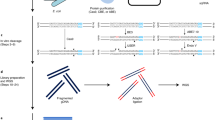
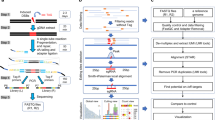
Abstract
Nucleotide excision repair is a versatile mechanism to repair a variety of bulky DNA adducts. We developed excision repair sequencing (XR-seq) to study nucleotide excision repair of DNA adducts in humans, mice, Arabidopsis thaliana, yeast and Escherichia coli. In this protocol, the excised oligomers, generated in the nucleotide excision repair reaction, are isolated by cell lysis and fractionation, followed by immunoprecipitation with damage- or repair factor–specific antibodies from the non-chromatin fraction. The single-stranded excised oligomers are ligated to adapters and re-immunoprecipitated with damage-specific antibodies. The DNA damage in the excised oligomers is then reversed by enzymatic or chemical reactions before being converted into a sequencing library by PCR amplification. Alternatively, the excised oligomers containing DNA damage, especially those containing irreversible DNA damage such as benzo[a]pyrene-induced DNA adducts, can be converted to a double-stranded DNA (dsDNA) form by using appropriate translesion DNA synthesis (TLS) polymerases and then can be amplified by PCR. The current genome-wide approaches for studying repair measure the loss of damage signal with time, which limits their resolution. By contrast, an advantage of XR-seq is that the repair signal is directly detected above a background of zero. An XR-seq library using the protocol described here can be obtained in 7–9 d.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Sancar, A. Mechanisms of DNA repair by photolyase and excision nuclease (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 55, 8502–8527 (2016).
Wood, R. D. Nucleotide excision repair in mammalian cells. J. Biol. Chem. 272, 23465–23468 (1997).
Hu, J., Selby, C. P., Adar, S., Adebali, O. & Sancar, A. Molecular mechanisms and genomic maps of DNA excision repair in Escherichia coli and humans. J. Biol. Chem. 292, 15588–15597 (2017).
Gong, F., Kwon, Y. & Smerdon, M. J. Nucleotide excision repair in chromatin and the right of entry. DNA Repair (Amst) 4, 884–896 (2005).
Hanawalt, P. C. & Spivak, G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell. Biol. 9, 958–970 (2008).
Mao, P., Wyrick, J. J., Roberts, S. A. & Smerdon, M. J. UV-induced DNA damage and mutagenesis in chromatin. Photochem. Photobiol. 93, 216–228 (2017).
Teng, Y. et al. A novel method for the genome-wide high resolution analysis of DNA damage. Nucleic Acids Res. 39, e10 (2011).
Bryan, D. S., Ransom, M., Adane, B., York, K. & Hesselberth, J. R. High resolution mapping of modified DNA nucleobases using excision repair enzymes. Genome Res. 24, 1534–1542 (2014).
Powell, J. R. et al. 3D-DIP-Chip: a microarray-based method to measure genomic DNA damage. Sci. Rep. 5, 7975 (2015).
Hu, J., Lieb, J. D., Sancar, A. & Adar, S. Cisplatin DNA damage and repair maps of the human genome at single-nucleotide resolution. Proc. Natl. Acad. Sci. USA 113, 11507–11512 (2016).
Mao, P., Smerdon, M. J., Roberts, S. A. & Wyrick, J. J. Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution. Proc. Natl. Acad. Sci. USA 113, 9057–9062 (2016).
Garcia-Nieto, P. E. et al. Carcinogen susceptibility is regulated by genome architecture and predicts cancer mutagenesis. EMBO J. 36, 2829–2843 (2017).
Hu, J., Adar, S., Selby, C. P., Lieb, J. D. & Sancar, A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 29, 948–960 (2015).
Adar, S., Hu, J., Lieb, J. D. & Sancar, A. Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis. Proc. Natl. Acad. Sci. USA 113, E2124–E2133 (2016).
Hu, J., Adebali, O., Adar, S. & Sancar, A. Dynamic maps of UV damage formation and repair for the human genome. Proc. Natl. Acad. Sci. USA 114, 6758–6763 (2017).
Chiou, Y. Y., Hu, J., Sancar, A. & Selby, C. P. RNA polymerase II is released from the DNA template during transcription-coupled repair in mammalian cells. J. Biol. Chem. 293, 2476–2486 (2018).
Yang, Y. et al. Cisplatin-DNA adduct repair of transcribed genes is controlled by two circadian programs in mouse tissues. Proc. Natl. Acad. Sci. USA 115, E4777–E4785 (2018).
Oztas, O., Selby, C. P., Sancar, A. & Adebali, O. Genome-wide excision repair in Arabidopsis is coupled to transcription and reflects circadian gene expression patterns. Nat. Commun. 9, 1503 (2018).
Li, W., Adebali, O., Yang, Y., Selby, C. P. & Sancar, A. Single-nucleotide resolution dynamic repair maps of UV damage in Saccharomyces cerevisiae genome. Proc. Natl. Acad. Sci. USA 115, E3408–E3415 (2018).
Adebali, O., Chiou, Y. Y., Hu, J., Sancar, A. & Selby, C. P. Genome-wide transcription-coupled repair in Escherichia coli is mediated by the Mfd translocase. Proc. Natl. Acad. Sci. USA 114, E2116–E2125 (2017).
Adebali, O., Sancar, A. & Selby, C. P. Mfd translocase is necessary and sufficient for transcription-coupled repair in Escherichia coli. J. Biol. Chem. 292, 18386–18391 (2017).
Reardon, J. T. & Sancar, A. Nucleotide excision repair. Prog. Nucleic Acid Res. Mol. Biol. 79, 183–235 (2005).
Truglio, J. J., Croteau, D. L., Van Houten, B. & Kisker, C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 106, 233–252 (2006).
Canturk, F. et al. Nucleotide excision repair by dual incisions in plants. Proc. Natl. Acad. Sci. USA 113, 4706–4710 (2016).
Kemp, M. G., Reardon, J. T., Lindsey-Boltz, L. A. & Sancar, A. Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair. J. Biol. Chem. 287, 22889–22899 (2012).
Hu, J. et al. Nucleotide excision repair in human cells: fate of the excised oligonucleotide carrying DNA damage in vivo. J. Biol. Chem. 288, 20918–20926 (2013).
Choi, J. H., Kim, S. Y., Kim, S. K., Kemp, M. G. & Sancar, A. An integrated approach for analysis of the DNA damage response in mammalian cells: nucleotide excision repair, DNA damage checkpoint, and apoptosis. J. Biol. Chem. 290, 28812–28821 (2015).
Baek, S., Han, S., Kang, D., Kemp, M. G. & Choi, J. H. Simultaneous detection of nucleotide excision repair events and apoptosis-induced DNA fragmentation in genotoxin-treated cells. Sci. Rep. 8, 2265 (2018).
Li, W. et al. Human genome-wide repair map of DNA damage caused by the cigarette smoke carcinogen benzo[a]pyrene. Proc. Natl. Acad. Sci. USA 114, 6752–6757 (2017).
ENCODE Project Consortium.. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Wade, J. T. & Grainger, D. C. Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol. 12, 647–653 (2014).
Thomason, M. K. et al. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J. Bacteriol. 197, 18–28 (2015).
Llorens-Rico, V. et al. Bacterial antisense RNAs are mainly the product of transcriptional noise. Sci. Adv. 2, e1501363 (2016).
Kang, T. H., Lindsey-Boltz, L. A., Reardon, J. T. & Sancar, A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 107, 4890–4895 (2010).
Gaddameedhi, S., Selby, C. P., Kaufmann, W. K., Smart, R. C. & Sancar, A. Control of skin cancer by the circadian rhythm. Proc. Natl. Acad. Sci. USA 108, 18790–18795 (2011).
Perera, D. et al. Differential DNA repair underlies mutation hotspots at active promoters in cancer genomes. Nature 532, 259–263 (2016).
Sabarinathan, R., Mularoni, L., Deu-Pons, J., Gonzalez-Perez, A. & Lopez-Bigas, N. Nucleotide excision repair is impaired by binding of transcription factors to DNA. Nature 532, 264–267 (2016).
Poulos, R. C. et al. Functional mutations form at CTCF-cohesin binding sites in melanoma due to uneven nucleotide excision repair across the motif. Cell Rep. 17, 2865–2872 (2016).
Lim, B., Mun, J., Kim, Y. S. & Kim, S. Y. Variability in chromatin architecture and associated DNA repair at genomic positions containing somatic mutations. Cancer Res. 77, 2822–2833 (2017).
Lavigne, M. D., Konstantopoulos, D., Ntakou-Zamplara, K. Z., Liakos, A. & Fousteri, M. Global unleashing of transcription elongation waves in response to genotoxic stress restricts somatic mutation rate. Nat. Commun. 8, 2076 (2017).
Klaassen, C. D., Casarett, L. J. & Doull, J. Casarett and Doull’s Toxicology: The Basic Science of Poisons. 8th edn (McGraw-Hill Education, New York, 2013).
Bohr, V. A., Smith, C. A., Okumoto, D. S. & Hanawalt, P. C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 40, 359–369 (1985).
Besaratinia, A. & Pfeifer, G. P. Measuring the formation and repair of UV damage at the DNA sequence level by ligation-mediated PCR. Methods Mol. Biol. 920, 189–202 (2012).
Li, S., Waters, R. & Smerdon, M. J. Low- and high-resolution mapping of DNA damage at specific sites. Methods 22, 170–179 (2000).
Yu, S. et al. Global genome nucleotide excision repair is organized into domains that promote efficient DNA repair in chromatin. Genome Res. 26, 1376–1387 (2016).
Shu, X., Xiong, X., Song, J., He, C. & Yi, C. Base-resolution analysis of cisplatin-DNA adducts at the genome scale. Angew. Chem. Int. Ed. Engl. 55, 14246–14249 (2016).
Kang, T. H., Reardon, J. T., Kemp, M. & Sancar, A. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc. Natl. Acad. Sci. USA 106, 2864–2867 (2009).
Pfeifer, G. P., Drouin, R., Riggs, A. D. & Holmquist, G. P. In vivo mapping of a DNA adduct at nucleotide resolution: detection of pyrimidine (6-4) pyrimidone photoproducts by ligation-mediated polymerase chain reaction. Proc. Natl. Acad. Sci. USA 88, 1374–1378 (1991).
Pfeifer, G. P., Drouin, R., Riggs, A. D. & Holmquist, G. P. Binding of transcription factors creates hot spots for UV photoproducts in vivo. Mol. Cell. Biol. 12, 1798–1804 (1992).
Wellinger, R. E. & Thoma, F. Taq DNA polymerase blockage at pyrimidine dimers. Nucleic Acids Res. 24, 1578–1579 (1996).
Wellinger, R. E. & Thoma, F. Nucleosome structure and positioning modulate nucleotide excision repair in the non-transcribed strand of an active gene. EMBO J. 16, 5046–5056 (1997).
Kunala, S. & Brash, D. E. Excision repair at individual bases of the Escherichia coli lacI gene: relation to mutation hot spots and transcription coupling activity. Proc. Natl. Acad. Sci. USA 89, 11031–11035 (1992).
Hirt, B. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26, 365–369 (1967).
Heffernan, T. P. et al. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol. Cell. Biol. 22, 8552–8561 (2002).
Selby, C. P. & Sancar, A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc. Natl. Acad. Sci. USA 103, 17696–17700 (2006).
Selby, C. P. & Sancar, A. The second chromophore in Drosophila photolyase/cryptochrome family photoreceptors. Biochemistry 51, 167–171 (2012).
Kodama, Y., Shumway, M. & Leinonen, R., International Nucleotide Sequence Database Collaboration. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 40, D54–D56 (2012).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Hubbard, T. et al. The Ensembl genome database project. Nucleic Acids Res. 30, 38–41 (2002).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Quinlan, A. R. BEDTools: the Swiss-Army Tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 11.12.1–11.12.34 (2014).
Kuhn, R. M., Haussler, D. & Kent, W. J. The UCSC genome browser and associated tools. Brief. Bioinform. 14, 144–161 (2013).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, New York, 2016).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Acknowledgements
We thank A. Kakoki for critical reading of the manuscript. This work was supported by National Institutes of Health grants GM118102 and ES027255 (to A.S.) and Scientific and Technological Research Council of Turkey grant 118C023 (to O.A.).
Author information
Authors and Affiliations
Contributions
J.H., W.L., O.A., Y.Y., O.O. and C.P.S. performed the experiments and analyzed the data described in the protocol. W.L. wrote the manuscript with assistance from C.P.S, J.H., O.A., Y.Y., O.O. and A.S. All authors contributed to, reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Hu, J., Adar, S., Selby, C. P., Lieb, J. D. & Sancar, A. Genes Dev. 29, 948–960 (2015): http://www.genesdev.org/cgi/doi/10.1101/gad.261271.115
Li, W. et al. Proc. Natl. Acad. Sci. USA 114, 6752–6757 (2017): https://doi.org/10.1073/pnas.1706021114
Adebali, O., Chiou, Y.-Y., Hu, J., Sancar, A. & Selby, C. P. Proc. Natl. Acad. Sci. USA 114, E2116–E2125 (2017): https://doi.org/10.1073/pnas.1700230114
Yang, Y. et al. Proc. Natl. Acad. Sci. USA 115, E4777–E4785 (2018): https://doi.org/10.1073/pnas.1804493115
Supplementary information
Rights and permissions
About this article
Cite this article
Hu, J., Li, W., Adebali, O. et al. Genome-wide mapping of nucleotide excision repair with XR-seq. Nat Protoc 14, 248–282 (2019). https://doi.org/10.1038/s41596-018-0093-7
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-018-0093-7
This article is cited by
-
Global repair is the primary nucleotide excision repair subpathway for the removal of pyrimidine-pyrimidone (6-4) damage from the Arabidopsis genome
Scientific Reports (2024)
-
Sequence dependent UV damage of complete pools of oligonucleotides
Scientific Reports (2023)
-
Comparative analyses of two primate species diverged by more than 60 million years show different rates but similar distribution of genome-wide UV repair events
BMC Genomics (2021)
-
Genome-wide mapping of genomic DNA damage: methods and implications
Cellular and Molecular Life Sciences (2021)
-
The cooperative action of CSB, CSA, and UVSSA target TFIIH to DNA damage-stalled RNA polymerase II
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.