections observed in the clinic. Specifically, we describe a platform that can be applied to the preparation of medium that mimics lung and wound exudate or blood conditions for the growth and susceptibility testing of bacteria, including ESKAPE pathogens. This protocol can also be applied to most physiologically relevant liquid medium and aerobic pathogens, and takes 3–4 d to complete.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Wiegand, I., Hilpert, K. & Hancock, R. E. W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175 (2008).
Clinical Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing (CLSI document M100, 2020).
Nizet, V. The accidental orthodoxy of Drs. Mueller and Hinton. EBioMedicine 22, 26–27 (2017).
Cornforth, D. M., Diggle, F. L., Melvin, J. A., Bomberger, J. M. & Whiteley, M. Quantitative framework for model evaluation in microbiology research using Pseudomonas aeruginosa and cystic fibrosis infection as a test case. mBio 11, e03042-19 (2020).
Tata, M. et al. RNASeq based transcriptional profiling of Pseudomonas aeruginosa PA14 after short- and long-term anoxic cultivation in synthetic cystic fibrosis sputum medium. PLoS One 11, e0147811 (2016).
Palmer, K. L., Mashburn, L. M., Singh, P. K. & Whiteley, M. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol 187, 5267–5277 (2005).
Kruczek, C. et al. Major transcriptome changes accompany the growth of Pseudomonas aeruginosa in blood from patients with severe thermal injuries. PLoS One 11, e0149229 (2016).
Umland, T. C. et al. In vivo-validated essential genes identified in Acinetobacter baumannii by using human ascites overlap poorly with essential genes detected on laboratory media. mBio 3, e00113-12 (2012).
Thulin, E., Thulin, M. & Andersson, D. I. Reversion of high-level mecillinam resistance to susceptibility in Escherichia coli during growth in urine. EBioMedicine 23, 111–118 (2017).
Lee, S. A. et al. General and condition-specific essential functions of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 112, 5189–5194 (2015).
Ersoy, S. C. et al. Correcting a fundamental flaw in the paradigm for antimicrobial susceptibility testing. EBioMedicine 20, 173–181 (2017).
Hancock, R. E. W. Rethinking the antibiotic discovery paradigm. EBioMedicine 2, 629–630 (2015).
Kubicek-Sutherland, J. Z. et al. Host-dependent induction of transient antibiotic resistance: a prelude to treatment failure. EBioMedicine 2, 1169–1178 (2015).
World Health Organization. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. http://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/ (WHO, 2017).
De Oliveira, D. M. P. et al. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 33, e00181-19 (2020).
Turner, K. H., Wessel, A. K., Palmer, G. C., Murray, J. L. & Whiteley, M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl Acad. Sci. 112, 4110–4115 (2015).
Malachowa, N. et al. Global changes in Staphylococcus aureus gene expression in human blood. PLoS One 6, e18617 (2011).
Quinn, B. et al. Human serum albumin alters specific genes that can play a role in survival and persistence in Acinetobacter baumannii. Sci. Rep. 8, 14741 (2018).
Mäder, U. et al. Staphylococcus aureus transcriptome architecture: from laboratory to infection-mimicking conditions. PLoS Genet 12, e1005962 (2016).
Belanger, C. R. et al. Identification of novel targets of azithromycin activity against Pseudomonas aeruginosa grown in physiologically relevant media. Proc. Natl Acad. Sci. 117, 33519–33529 (2020).
Winsor, G. L. et al. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44, D646–D653 (2016).
Fung, C. et al. Gene expression of Pseudomonas aeruginosa in a mucin-containing synthetic growth medium mimicking cystic fibrosis lung sputum. J. Med. Microbiol. 59, 1089–1100 (2010).
Lin, L. et al. Azithromycin synergizes with cationic antimicrobial peptides to exert bactericidal and therapeutic activity against highly multidrug-resistant Gram-negative bacterial pathogens. EBioMedicine 2, 690–698 (2015).
Colquhoun, J. M., Wozniak, R. A. F. & Dunman, P. M. Clinically relevant growth conditions alter Acinetobacter baumannii antibiotic susceptibility and promote identification of novel antibacterial agents. PLoS One 10, e0143033 (2015).
Yeaman, M. R., Gank, K. D., Bayer, A. S. & Brass, E. P. Synthetic peptides that exert antimicrobial activities in whole blood and blood-derived matrices. Antimicrob. Agents Chemother. 46, 3883–3891 (2002).
Weber, B. S. et al. Genetic and chemical screening in human blood serum reveals unique antibacterial targets and compounds against Klebsiella pneumoniae. Cell Rep 32, 107927 (2020).
Palmer, K. L., Aye, L. M. & Whiteley, M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189, 8079–8087 (2007).
Kirchner, S. et al. Use of artificial sputum medium to test antibiotic efficacy against Pseudomonas aeruginosa in conditions more relevant to the cystic fibrosis lung. J. Vis. Exp. https://doi.org/10.3791/3857 (2012).
Sriramulu, D. D., Lünsdorf, H., Lam, J. S. & Römling, U. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 54, 667–676 (2005).
Yeung, A. T. Y., Parayno, A. & Hancock, R. E. W. Mucin promotes rapid surface motility in Pseudomonas aeruginosa. mBio 3, e00073–12 (2012).
Kreda, S. M., Davis, C. W. & Rose, M. C. CFTR, mucins, and mucus obstruction in cystic fibrosis. Cold Spring Harb. Perspect. Med. 2, a009589 (2012).
Kirchner, K. K., Wagener, J. S., Khan, T. Z., Copenhaver, S. C. & Accurso, F. J. Increased DNA levels in bronchoalveolar lavage fluid obtained from infants with cystic fibrosis. Am. J. Respir. Crit. Care Med. 154, 1426–1429 (1996).
Haney, E., Trimble, M., Cheng, J., Vallé, Q. & Hancock, R. Critical assessment of methods to quantify biofilm growth and evaluate antibiofilm activity of host defence peptides. Biomolecules 8, 29 (2018).
Acknowledgements
Our own antibiotics research is funded by the Canadian Institutes for Health Research Foundation grant FDN-154287. C.R.B. received a Doctoral Studentship Award from Cystic Fibrosis Canada. R.E.W.H. holds a Canada Research Chair in Health and Genomics and a UBC Killam Professorship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Fekade Sime and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Belanger C. R. et al. Proc. Natl Acad. Sci. USA 117, 33519–33529 (2020): https://doi.org/10.1073/pnas.2007626117
This protocol is an extension to: Nat. Protoc. 3, 163–175 (2008): https://doi.org/10.1038/nprot.2007.521
Supplementary information
Supplementary Information
Supplementary Table 1.
Rights and permissions
About this article
Cite this article
Belanger, C.R., Hancock, R.E.W. Testing physiologically relevant conditions in minimal inhibitory concentration assays. Nat Protoc 16, 3761–3774 (2021). https://doi.org/10.1038/s41596-021-00572-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00572-8
This article is cited by
-
Gut microbes predominantly act as living beneficial partners rather than raw nutrients
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.