Abstract
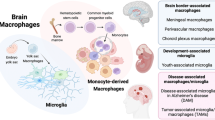
Myeloid cells in the central nervous system (CNS), such as microglia, CNS-associated macrophages (CAMs), dendritic cells and monocytes, are vital for steady-state immune homeostasis as well as the resolution of tissue damage during brain development or disease-related pathology. The complementary usage of multimodal high-throughput and high-dimensional single-cell technologies along with recent advances in cell-fate mapping has revealed remarkable myeloid cell heterogeneity in the CNS. Despite the establishment of extensive expression profiles revealing myeloid cell multiplicity, the local anatomical conditions for the temporal- and spatial-dependent cellular engraftment are poorly understood. Here we highlight recent discoveries of the context-dependent mechanisms of myeloid cell migration and settlement into distinct subtissular structures in the CNS. These insights offer better understanding of the factors needed for compartment-specific myeloid cell recruitment, integration and residence during development and perturbation, which may lead to better treatment of CNS diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ginhoux, F. et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010). In this seminal work, the YS origin of microglia was established using fate-mapping mouse models. This marked the beginning of the discovery of the embryonic origin of many tissue-resident macrophages.
Goldmann, T. et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol. 17, 797–805 (2016). This paper demonstrated that leptomeningeal and perivascular macrophages are YS derived and are not replaced by HSC-derived cells in mice.
Stremmel, C. et al. Yolk sac macrophage progenitors traffic to the embryo during defined stages of development. Nat. Commun. 9, 75 (2018).
Kierdorf, K. et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 16, 273–280 (2013).
Ajami, B., Bennett, J. L., Krieger, C., Tetzlaff, W. & Rossi, F. M. V. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 10, 1538–1543 (2007).
Mildner, A. et al. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat. Neurosci. 10, 1544–1553 (2007).
Van Hove, H. et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci. 22, 1021–1035 (2019). This study is an important resource in the field as it provides a very detailed and thorough analysis of the transcription profiles of various CNS macrophage subsets. It also describes how Kolmer’s epiplexus cells share many features of microglia.
Prinz, M., Masuda, T., Wheeler, M. A. & Quintana, F. J. Microglia and central nervous system-associated macrophages—from origin to disease modulation. Annu. Rev. Immunol. 39, 251–277 (2021).
Morrison, S. J. & Scadden, D. T. The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334 (2014).
Guilliams, M. & Scott, C. L. Does niche competition determine the origin of tissue-resident macrophages? Nat. Rev. Immunol. 17, 451–460 (2017).
Bouchaud, C. & Bosler, O. The circumventricular organs of the mammalian brain with special reference to monoaminergic innervation. In International Review of Cytology Vol. 105, 283–327 (Elsevier, 1986).
Daneman, R. & Prat, A. The blood–brain barrier. Cold Spring Harb. Perspect. Biol. 7, a020412 (2015).
Weller, R. O., Sharp, M. M., Christodoulides, M., Carare, R. O. & Møllgård, K. The meninges as barriers and facilitators for the movement of fluid, cells and pathogens related to the rodent and human CNS. Acta Neuropathol. 135, 363–385 (2018).
Engelhardt, B., Vajkoczy, P. & Weller, R. O. The movers and shapers in immune privilege of the CNS. Nat. Immunol. 18, 123–131 (2017).
Owens, T., Bechmann, I. & Engelhardt, B. Perivascular spaces and the two steps to neuroinflammation. J. Neuropathol. Exp. Neurol. 67, 1113–1121 (2008).
Munro, D. A. D., Movahedi, K. & Priller, J. Macrophage compartmentalization in the brain and cerebrospinal fluid system. Sci. Immunol. 7, eabk0391 (2022).
Guilliams, M., Thierry, G. R., Bonnardel, J. & Bajenoff, M. Establishment and maintenance of the macrophage niche. Immunity 52, 434–451 (2020). This review article nicely illustrates the concept of the macrophage niche and provides several examples of known niche factors and cellular sources of such factors.
Derk, J., Jones, H. E., Como, C., Pawlikowski, B. & Siegenthaler, J. A. Living on the edge of the CNS: meninges cell diversity in health and disease. Front. Cell. Neurosci. 15, 703944 (2021).
Louveau, A. et al. Structural and functional features of central nervous system lymphatic vessels. Nature 523, 337–341 (2015). This paper shows that lymphatic vessels in the dura mater drain CNS-derived antigens into the cervical lymph nodes, provoking a paradigm shift in our understanding of CNS immune reactions and CNS immune privilege.
Mrdjen, D. et al. High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 48, 380–395 (2018).
Rustenhoven, J. et al. Functional characterization of the dural sinuses as a neuroimmune interface. Cell 184, 1000–1016 (2021).
Sato, T., Konishi, H., Tamada, H., Nishiwaki, K. & Kiyama, H. Morphology, localization, and postnatal development of dural macrophages. Cell Tissue Res. 384, 49–58 (2021).
Brioschi, S. et al. Heterogeneity of meningeal B cells reveals a lymphopoietic niche at the CNS borders. Science 373, eabf9277 (2021).
Merlini, A. et al. Distinct roles of the meningeal layers in CNS autoimmunity. Nat. Neurosci. 25, 887–899 (2022).
Jordão, M. J. C. et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363, eaat7554 (2019).
Ransohoff, R. M. & Engelhardt, B. The anatomical and cellular basis of immune surveillance in the central nervous system. Nat. Rev. Immunol. 12, 623–635 (2012).
Zhang, E. T., Inman, C. B. & Weller, R. O. Interrelationships of the pia mater and the perivascular (Virchow–Robin) spaces in the human cerebrum. J. Anat. 170, 111–123 (1990).
Masuda, T. et al. Specification of CNS macrophage subsets occurs postnatally in defined niches. Nature 604, 740–748 (2022).
Faraco, G. et al. Perivascular macrophages mediate the neurovascular and cognitive dysfunction associated with hypertension. J. Clin. Invest. 126, 4674–4689 (2016).
Lun, M. P., Monuki, E. S. & Lehtinen, M. K. Development and functions of the choroid plexus–cerebrospinal fluid system. Nat. Rev. Neurosci. 16, 445–457 (2015).
Dani, N. et al. A cellular and spatial map of the choroid plexus across brain ventricles and ages. Cell 184, 3056–3074 (2021).
Tischfield, M. A. et al. Cerebral vein malformations result from loss of Twist1 expression and BMP signaling from skull progenitor cells and dura. Dev. Cell 42, 445–461 (2017).
Dasgupta, K., Chung, J. U., Asam, K. & Jeong, J. Molecular patterning of the embryonic cranial mesenchyme revealed by genome-wide transcriptional profiling. Dev. Biol. 455, 434–448 (2019).
DeSisto, J. et al. Single-cell transcriptomic analyses of the developing meninges reveal meningeal fibroblast diversity and function. Dev. Cell 54, 43–59 (2020).
McLone, D. G. & Bondareff, W. Developmental morphology of the subarachnoid space and contiguous structures in the mouse. Am. J. Anat. 142, 273–293 (1975).
Dasgupta, K. & Jeong, J. Developmental biology of the meninges. Genesis 57, e23288 (2019).
Gupta, A., Rarick, K. R. & Ramchandran, R. Established, new and emerging concepts in brain vascular development. Front. Physiol. 12, 636736 (2021).
Blanchette, M. & Daneman, R. Formation and maintenance of the BBB. Mech. Dev. 138, 8–16 (2015).
Daneman, R., Zhou, L., Kebede, A. A. & Barres, B. A. Pericytes are required for blood–brain barrier integrity during embryogenesis. Nature 468, 562–566 (2010).
Yang, Y., Higashimori, H. & Morel, L. Developmental maturation of astrocytes and pathogenesis of neurodevelopmental disorders. J. Neurodev. Disord. 5, 22 (2013).
Hoeffel, G. et al. C-Myb+ erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–678 (2015).
Vanlandewijck, M. et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018). An important work that provides a valuable resource of single-cell RNA-sequencing data from diverse CNS cell types. It shows heterogeneity of cell types along the brain vasculature tree (zonation) and describes perivascular fibroblast-like cells.
Saunders, A. et al. Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015–1030 (2018).
Garcia, F. J. et al. Single-cell dissection of the human brain vasculature. Nature 603, 893–899 (2022).
Yang, A. C. et al. A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature 603, 885–892 (2022).
Månberg, A. et al. Altered perivascular fibroblast activity precedes ALS disease onset. Nat. Med. 27, 640–646 (2021).
Hupe, M. et al. Gene expression profiles of brain endothelial cells during embryonic development at bulk and single-cell levels. Sci. Signal. 10, eaag2476 (2017).
Dorrier, C. E., Jones, H. E., Pintarić, L., Siegenthaler, J. A. & Daneman, R. Emerging roles for CNS fibroblasts in health, injury and disease. Nat. Rev. Neurosci. 23, 23–34 (2022).
Krausgruber, T. et al. Structural cells are key regulators of organ-specific immune responses. Nature 583, 296–302 (2020).
Mass, E. Delineating the origins, developmental programs and homeostatic functions of tissue-resident macrophages. Int. Immunol. 30, 493–501 (2018).
Yona, S. et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013).
Utz, S. G. et al. Early fate defines microglia and non-parenchymal brain macrophage development. Cell 181, 557–573 (2020).
Mass, E. et al. Specification of tissue-resident macrophages during organogenesis. Science 353, aaf4238 (2016).
Schulz, C. et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012).
Swinnen, N. et al. Complex invasion pattern of the cerebral cortex by microglial cells during development of the mouse embryo. Glia 61, 150–163 (2013).
Ransohoff, R. M. & Cardona, A. E. The myeloid cells of the central nervous system parenchyma. Nature 468, 253–262 (2010).
Lelli, A. et al. The NADPH oxidase Nox2 regulates VEGFR1/CSF-1R-mediated microglial chemotaxis and promotes early postnatal infiltration of phagocytes in the subventricular zone of the mouse cerebral cortex. Glia 61, 1542–1555 (2013).
Arnò, B. et al. Neural progenitor cells orchestrate microglia migration and positioning into the developing cortex. Nat. Commun. 5, 5611 (2014).
Thion, M. S. et al. Microbiome influences prenatal and adult microglia in a sex-specific manner. Cell 172, 500–516 (2018).
Werner, Y. et al. Cxcr4 distinguishes HSC-derived monocytes from microglia and reveals monocyte immune responses to experimental stroke. Nat. Neurosci. 23, 351–362 (2020).
Calderwood, D. A., Campbell, I. D. & Critchley, D. R. Talins and kindlins: partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 14, 503–517 (2013).
Domenga, V. et al. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 18, 2730–2735 (2004).
Munro, D. A. D. et al. CNS macrophages differentially rely on an intronic Csf1r enhancer for their development. Development 147, dev194449 (2020).
Cugurra, A. et al. Skull and vertebral bone marrow are myeloid cell reservoirs for the meninges and CNS parenchyma. Science 373, eabf7844 (2021).
Gautier, E. L. et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128 (2012).
Gosselin, D. et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014).
Lavin, Y. et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).
Matcovitch-Natan, O. et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670 (2016).
Bennett, F. C. et al. A combination of ontogeny and CNS environment establishes microglial identity. Neuron 98, 1170–1183 (2018).
Buttgereit, A. et al. Sall1 is a transcriptional regulator defining microglia identity and function. Nat. Immunol. 17, 1397–1406 (2016).
Scott, C. L. et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 7, 10321 (2016).
van de Laar, L. et al. Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity 44, 755–768 (2016).
Cronk, J. C. et al. Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. J. Exp. Med. 215, 1627–1647 (2018).
Lund, H. et al. Competitive repopulation of an empty microglial niche yields functionally distinct subsets of microglia-like cells. Nat. Commun. 9, 4845 (2018).
Shemer, A. et al. Engrafted parenchymal brain macrophages differ from microglia in transcriptome, chromatin landscape and response to challenge. Nat. Commun. 9, 5206 (2018).
Green, K. N., Crapser, J. D. & Hohsfield, L. A. To kill a microglia: a case for CSF1R inhibitors. Trends Immunol. 41, 771–784 (2020).
Masuda, T. et al. Novel Hexb-based tools for studying microglia in the CNS. Nat. Immunol. 21, 802–815 (2020).
Nicholson, A. M. et al. CSF1R mutations link POLD and HDLS as a single disease entity. Neurology 80, 1033–1040 (2013).
Oosterhof, N. et al. Homozygous mutations in CSF1R cause a pediatric-onset leukoencephalopathy and can result in congenital absence of microglia. Am. J. Hum. Genet. 104, 936–947 (2019).
Kana, V. et al. CSF-1 controls cerebellar microglia and is required for motor function and social interaction. J. Exp. Med. 216, 2265–2281 (2019).
Chitu, V., Gokhan, Ş., Nandi, S., Mehler, M. F. & Stanley, E. R. Emerging roles for CSF-1 receptor and its ligands in the nervous system. Trends Neurosci. 39, 378–393 (2016).
Abboud, S. L., Bunegin, M., Ghosh-Choudhury, N. & Woodruff, K. Analysis of the mouse CSF-1 gene promoter in a transgenic mouse model. J. Histochem. Cytochem. 51, 941–949 (2003).
Easley-Neal, C., Foreman, O., Sharma, N., Zarrin, A. A. & Weimer, R. M. CSF1R ligands IL-34 and CSF1 are differentially required for microglia development and maintenance in white and gray matter brain regions. Front. Immunol. 10, 2199 (2019).
Greter, M. et al. Stroma-derived interleukin-34 controls the development and maintenance of Langerhans cells and the maintenance of microglia. Immunity 37, 1050–1060 (2012).
Wang, Y. et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol. 13, 753–760 (2012).
Rojo, R. et al. Deletion of a Csf1r enhancer selectively impacts CSF1R expression and development of tissue macrophage populations. Nat. Commun. 10, 3215 (2019).
Butovsky, O. et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143 (2014). This paper established TGF-β as an important niche factor for microglial cell identity and function.
Zöller, T. et al. Silencing of TGFβ signalling in microglia results in impaired homeostasis. Nat. Commun. 9, 4011 (2018).
Robertson, I. B. & Rifkin, D. B. Regulation of the bioavailability of TGF-β and TGF-β-related proteins. Cold Spring Harb. Perspect. Biol. 8, a021907 (2016).
Wrana, J. L., Attisano, L., Wieser, R., Ventura, F. & Massagué, J. Mechanism of activation of the TGF-β receptor. Nature 370, 341–347 (1994).
Qin, Y. et al. A milieu molecule for TGF-β required for microglia function in the nervous system. Cell 174, 156–171 (2018).
Wong, K. et al. Mice deficient in NRROS show abnormal microglial development and neurological disorders. Nat. Immunol. 18, 633–641 (2017).
Arnold, T. D. et al. Impaired αVβ8 and TGFβ signaling lead to microglial dysmaturation and neuromotor dysfunction. J. Exp. Med. 216, 900–915 (2019).
Abutbul, S. et al. TGF-β signaling through SMAD2/3 induces the quiescent microglial phenotype within the CNS environment. Glia 60, 1160–1171 (2012).
Smith, C. et al. Biallelic mutations in NRROS cause an early onset lethal microgliopathy. Acta Neuropathol. 139, 947–951 (2020).
Otero, K. et al. Macrophage colony-stimulating factor induces the proliferation and survival of macrophages via a pathway involving DAP12 and β-catenin. Nat. Immunol. 10, 734–743 (2009).
Nataf, S. et al. Brain and bone damage in KARAP/DAP12 loss-of-function mice correlate with alterations in microglia and osteoclast lineages. Am. J. Pathol. 166, 275–286 (2005).
Erny, D. et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 18, 965–977 (2015). This study shows that the presence of microbiota is essential for microglia to acquire their adult phenotype and to be able to initiate proper immune reactions.
Erny, D. et al. Microbiota-derived acetate enables the metabolic fitness of the brain innate immune system during health and disease. Cell Metab. 33, 2260–2276 (2021).
Mezö, C. et al. Different effects of constitutive and induced microbiota modulation on microglia in a mouse model of Alzheimer’s disease. Acta Neuropathol. Commun. 8, 119 (2020).
Mossad, O. et al. Gut microbiota drives age-related oxidative stress and mitochondrial damage in microglia via the metabolite N6-carboxymethyllysine. Nat. Neurosci. 25, 295–305 (2022).
Boehme, M. et al. Microbiota from young mice counteracts selective age-associated behavioral deficits. Nat. Aging 1, 666–676 (2021).
Sankowski, R. et al. Commensal microbiota divergently affect myeloid subsets in the mammalian central nervous system during homeostasis and disease. EMBO J. 40, e108605 (2021).
Mossad, O. & Erny, D. The microbiota–microglia axis in central nervous system disorders. Brain Pathol. 30, 1159–1177 (2020).
Keshavarzian, A., Engen, P., Bonvegna, S. & Cilia, R. The gut microbiome in Parkinson’s disease: a culprit or a bystander? In Progress in Brain Research (eds Björklund, A. & Cenci, M. A.) Vol. 252, 357–450 (Elsevier, 2020).
Sampson, T. R. et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167, 1469–1480 (2016).
Abdel-Haq, R. et al. A prebiotic diet modulates microglial states and motor deficits in α-synuclein overexpressing mice. eLife 11, e81453 (2022).
Ajami, B., Bennett, J. L., Krieger, C., McNagny, K. M. & Rossi, F. M. V. Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat. Neurosci. 14, 1142–1149 (2011).
De Vlaminck, K. et al. Differential plasticity and fate of brain-resident and recruited macrophages during the onset and resolution of neuroinflammation. Immunity 55, 2085–2102 (2022).
Ransohoff, R. M., Kivisäkk, P. & Kidd, G. Three or more routes for leukocyte migration into the central nervous system. Nat. Rev. Immunol. 3, 569–581 (2003).
Schulz, M. & Engelhardt, B. The circumventricular organs participate in the immunopathogenesis of experimental autoimmune encephalomyelitis. Cerebrospinal Fluid Res. 2, 8 (2005).
Herisson, F. et al. Direct vascular channels connect skull bone marrow and the brain surface enabling myeloid cell migration. Nat. Neurosci. 21, 1209–1217 (2018).
Pulous, F. E. et al. Cerebrospinal fluid can exit into the skull bone marrow and instruct cranial hematopoiesis in mice with bacterial meningitis. Nat. Neurosci. 25, 567–576 (2022).
Mazzitelli, J. A. et al. Cerebrospinal fluid regulates skull bone marrow niches via direct access through dural channels. Nat. Neurosci. 25, 555–560 (2022). These two studies suggest that CSF can access the skull bone marrow niche, which allows CNS-derived factors to directly manipulate the local HSC pool.
Tay, T. L. et al. A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat. Neurosci. 20, 793–803 (2017).
Wu, X., Saito, T., Saido, T. C., Barron, A. M. & Ruedl, C. Microglia and CD206+ border-associated mouse macrophages maintain their embryonic origin during Alzheimer’s disease. eLife 10, e71879 (2021).
Reed-Geaghan, E. G., Croxford, A. L., Becher, B. & Landreth, G. E. Plaque-associated myeloid cells derive from resident microglia in an Alzheimer’s disease model. J. Exp. Med. 217, e20191374 (2020).
Mildner, A. et al. Distinct and non-redundant roles of microglia and myeloid subsets in mouse models of Alzheimer’s disease. J. Neurosci. 31, 11159–11171 (2011).
Silvin, A. et al. Dual ontogeny of disease-associated microglia and disease inflammatory macrophages in aging and neurodegeneration. Immunity 55, 1448–1465 (2022).
Yan, P. et al. Peripheral monocyte-derived cells counter amyloid plaque pathogenesis in a mouse model of Alzheimer’s disease. J. Clin. Invest. 132, e152565 (2022).
King, I. L., Dickendesher, T. L. & Segal, B. M. Circulating Ly-6C+ myeloid precursors migrate to the CNS and play a pathogenic role during autoimmune demyelinating disease. Blood 113, 3190–3197 (2009).
Mildner, A. et al. CCR2+Ly-6Chi monocytes are crucial for the effector phase of autoimmunity in the central nervous system. Brain 132, 2487–2500 (2009).
Amorim, A. et al. IFNγ and GM-CSF control complementary differentiation programs in the monocyte-to-phagocyte transition during neuroinflammation. Nat. Immunol. 23, 217–228 (2022).
Rua, R. et al. Infection drives meningeal engraftment by inflammatory monocytes that impairs CNS immunity. Nat. Immunol. 20, 407–419 (2019).
Hawkes, C. A. & McLaurin, J. Selective targeting of perivascular macrophages for clearance of β-amyloid in cerebral amyloid angiopathy. Proc. Natl Acad. Sci. USA 106, 1261–1266 (2009).
El Khoury, J. et al. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat. Med. 13, 432–438 (2007).
Michaud, J.-P., Bellavance, M.-A., Préfontaine, P. & Rivest, S. Real-time in vivo imaging reveals the ability of monocytes to clear vascular amyloid beta. Cell Rep. 5, 646–653 (2013).
Cui, J. et al. Inflammation of the embryonic choroid plexus barrier following maternal immune activation. Dev. Cell 55, 617–628 (2020).
Waisman, A., Ginhoux, F., Greter, M. & Bruttger, J. Homeostasis of microglia in the adult brain: review of novel microglia depletion systems. Trends Immunol. 36, 625–636 (2015).
Parkhurst, C. N. et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155, 1596–1609 (2013).
Heppner, F. L. et al. Experimental autoimmune encephalomyelitis repressed by microglial paralysis. Nat. Med. 11, 146–152 (2005).
Rooijen, N. V. & Sanders, A. Liposome mediated depletion of macrophages: mechanism of action, preparation of liposomes and applications. J. Immunol. Methods 174, 83–93 (1994).
Elmore, M. R. P. et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82, 380–397 (2014).
Dagher, N. N. et al. Colony-stimulating factor 1 receptor inhibition prevents microglial plaque association and improves cognition in 3xTg-AD mice. J. Neuroinflammation 12, 139 (2015).
Huang, Y. et al. Repopulated microglia are solely derived from the proliferation of residual microglia after acute depletion. Nat. Neurosci. 21, 530–540 (2018).
Zhan, L. et al. Proximal recolonization by self-renewing microglia re-establishes microglial homeostasis in the adult mouse brain. PLoS Biol. 17, e3000134 (2019).
Hagemeyer, N. et al. Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol. 134, 441–458 (2017).
Bruttger, J. et al. Genetic cell ablation reveals clusters of local self-renewing microglia in the mammalian central nervous system. Immunity 43, 92–106 (2015).
Hohsfield, L. A. et al. Effects of long-term and brain-wide colonization of peripheral bone marrow-derived myeloid cells in the CNS. J. Neuroinflammation 17, 279 (2020).
Shibuya, Y. et al. Treatment of a genetic brain disease by CNS-wide microglia replacement. Sci. Transl. Med. 14, eabl9945 (2022). In a proof-of-concept approach, this study provides evidence that microglia can be fully replaced by circulating progenitors through a combination of bone marrow transfer and depletion and to treat a detrimental brain disease in mice.
Xu, Z. et al. Efficient strategies for microglia replacement in the central nervous system. Cell Rep. 32, 108041 (2020).
Rubino, S. J. et al. Acute microglia ablation induces neurodegeneration in the somatosensory system. Nat. Commun. 9, 4578 (2018).
Peng, J., Zou, Q., Chen, M.-J., Ma, C.-L. & Li, B.-M. Motor deficits seen in microglial ablation mice could be due to non-specific damage from high dose diphtheria toxin treatment. Nat. Commun. 13, 3874 (2022).
Willis, E. F. et al. Repopulating microglia promote brain repair in an IL-6-dependent manner. Cell 180, 833–846 (2020).
Chokr, S. M., Milinkeviciute, G., Jimenez, G. A., Abubakr, H. & Cramer, K. S. Long-term microglia depletion impairs synapse elimination and auditory brainstem function. Sci. Rep. 12, 18521 (2022).
Acknowledgements
T.M. was supported by the AMED (JP20gm6310016 (PRIME), JP21wm0425001) and the JSPS (KAKENHI JP21H02752, JP21H00204, JP22H05062). M.P. is supported by the Sobek Foundation, the Ernst Jung Foundation, the German Research Foundation (SFB 992, SFB 1160, SFB 1479, TRR 167, TRR 359, Reinhart Koselleck grant, Gottfried Wilhelm Leibniz prize) and the Ministry of Science, Research and Arts, Baden-Wuerttemberg (Sonderlinie ‘Neuroinflammation’) and by the BMBF-funded competence network of multiple sclerosis (KKNMS). This study was supported by the German Research Foundation (DFG) under Germany’s Excellence Strategy (CIBSS, EXC-2189, project ID 390939984).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Mariko Bennett, Marco Colonna and Mikael Simons for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Amann, L., Masuda, T. & Prinz, M. Mechanisms of myeloid cell entry to the healthy and diseased central nervous system. Nat Immunol 24, 393–407 (2023). https://doi.org/10.1038/s41590-022-01415-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-022-01415-8
This article is cited by
-
Microbiota–gut–brain axis and its therapeutic applications in neurodegenerative diseases
Signal Transduction and Targeted Therapy (2024)
-
Endothelial cells and macrophages as allies in the healthy and diseased brain
Acta Neuropathologica (2024)
-
The niche matters: origin, function and fate of CNS-associated macrophages during health and disease
Acta Neuropathologica (2024)
-
Prenatal Programming of Monocyte Chemotactic Protein-1 Signaling in Autism Susceptibility
Molecular Neurobiology (2024)
-
Border-associated macrophages promote cerebral amyloid angiopathy and cognitive impairment through vascular oxidative stress
Molecular Neurodegeneration (2023)