Abstract
Current models suggest that DNA double-strand breaks (DSBs) can move to the nuclear periphery for repair. It is unclear to what extent human DSBs display such repositioning. Here we show that the human nuclear envelope localizes to DSBs in a manner depending on DNA damage response (DDR) kinases and cytoplasmic microtubules acetylated by α-tubulin acetyltransferase-1 (ATAT1). These factors collaborate with the linker of nucleoskeleton and cytoskeleton complex (LINC), nuclear pore complex (NPC) protein NUP153, nuclear lamina and kinesins KIF5B and KIF13B to generate DSB-capturing nuclear envelope tubules (dsbNETs). dsbNETs are partly supported by nuclear actin filaments and the circadian factor PER1 and reversed by kinesin KIFC3. Although dsbNETs promote repair and survival, they are also co-opted during poly(ADP-ribose) polymerase (PARP) inhibition to restrain BRCA1-deficient breast cancer cells and are hyper-induced in cells expressing the aging-linked lamin A mutant progerin. In summary, our results advance understanding of nuclear structure–function relationships, uncover a nuclear–cytoplasmic DDR and identify dsbNETs as critical factors in genome organization and stability.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data are in the article, in the 22 Source Data files related to Figs. 1–5 and Extended Data Figs. 1–10 and in Supplementary Videos 1–5. All bioinformatics and human tumor datasets are in public databases as described in the Methods. All data and materials are available upon reasonable request. Source data are provided with this paper.
References
Hustedt, N. & Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 19, 1–9 (2016).
Nagai, S. et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322, 597–602 (2008).
Mekhail, K., Seebacher, J., Gygi, S. P. & Moazed, D. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature 456, 667–670 (2008).
Chiolo, I. et al. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144, 732–744 (2011).
Dion, V., Kalck, V., Horigome, C., Towbin, B. D. & Gasser, S. M. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat. Cell Biol. 14, 502–509 (2012).
Mine-Hattab, J. & Rothstein, R. Increased chromosome mobility facilitates homology search during recombination. Nat. Cell Biol. 14, 510–517 (2012).
Chung, D. K. et al. Perinuclear tethers license telomeric DSBs for a broad kinesin- and NPC-dependent DNA repair process. Nat. Commun. 6, 7742 (2015).
Ryu, T. et al. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat. Cell Biol. 17, 1401–1411 (2015).
Oshidari, R. et al. Nuclear microtubule filaments mediate non-linear directional motion of chromatin and promote DNA repair. Nat. Commun. 9, 2567 (2018).
Caridi, C. P. et al. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 559, 54–60 (2018).
Oshidari, R., Mekhail, K. & Seeber, A. Mobility and repair of damaged DNA: random or directed? Trends Cell Biol. 30, 144–156 (2020).
Shokrollahi, M. & Mekhail, K. Interphase microtubules in nuclear organization and genome maintenance. Trends Cell Biol. 31, 721–731 (2021).
Stanic, M. & Mekhail, K. Integration of DNA damage responses with dynamic spatial genome organization. Trends Genet. 38, 290–304 (2022).
Oshidari, R. et al. DNA repair by Rad52 liquid droplets. Nat. Commun. 11, 695 (2020).
Lottersberger, F., Karssemeijer, R. A., Dimitrova, N. & de Lange, T. 53BP1 and the LINC complex promote microtubule-dependent DSB mobility and DNA repair. Cell 163, 880–893 (2015).
Jakob, B., Splinter, J., Durante, M. & Taucher-Scholz, G. Live cell microscopy analysis of radiation-induced DNA double-strand break motion. Proc. Natl Acad. Sci. USA 106, 3172–3177 (2009).
Arnould, C. et al. Chromatin compartmentalization regulates the response to DNA damage. Nature 623, 183–192 (2023).
Zagelbaum, J. et al. Multiscale reorganization of the genome following DNA damage facilitates chromosome translocations via nuclear actin polymerization. Nat. Struct. Mol. Biol. 30, 99–106 (2023).
Soutoglou, E. et al. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 9, 675–682 (2007).
Misteli, T. & Soutoglou, E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat. Rev. Mol. Cell Biol. 10, 243–254 (2009).
Tsouroula, K. et al. Temporal and spatial uncoupling of DNA double strand break repair pathways within mammalian heterochromatin. Mol. Cell 63, 293–305 (2016).
Lemaitre, C. et al. Nuclear position dictates DNA repair pathway choice. Genes Dev. 28, 2450–2463 (2014).
Chen, B. et al. Transmembrane nuclease NUMEN/ENDOD1 regulates DNA repair pathway choice at the nuclear periphery. Nat. Cell Biol. 25, 1004–1016 (2023).
Garvin, A. J. et al. The deSUMOylase SENP2 coordinates homologous recombination and nonhomologous end joining by independent mechanisms. Genes Dev. 33, 333–347 (2019).
Hang, J. & Dasso, M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277, 19961–19966 (2002).
Zhang, H., Saitoh, H. & Matunis, M. J. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 22, 6498–6508 (2002).
Mekhail, K. & Moazed, D. The nuclear envelope in genome organization, expression and stability. Nat. Rev. Mol. Cell Biol. 11, 317–328 (2010).
Malhas, A., Goulbourne, C. & Vaux, D. J. The nucleoplasmic reticulum: form and function. Trends Cell Biol. 21, 362–373 (2011).
Hartlerode, A. J., Morgan, M. J., Wu, Y., Buis, J. & Ferguson, D. O. Recruitment and activation of the ATM kinase in the absence of DNA-damage sensors. Nat. Struct. Mol. Biol. 22, 736–743 (2015).
Pascual-Garcia, P. & Capelson, M. The nuclear pore complex and the genome: organizing and regulatory principles. Curr. Opin. Genet. Dev. 67, 142–150 (2021).
Radu, A., Moore, M. S. & Blobel, G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell 81, 215–222 (1995).
Puthiyaveetil, A. G., Reilly, C. M., Pardee, T. S. & Caudell, D. L. Non-homologous end joining mediated DNA repair is impaired in the NUP98-HOXD13 mouse model for myelodysplastic syndrome. Leuk. Res. 37, 112–116 (2013).
Moudry, P. et al. Nucleoporin NUP153 guards genome integrity by promoting nuclear import of 53BP1. Cell Death Differ. 19, 798–807 (2012).
Dammermann, A., Desai, A. & Oegema, K. The minus end in sight. Curr. Biol. 13, R614–R624 (2003).
Hirokawa, N., Noda, Y., Tanaka, Y. & Niwa, S. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682–696 (2009).
Wiesner, C., Faix, J., Himmel, M., Bentzien, F. & Linder, S. KIF5B and KIF3A/KIF3B kinesins drive MT1-MMP surface exposure, CD44 shedding, and extracellular matrix degradation in primary macrophages. Blood 116, 1559–1569 (2010).
Portran, D., Schaedel, L., Xu, Z., Thery, M. & Nachury, M. V. Tubulin acetylation protects long-lived microtubules against mechanical ageing. Nat. Cell Biol. 19, 391–398 (2017).
Xu, Z. et al. Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science 356, 328–332 (2017).
Reed, N. A. et al. Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 16, 2166–2172 (2006).
Serra-Marques, A. et al. Concerted action of kinesins KIF5B and KIF13B promotes efficient secretory vesicle transport to microtubule plus ends. eLife 9, e61302 (2020).
Le Bozec, B. et al. Circadian PERIOD complex regulates TC-DSB repair through anchoring to the nuclear envelope. Preprint at bioRxiv https://doi.org/10.1101/2023.05.11.540338 (2023).
Lisby, M., Mortensen, U. H. & Rothstein, R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat. Cell Biol. 5, 572–577 (2003).
Aten, J. A. et al. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 303, 92–95 (2004).
Arnould, C. et al. Loop extrusion as a mechanism for formation of DNA damage repair foci. Nature 590, 660–665 (2021).
Shanbhag, N. M., Rafalska-Metcalf, I. U., Balane-Bolivar, C., Janicki, S. M. & Greenberg, R. A. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141, 970–981 (2010).
Tang, J. et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 20, 317–325 (2013).
Schrank, B. R. et al. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature 559, 61–66 (2018).
Sosa, B. A., Rothballer, A., Kutay, U. & Schwartz, T. U. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell 149, 1035–1047 (2012).
Reid, D. A. et al. Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc. Natl Acad. Sci. USA 112, E2575–E2584 (2015).
Zhao, B. et al. The essential elements for the noncovalent association of two DNA ends during NHEJ synapsis. Nat. Commun. 10, 3588 (2019).
Gioia, U. et al. Pharmacological boost of DNA damage response and repair by enhanced biogenesis of DNA damage response RNAs. Sci. Rep. 9, 6460 (2019).
Gunn, A. & Stark, J. M. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 920, 379–391 (2012).
St Claire, K. et al. Langerhans cell histiocytosis: a great imitator. Clin. Dermatol. 38, 223–234 (2020).
Omori, M. et al. Cytologic features of ovarian granulosa cell tumors in pleural and ascitic fluids. Diagn. Cytopathol. 43, 581–584 (2015).
Bunting, S. F. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010).
Dias, M. P., Moser, S. C., Ganesan, S. & Jonkers, J. Understanding and overcoming resistance to PARP inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 18, 773–791 (2021).
Larrieu, D., Britton, S., Demir, M., Rodriguez, R. & Jackson, S. P. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science 344, 527–532 (2014).
Scaffidi, P. & Misteli, T. Reversal of the cellular phenotype in the premature aging disease Hutchinson–Gilford progeria syndrome. Nat. Med. 11, 440–445 (2005).
Balmus, G. et al. Targeting of NAT10 enhances healthspan in a mouse model of human accelerated aging syndrome. Nat. Commun. 9, 1700 (2018).
Dellaire, G., Kepkay, R. & Bazett-Jones, D. P. High resolution imaging of changes in the structure and spatial organization of chromatin, γ-H2A.X and the MRN complex within etoposide-induced DNA repair foci. Cell Cycle 8, 3750–3769 (2009).
Viita, T. et al. Nuclear actin interactome analysis links actin to KAT14 histone acetyl transferase and mRNA splicing. J. Cell Sci. 132, jcs226852 (2019).
Muslimovic, A., Nystrom, S., Gao, Y. & Hammarsten, O. Numerical analysis of etoposide induced DNA breaks. PLoS ONE 4, e5859 (2009).
Hoa, N. N. et al. Mre11 is essential for the removal of lethal topoisomerase 2 covalent cleavage complexes. Mol. Cell 64, 1010 (2016).
Quennet, V., Beucher, A., Barton, O., Takeda, S. & Lobrich, M. CtIP and MRN promote non-homologous end-joining of etoposide-induced DNA double-strand breaks in G1. Nucleic Acids Res. 39, 2144–2152 (2011).
Abraham, K. J. et al. Nucleolar RNA polymerase II drives ribosome biogenesis. Nature 585, 298–302 (2020).
Lukinavicius, G. et al. Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat. Methods 11, 731–733 (2014).
Maass, P. G. et al. Spatiotemporal allele organization by allele-specific CRISPR live-cell imaging (SNP-CLING). Nat. Struct. Mol. Biol. 25, 176–184 (2018).
Wu, X. & Hammer, J. A. ZEISS Airyscan: optimizing usage for fast, gentle, super-resolution imaging. Methods Mol. Biol. 2304, 111–130 (2021).
Chan, J. N. et al. Perinuclear cohibin complexes maintain replicative life span via roles at distinct silent chromatin domains. Dev. Cell 20, 867–879 (2011).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Arzt, M. et al. LABKIT: labeling and segmentation toolkit for big image data. Front. Comput. Sci. 4, 777728 (2022).
Beleon, A. et al. CometAnalyser: a user-friendly, open-source deep-learning microscopy tool for quantitative comet assay analysis. Comput. Struct. Biotechnol. J. 20, 4122–4130 (2022).
Patel, P. S. et al. Excessive transcription-replication conflicts are a vulnerability of BRCA1-mutant cancers. Nucleic Acids Res. 51, 4341–4362 (2023).
Hmeljak, J. et al. Integrative molecular characterization of malignant pleural mesothelioma. Cancer Discov. 8, 1548–1565 (2018).
Thennavan, A. et al. Molecular analysis of TCGA breast cancer histologic types. Cell Genom. 1, 100067 (2021).
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Knijnenburg, T. A. et al. Genomic and molecular landscape of DNA damage repair deficiency across The Cancer Genome Atlas. Cell Rep. 23, 239–254 (2018).
Liu, Q. et al. Loss of TGFβ signaling increases alternative end-joining DNA repair that sensitizes to genotoxic therapies across cancer types. Sci. Transl. Med. 13, eabc4465 (2021).
Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013).
Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 (2014).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Ghandi, M. et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019).
Szklarczyk, D. et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646 (2023).
Acknowledgements
We thank K. Lau and P. Paroutis at the SickKids Children’s Hospital Imaging Facility; F. Xu at the Advanced Optical Microscopy Facility of the Princess Margaret Cancer Research Tower; and the Imaris Bitplane support team. We thank R. A. Greenberg (University of Pennsylvania) for the FokI-DSB system; J. Stark (City of Hope) for DR-GFP and EJ5-GFP; J. Lammerding (Cornell University) for the LMNB1 plasmid; and S. Harding (University of Toronto) for the NBS1 antibody. We thank members of the K. Mekhail, R. Hakem and M. Ohh laboratories for support. We thank D. Durocher and I. Chiolo for discussions. Representative images from human breast carcinoma cases were from the TCGA Research Network (https://www.cancer.gov/tcga). M. Shokrollahi was supported by an Ontario Graduate Scholarship (OGS). A. Hundal was supported by an Ontario Women’s Health Scholars Award, a Canadian Institutes of Health Research (CIHR) scholarship and an OGS. D.U. is supported by an OGS. J.H. was supported by the Princess Margaret Cancer Foundation and a CIHR fellowship (187888). P.G.M. was supported by the CIHR (173542). M.A.P. was supported by the Instituto Salud Carlos III (PI21/01306 and CIBERES, co-funded by the European Regional Development Fund), Generalitat de Catalunya (SGR 2017-449; PERIS PFI-Salut SLT017-20-000076 to R.E.) and the CERCA Program to IDIBELL. R.H. was supported by funds from the CIHR (143214, 183597 and 186072) and holds the Lau Chair in Breast Cancer Research at the University of Toronto and Princess Margaret Hospital. This research was primarily supported by CIHR funds (156382, 159676, 180469 and 190143) to K.M., who is supported by the Royal Society of Canada. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
M. Shokrollahi, M. Stanic, A. Hundal and K.M. designed the mechanistic studies. A. Hundal, A. Hakem, M.A.P., R.H. and K.M. designed the cancer studies. M. Shokrollahi designed, conducted and analyzed the super-resolution imaging, reconstruction and related analyses; devised most 3D and 2D tubule scoring approaches; and created reconstruction and other videos. M. Stanic designed, conducted and analyzed fluorescent targeted DSB experiments, ChIP, enoxacin and cell cycle analyses, with assistance from C.A.J. A. Hundal designed, conducted and analyzed imaging, proximity, comet, metaphase, growth, co-IP and senescence analyses, with contributions from D.U. to manual and automated image analyses, from R.K. to repair assays, from M.P.H to metaphase, from A. Hakem to colony assays and from A. Hakem and J.H. to xenograft assays. M. Shokrollahi, M. Stanic, A. Hundal and J.N.Y.C. performed knockdowns and related imaging and immunoblotting. J.N.Y.C. assisted K.M. in project coordination, conducted co-IPs and designed, engineered and validated the SUN1 chimeras, which were imaged and analyzed by M. Stanic. The bioinformatic analyses were performed by R.E. and M.A.P., and J.H. analyzed CRISPR screen datasets. P.G.M. provided critical imaging tools and resources. B.D. retrieved and analyzed the human tumor images. K.M. supervised M. Shokrollahi, M. Stanic, A. Hundal, J.N.Y.C., D.U. and C.A.J.; R.H. supervised A. Hundal, A. Hakem, J.H. and R.K.; and M.A.P. supervised R.E. K.M., together with R.H., conceptualized the breast cancer studies. K.M. conceived and coordinated the entire project, obtained the main funding, handled editorial communications and wrote the main text. All authors contributed to writing or editing.
Corresponding authors
Ethics declarations
Competing interests
K.M., R.H., M. Shokrollahi, M. Stanic and A. Hundal are listed as inventors on patent applications related to this work. The other authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Dimitris Typas was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
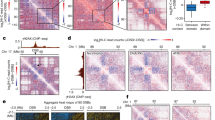
Extended Data Fig. 1 Characterization of etoposide-induced LMNB1 tubules.
a, Treatment regimens used. b, LMNB1 imaging, reconstruction, and measurement of tubules via dividing the total LMNB1 surface area (boundary + tubular) by the total surface area of DAPI-marked DNA yielding the tubular score (see Supplementary Videos 1–3). Scale bar, 3 μm. c, LMNB1 visualized three-dimensionally (3D) and two-dimensionally. Scale bar, 3 μm. d, Representative two-dimensional planes from nuclei negative (top) or positive (bottom) for LMNB1-tubules used to calculate the percentage of cells with tubules, their width, and the number of tubules crossing the midplane. Scale bars, 3 μm (top), 5 μm (bottom). e, Types of nuclear envelope invaginations. f, The number of tubules crossing the nuclear midplane showing etoposide-induced intranuclear LMNB1 tubules and their reversal upon drug removal (wash) or prevention by nocodazole. g-h, Effect of nocodazole on LMNB1 tubules assessed by percentage of tubules-positive cells (g) or tubular width (h). Nocodazole pre-treatment prevented etoposide from inducing tubules whether nocodazole was kept or removed from the media (g). i, Cell cycle staging following the indicated treatments. j, Live-cell imaging shows the induction of SIR-tubulin (SRT)-stained microtubules displacing chromatin (arrows) following etoposide treatment, their reversal upon etoposide removal, and their prevention by nocodazole. Scale bar, 5 μm. k, Fixed-cell immunofluorescence showing etoposide-induced endogenous Tubulin (TUB)-positive filaments infiltrating chromatin (arrows) are prevented by nocodazole. Scale bars, 7 μm (left), 3.5 μm (right). l, Time-lapse imaging shows microtubule plus-ends (EB1) in a growing LMNB1 tubule (arrows). EB1 filaments disappeared before LMNB1 tubule reversal. Imaging was initiated immediately after a 1 h ETP treatment. Insets show contrasted magnifications of the EB1-marked LMNB1 tubules, facilitating visualization. Scale bar, 5 μm. m, Effects of nocodazole and vinblastine on etoposide-induced LMNB1 tubules. b-d,f-m, U2OS cells; standard etoposide treatment was 100 μM for 1 h; ETP, etoposide; NOC, nocodazole; VNBL, Vinblastine; MTs, microtubules; (f-i, j, m) Data are mean ± s.d.; n = 3 biologically independent replicates, two-way ANOVA with Siddak’s (f-h, j, m) or Dunnett’s (i) multiple comparisons tests.
Extended Data Fig. 2 Characterization of LMNB1 tubules-DSB links.
a, 53BP1 foci associate with etoposide-induced LMNB1 tubules. Scale bars, 3 μm (left) and 1 μm (right). b, Co-immunoprecipitation showing etoposide-induced interactions between endogenous 53BP1 and LMNB1. c, Time-lapse images and quantifications indicating the faster resolution of 53BP1 foci associated with LMNB1 tubules (two left arrows) compared to non-tubule-associated foci (two right arrows). Cells were treated with ETP for 1 h, subjected to a 1 h wash, and imaged. Scale bar, 5 μm. d, Live-cell time-lapse imaging shows the natural reversal of an etoposide-induced LMNB1 tubule (arrow) in the presence of the drug (see Supplementary Videos 4 and 5). Scale bar, 3 μm. e, Treatment with nocodazole (NOC) or vinblastine (VINBL) induces 53BP1 foci further in the presence of etoposide. f, RAD51 foci associate with etoposide-induced LMNB1 tubules. Scale bars, 2 μm. g, STRING analysis shows connections between nuclear envelope (top), kinesin (bottom), and DDR kinase (right) proteins. Shown is a full STRING network, including physical and functional evidence, with the connections’ thickness reflecting the confidence level. h, Immunoblotting confirms the induction of γH2AX in etoposide-treated cells. Vinculin served as loading control. i, Immunoblots confirming the activation of the DDR kinases ATR (pCHK1), ATM (pCHK2), and DNAPK (pDNAPK) by etoposide and the inhibition of the activation using DDR kinase inhibitors (ATRi, ATMi, or DNAPKi). j-m, Effect of knocking down KU70 (j) or RNF8, RAD50, or NBS1 (k) on tubule formation (j,k) with immunoblots confirming knockdown for KU70 (l) and the other factors indicated (m). DNA repair factors on LMNB1 tubules. a-f,h-m, U2OS cells; standard etoposide treatment was 100 μM for 1 h; Data shown the mean (c) or mean ± s.d. (e, j-k); n = 3 biologically independent replicates (e, j-k) or n = 8 foci per type and timepoint from 4 independent biological replicates (c); multiple unpaired t-tests with Welch’s correction (c, above graph), two-tailed Wilcoxon matched-pairs signed rank test (c, on graph) and two-way ANOVA with Siddak’s multiple comparisons test (e, j-k).
Extended Data Fig. 3 Induction of LMNB1 tubules under different experimental conditions and in diverse cell types.
a-b, Levels of γH2AX (a) and LMNB1 tubules (b) in cells treated with etoposide at the indicated concentrations. c-f, Representative images (c) and quantifications (d-f) showed that etoposide induces LMNB1 tubules as assessed by the tubular score (d), number of tubules crossing the nuclear midplane (e), and tubular width (f) in the indicated cell lines. Scale bar, 5 μm (c). g-h, Quantification of total 53BP1 levels (g) and LMNB1 tubules (h) in cells treated with the DNA damaging agent etoposide, microtubule inhibitor nocodazole (NOC), CDK/transcription-inhibiting flavopiridol (FVP), or proteasome inhibitor MG132. a-h, U2OS cells unless otherwise indicated; standard etoposide treatment was 100 μM for 1 h; data are shown as mean ± s.d.; n = 4 (a,b) or n = 3 (d-g) biologically independent replicates, or n = 3 biologically independent replicates each with three technical replicates (h), two-way ANOVA with Dunnett’s multiple comparisons test (a-b), two-tailed Mann-Whitney test (d, f), two-tailed unpaired t-test (e), or one-way ANOVA with Siddak’s multiple comparisons test (g-h).
Extended Data Fig. 4 Controls related to the characterization of factors modulating the tubules.
a, Immunoblots indicating the knockdown of indicated factors using small-interfering RNAs (siRNA). Actin served as loading control. b, Proximity ligation assay (PLA) images showing the effect of etoposide treatment on the proximity of 53BP1 to LMNB1 or SUN1. Nuclear PLA signals are quantified in Fig. 2c, and internal/boundary PLA ratios from nuclear midplanes are shown here. c-e, Immunofluorescence showing that NUP153 knockdown (c) does not alter SUN1 levels (d) or localization (e). f-g, Effect of NUP50 knockdown (f) on the formation of LMNB1 tubules (g). h-i, Effect of LMNB1 knockdown (h) on the tubules as marked by an antibody for LMNA/C (i). j, Immunoblots confirming the expression of exogenous wild-type and mutant KIF5B. k, Representative images showing the localization of LMNB1 together with wild-type or mutant KIF5B. Tubules were observed in etoposide-treated cells expressing wild-type but not mutant KIF5B. Wild-type KIF5B localized to the LMNB1 tubules (arrows). l, Quantification of etoposide-dependent LMNB1 tubules formation (left) and 53BP1 foci (right) in cells expressing wild-type or mutant KIF5B. m, Representative images of α-Tubulin-positive LMNB1 tubules (arrows) and quantifications showing the effect of DDR kinase inhibitors on the ability of etoposide to induce LMNB1 tubules. ETP-dependent changes are shown in the bottom row of percentages, and DDR kinase inhibitor-dependent changes to the induction compared to control (CTL) are shown in the top row of percentages. a-m, U2OS cells; standard etoposide treatment was 100 μM for 1 h; scale bars, 5 μm (b, e, k, m) or 10 μm (c, i); data are shown as the mean ± s.d.; n = 3 (a-l) or n = 3 biologically independent replicates each with two technical replicates (m), two-tailed paired t-test (b), two-tailed unpaired t-test (c, f, h), two-way ANOVA with Dunnett’s (d) or Siddak’s (g, i, l-m) multiple comparisons test.
Extended Data Fig. 5 Controls related to the characterization of factors modulating nuclear envelope tubules, repair kinetics, and repair foci behaviors.
a, Immunoblots showing the effect of etoposide, etoposide removal (ETP + wash), and ATAT1 knockdown on the levels of Ac-αTub and αTub with Vinculin control. Data are representative of two independent experiments. b-c, Knockdown of the indicated factors, confirmed using RT-qPCR (b), did not alter the distribution of cells in the cell cycle (c). d, Quantification and representative images from neutral comet assays show that the knockdown of ATAT1, KIF5B, or NUP153 increases the levels of ETP-induced DNA damage and decreases the rate of DNA repair. Scale bar, 100 μm. e-f, Quantification of 53BP1 foci numbers (e) and sizes (f) over time show SUN1 knockdown increases the number and size of etoposide-induced DNA damage and decreases the rate of DNA repair. g, Scatter plot showing the 53BP1 foci number (Nb.) and size (S) distribution in control and SUN1 knockdown cells for the ETP+wash(6 h) condition with means shown in the table above. a-g, U2OS cells; standard etoposide treatment was 100 μM for 1 h; data are mean ± s.d.; n = 3 biologically independent replicates, two-way ANOVA with Siddak’s (b, d-f) or Dunnett’s (c) multiple comparisons test.
Extended Data Fig. 6 Controls related to targeted DSB induction, tubule formation, and damage capture.
a, The ER-mCherry-LacI-FokI-DD (FokI-DSB) reporter system. b, Colocalization of the induced FokI-DSB with 53BP1 (arrows). Large arrows indicate the DSB shown in the zoomed images. Scale bar, 10 μm. c, Chromatin immunoprecipitation shows the enrichment of 53BP1 at the induced FokI-DSB. d, Phosphorylated CHK2 (pCHK2) immunoblotting indicates FokI-DSB induction or treatment with the drug enoxacin (ENO) activates the DDR. Vinculin served as loading control. e, Confirmation of siRNA-mediated protein knockdowns using immunofluorescence. f, Knockdown of SUN1 or SUN2 decreased the percentage of tubule-positive cells following FokI-DSB induction. g, Effect of disrupting LINC, NPC, or kinesin proteins on LMNB1 tubular diameter following FokI-DSB activation. h, Disruption of LINC, NPC, or kinesin proteins linked to dsbNET formation increases the distance between the FokI-DSB and nuclear edge. i, Cell cycle stage profiling of cells following the knockdown of indicated factors. j, Treatment with the histone deacetylase inhibitor SAHA is not redundant with SUN1 knockdown in terms of increasing the distance between FokI-DSB and the nuclear edge. k, Confirmation of nuclear actin repression following treatment with latrunculin-B (LATB). l,m, LATB efficiently repressed nuclear actin (l) and partly decreased etoposide-induced LMNB1 tubules (m). n, Expression of the nuclear actin-disrupting NLS-Actin-R62D, but not the control NLS-Actin, partially repressed etoposide-induced tubules. a-n, 2-6-5 cells (a-k) or U2OS cells (l-n; standard etoposide treatment was 100 μM for 1 h); data are shown as the mean ± s.d.; n = 3 (c, e-h, j-n) or n = 4 (i) biologically independent replicates; one-way ANOVA with Siddak’s multiple comparisons test (c), two-tailed unpaired t-test with Welch’s correction (e), two-way ANOVA with Siddak’s (f, k), Tukey’s (g, j, m-n) or Dunnett’s (h-i) multiple comparisons test, or two-tailed Mann Whitney test (l).
Extended Data Fig. 7 Chimeric SUN1 fusion protein constructs and changes to the expression of endogenous SUN1 and SUN2 following etoposide treatment.
a, SUN1 protein schematic showing the N-terminal lamina/chromatin-binding domain and the C-terminal SUN domain. Also shown are the confirmed post-translational modification sites of SUN1 from the UniProtein (middle) and PhosphoSite (bottom) databases. b, Immunoblot showing decreased endogenous SUN1 protein levels upon siRNA-mediated knockdown. c, Immunoblot demonstrating expression of the various SUN1 and control chimeric fusion proteins. Proteins of expected sizes are below the red asterisks. d, Schematic of fusion proteins used. e, Immunoblot showing decreased levels of all endogenous SUN1 forms following siRNA-mediated knockdown. f-g, Immunoblots of endogenous SUN1 (f) and SUN2 (g) proteins from vehicle or etoposide-treated cells. h-i, Etoposide treatment increases the phosphoserine signal (h) but not the ubiquitin signal (i) co-immunoprecipitating with SUN1 (SUN1 IP). The blots in (h) and (i) are part of the same experiment. b,c,e-i, blots are representative of two independent experiments. f-i, standard etoposide treatment was 100 μM for 1 h.
Extended Data Fig. 8 Controls related to DSB ends-tethering and repair.
a-b, Visualizing one end of the FokI-DSB (a) using sgRNAs (sgDSB) and GFP-dCas9 expression (b) confirmed the long and split FokI-DSB shapes represent less connected DSB ends. A non-targeting sgRNA (sgNT) served as control. Scale bar, 1 μm (a). c, Knockdown of the DSB ends-tethering NBS1 or RAD50, but not treatment with the silent chromatin-disrupting and histone deacetylase inhibitor SAHA, increases the percentage of cells with a split DSB. d, ChIP indicates that the knockdown of SUN1 or KIF5B decreases etoposide-induced increases in XRCC4 enrichment at FokI-DSB. e, XRCC4 levels are stable following the knockdown of SUN1 or KIF5B. f-h, Enoxacin enlarges the 53BP1 focus surrounding the FokI-DSB (f-g) and restores FokI-DSB’s colocalizatoin with 53BP1 (h). Scale bar, 1 μm. i, Effect of LINC protein knockdowns and enoxacin treatment on LMNB1 tubular diameter in cells without or with FokI-DSB activation. Data from Extended Data Fig. 6g are reshown here as they were part of the same experiments and for comparison. j, Effect of expressing the indicated proteins on FokI-DSB shape (long or split). k-l, Knockdown of KIF5B decreased DNA repair as assessed using chromosomally-integrated HR (DR-GFP) (k) or NHEJ (EJ5-GFP) (l) reporters. The knockdown of BRCA1 and XRCC4 served as positive control for HR and NHEJ repair, respectively. a-k, 2-6-5 (a-j) or U2OS (k-l) cells; data are shown as the mean ± s.d.; n = 3 biologically independent replicates each with three (a) or two (c) technical replicates, n = 3 biologically independent replicates (d, g-j), n = 9 (siCTL, siKIF5B) or n = 6 (siBRCA1) biologically independent replicates (k), or n = 3 technical replicates (l); two-way ANOVA with Dunnett’s (c, h, j), Siddak’s (i), or Tukey’s (d, k-l) multiple comparisons test, or two-tailed Mann-Whitney test (g).
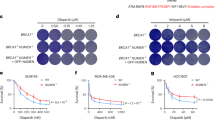
Extended Data Fig. 9 Analyses of KIF5B or KIFC3 co-expression with DNA repair pathway signatures across cancers, tumor sections from breast cancer patients, and dsbNETs and DSBs in etoposide- or olaparib-treated breast cancer cells.
a-c, The expression of KIFC3 and KIF5B negatively and positively correlated with the NHEJ (a) signature more than the HR (b) or altEJ (c) signatures across cancers, respectively. Forest plots show the Pearson Correlation Coefficient (PCC, square) and 95% confidence interval (bar) of defined gene-signature expression correlation analyses across The Cancer Genome Atlas (TCGA) cancer studies. Cancer acronyms are on the Y-axis per Supplementary Table 1, the number of primary tumors analyzed in each setting is indicated in parentheses, and nominally significant (P < 0.05) PCCs are indicated by filled squares, with the specific P values listed in the numerical Source Data file for this figure. Non-significant PCCs are indicated by open squares. The data for KIF5B and KIFC3 in panels a and b here are shown superimposed in Fig. 5c, d to facilitate comparison across cancers. d-e, Human breast cancer cases exhibit nuclear grooves. Representative images of cases of human breast carcinoma with indicated mutations from The Cancer Genome Atlas (TCGA, 400X magnification) (d). Arrows highlight nuclear grooves reminiscent of nuclear envelope tubules uncovered herein in cell culture. Scale bars, 50 μm (top) and 25 μm (bottom). Quantification reveals the number of grooves per nucleus in different human breast cancer cases (n = 10 cases for each of the BRCA1 and BRCA2 mutants) (e). f-h, Immunoblots confirm the knockdown of indicated factors in MDA-MB-231 or MDA-MB-436 TNBC cells. i-j, Effects of KIF5B or KIFC3 knockdown on LMNB1 tubules (i) and 53BP1 foci number (j) in MDA-MB-231 (231) or MDA-MB-436 (436) cells treated with etoposide (ETP, 100 μM, 1 h) or olaparib (PARPi, 2 μM, 24 h). Data are shown as mean ± s.d., n = 3 biologically independent replicates, two-way ANOVA with Tukey’s multiple comparisons test.
Extended Data Fig. 10 Cancer-related analyses of chromosomes, senescence, and growth, and assessment of a dsbNETs-Progerin link.
a, Representative images and quantifications showing the effect of KIF5B or KIFC3 knockdown on the levels of misjoined chromosome structures in MDA-MB-436 cells treated with vehicle or PARPi (2 μM, 16 h). Shown is the mean ± s.d., n = 50 metaphase spreads per condition. Scale bar, 10 μm. b, Quantifications and representative images showing the effect of KIF5B or KIFC3 knockdown on the level of senescence-associated β-galactosidase (SA-β-GAL) in MDA-MB-436 cells. Shown is the mean ± s.d., n = 3 biologically independent replicates. Scale bar, 100 μm. c-d, Effect of KIF5B or KIFC3 knockdown on MDA-MB-436 xenograft tumors in terms of volumetric growth (c) and the end-point tumor mass (d). Shown is the mean ± s.d., n = 5 mice per knockdown. e, Gene effect or score (mean ± s.d.) for the indicated genes when combined with BRCA1 wild-type (n = 68) or mutant (n = 8) breast and ovarian cancer cell lines. CRISPR screen data were accessed via DepMap (see Methods). f, Effect of LMNA or Progerin expression on LMNB1 tubulation and the number of 53BP1-marked DSBs in the absence or presence of etoposide (ETP, 100 μM, 1 h) in U2OS cells. a-f, Two-way ANOVA with Tukey’s (a-b, f(right)) or Siddak’s (f(left)) multiple comparisons test, two-tailed paired (c) or unpaired (d) t-test, or two-tailed Mann-Whitney test (e).
Supplementary information
Supplementary Information
Supplementary Discussion, Supplementary Tables 1–7 and Supplementary References.
Supplementary Video 1
3D reconstructed nucleus isolating the etoposide-induced intranuclear LMNB1 tubules associated with DSBs. LMNB1 tubules are shown associated with 53BP1 foci followed by 3D nuclear reconstruction separating the boundary (transparent/gray) and tubular (green) LMNB1 signal. 53BP1 foci at or away from LMNB1 tubules are color-coded magenta or cyan, respectively. Boundary LMNB1 is then removed, highlighting internal LMNB1 tubules associated with the 53BP1 foci.
Supplementary Video 2
3D reconstructed nucleus showing etoposide-induced LMNB1 tubules from around the nucleus associated with DSBs. Nuclear slicing shows LMNB1 tubules in the x–y plane with associated 53BP1 foci followed by 3D nuclear reconstruction separating boundary (transparent/gray) and tubular (green) LMNB1. 53BP1 foci at or away from LMNB1 tubules are color-coded magenta and cyan, respectively.
Supplementary Video 3
Detailed illustration of the 3D reconstruction process using a nucleus showing that DSB-associated LMNB1 tubules re-emerge after nocodazole washout and etoposide treatment. The 3D LMNB1 nuclear signal and associated 53BP1 foci were re-constructed in three dimensions. Nuclear slicing shows the LMNB1 tubules in x–y, x–z and y–z planes with associated 53BP1 signal. Subsequent reconstruction of LMNB1 and 53BP1 surfaces shows DSB foci at or away from LMNB1 tubules. The reconstructed boundary (transparent/gray) and tubular (green) LMNB1 structures are shown.
Supplementary Video 4
The natural reversal of etoposide-induced LMNB1 nuclear tubules in a live cell highlights the dynamic nature of these nuclear envelope structures. Shown is one plane representing slice 36 of 85 slices, which is deep into the nucleus. The LMNB1 tubule reversal process lasted approximately 15–24 min. See Supplementary Video 5 for the chromatin time-lapse imaging of the same nucleus.
Supplementary Video 5
Chromatin reorganization during the natural reversal of etoposide-induced LMNB1 tubules. Shown is one plane representing slice 36 of 85 slices, which is deep into the nucleus. Chromatin resealing in the wake of the reversing LMNB1 tubule lasted approximately 15–24 min (see Supplementary Video 4 for the matching LMNB1 signal). Together, Supplementary Videos 4 and 5 highlight the coordinated dynamics of LMNB1 tubules and chromatin inside the nucleus.
Source data
Source Data Fig. 1
Graphs source data.
Source Data Fig. 2
Graphs source data.
Source Data Fig. 3
Graphs source data.
Source Data Fig. 4
Graphs source data.
Source Data Fig. 5
Graphs source data.
Source Data Extended Data Fig. 1
Graphs source data.
Source Data Extended Data Fig. 2
Graphs source data.
Source Data Extended Data Fig. 3
Graphs source data.
Source Data Extended Data Fig. 4
Graphs source data.
Source Data Extended Data Fig. 5
Graphs source data.
Source Data Extended Data Fig. 6
Graphs source data.
Source Data Extended Data Fig. 8
Graphs source data.
Source Data Extended Data Fig. 9
Graphs source data.
Source Data Extended Data Fig. 10
Graphs source data.
Source Data Fig. 2
Uncropped blots source data.
Source Data Extended Data Fig. 2
Uncropped blots source data.
Source Data Extended Data Fig. 4
Uncropped blots source data.
Source Data Extended Data Fig. 5
Uncropped blots source data.
Source Data Extended Data Fig. 6
Uncropped blots source data.
Source Data Extended Data Fig. 7
Uncropped blots source data.
Source Data Extended Data Fig. 8
Uncropped blots source data.
Source Data Extended Data Fig. 9
Uncropped blots source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shokrollahi, M., Stanic, M., Hundal, A. et al. DNA double-strand break–capturing nuclear envelope tubules drive DNA repair. Nat Struct Mol Biol (2024). https://doi.org/10.1038/s41594-024-01286-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41594-024-01286-7