Abstract
Primary aldosteronism, a common cause of severe hypertension1, features constitutive production of the adrenal steroid aldosterone. We analyzed a multiplex family with familial hyperaldosteronism type II (FH-II)2 and 80 additional probands with unsolved early-onset primary aldosteronism. Eight probands had novel heterozygous variants in CLCN2, including two de novo mutations and four independent occurrences of a mutation encoding an identical p.Arg172Gln substitution; all relatives with early-onset primary aldosteronism carried the CLCN2 variant found in the proband. CLCN2 encodes a voltage-gated chloride channel expressed in adrenal glomerulosa that opens at hyperpolarized membrane potentials. Channel opening depolarizes glomerulosa cells and induces expression of aldosterone synthase, the rate-limiting enzyme for aldosterone biosynthesis. Mutant channels show gain of function, with higher open probabilities at the glomerulosa resting potential. These findings for the first time demonstrate a role of anion channels in glomerulosa membrane potential determination, aldosterone production and hypertension. They establish the cause of a substantial fraction of early-onset primary aldosteronism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Monticone, S. et al. Prevalence and clinical manifestations of primary aldosteronism encountered in primary care practice. J. Am. Coll. Cardiol. 69, 1811–1820 (2017).
Stowasser, M. et al. Familial hyperaldosteronism type II: five families with a new variety of primary aldosteronism. Clin. Exp. Pharmacol. Physiol. 19, 319–322 (1992).
NCD Risk Factor Collaboration (NCD-RisC). Worldwide trends in blood pressure from 1975 to 2015: a pooled analysis of 1479 population-based measurement studies with 19.1 million participants. Lancet 389, 37–55 (2017).
Lim, S. S. et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2224–2260 (2012).
Funder, J. W. et al. The management of primary aldosteronism: case detection, diagnosis, and treatment: an Endocrine Society Clinical Practice Guideline. J. Clin. Endocrinol. Metab. 101, 1889–1916 (2016).
Choi, M. et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science 331, 768–772 (2011).
Scholl, U. I. et al. Somatic and germline CACNA1D calcium channel mutations in aldosterone-producing adenomas and primary aldosteronism. Nat. Genet. 45, 1050–1054 (2013).
Azizan, E. A. et al. Somatic mutations in ATP1A1 and CACNA1D underlie a common subtype of adrenal hypertension. Nat. Genet. 45, 1055–1060 (2013).
Beuschlein, F. et al. Somatic mutations in ATP1A1 and ATP2B3 lead to aldosterone-producing adenomas and secondary hypertension. Nat. Genet. 45, 440–444 (2013).
Fernandes-Rosa, F. L. et al. Genetic spectrum and clinical correlates of somatic mutations in aldosterone-producing adenoma. Hypertension 64, 354–361 (2014).
Lifton, R. P. et al. A chimaeric 11β-hydroxylase/aldosterone synthase gene causes glucocorticoid-remediable aldosteronism and human hypertension. Nature 355, 262–265 (1992).
Scholl, U. I. et al. Hypertension with or without adrenal hyperplasia due to different inherited mutations in the potassium channel KCNJ5. Proc. Natl. Acad. Sci. USA 109, 2533–2538 (2012).
Scholl, U. I. et al. Recurrent gain of function mutation in calcium channel CACNA1H causes early-onset hypertension with primary aldosteronism. eLife 4, e06315 (2015).
Korah, H. E. & Scholl, U. I. An update on familial hyperaldosteronism. Horm. Metab. Res. 47, 941–946 (2015).
Spät, A. & Hunyady, L. Control of aldosterone secretion: a model for convergence in cellular signaling pathways. Physiol. Rev. 84, 489–539 (2004).
Carss, K. J., Stowasser, M., Gordon, R. D. & O’Shaughnessy, K. M. Further study of chromosome 7p22 to identify the molecular basis of familial hyperaldosteronism type II. J. Hum. Hypertens. 25, 560–564 (2011).
Dong, C. et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 24, 2125–2137 (2015).
Genin, E., Tullio-Pelet, A., Begeot, F., Lyonnet, S. & Abel, L. Estimating the age of rare disease mutations: the example of Triple-A syndrome. J. Med. Genet. 41, 445–449 (2004).
Dogan, R. I., Getoor, L., Wilbur, W. J. & Mount, S. M. SplicePort—an interactive splice-site analysis tool. Nucleic Acids Res. 35, W285–W291 (2007).
Thiemann, A., Gründer, S., Pusch, M. & Jentsch, T. J. A chloride channel widely expressed in epithelial and non-epithelial cells. Nature 356, 57–60 (1992).
Untiet, V. et al. Glutamate transporter–associated anion channels adjust intracellular chloride concentrations during glial maturation. Glia 65, 388–400 (2017).
Hu, C., Rusin, C. G., Tan, Z., Guagliardo, N. A. & Barrett, P. Q. Zona glomerulosa cells of the mouse adrenal cortex are intrinsic electrical oscillators. J. Clin. Invest. 122, 2046–2053 (2012).
Jeworutzki, E. et al. GlialCAM, a protein defective in a leukodystrophy, serves as a ClC-2 Cl– channel auxiliary subunit. Neuron 73, 951–961 (2012).
Stölting, G. et al. Regulation of ClC-2 gating by intracellular ATP. Pflugers Arch. 465, 1423–1437 (2013).
Niemeyer, M. I., Cid, L. P., Zúñiga, L., Catalán, M. & Sepúlveda, F. V. A conserved pore-lining glutamate as a voltage- and chloride-dependent gate in the ClC-2 chloride channel. J. Physiol. 553, 873–879 (2003).
Jentsch, T. J. Discovery of CLC transport proteins: cloning, structure, function and pathophysiology. J. Physiol. 593, 4091–4109 (2015).
Rainey, W. E., Bird, I. M. & Mason, J. I. The NCI-H295 cell line: a pluripotent model for human adrenocortical studies. Mol. Cell. Endocrinol. 100, 45–50 (1994).
Bassett, M. H., Suzuki, T., Sasano, H., White, P. C. & Rainey, W. E. The orphan nuclear receptors NURR1 and NGFIB regulate adrenal aldosterone production. Mol. Endocrinol. 18, 279–290 (2004).
Romero, D. G. et al. Regulators of G-protein signaling 4 in adrenal gland: localization, regulation, and role in aldosterone secretion. J. Endocrinol. 194, 429–440 (2007).
Garcia-Olivares, J. et al. Gating of human ClC-2 chloride channels and regulation by carboxy-terminal domains. J. Physiol. 586, 5325–5336 (2008).
Tauber, P. et al. Cellular pathophysiology of an adrenal adenoma–associated mutant of the plasma membrane Ca2+-ATPase ATP2B3. Endocrinology 157, 2489–2499 (2016).
Lafferty, A. R. et al. A novel genetic locus for low renin hypertension: familial hyperaldosteronism type II maps to chromosome 7 (7p22). J. Med. Genet. 37, 831–835 (2000).
Depienne, C. et al. Brain white matter oedema due to ClC-2 chloride channel deficiency: an observational analytical study. Lancet Neurol. 12, 659–668 (2013).
Blanz, J. et al. Leukoencephalopathy upon disruption of the chloride channel ClC-2. J. Neurosci. 27, 6581–6589 (2007).
Stowasser, M. et al. Clinical, biochemical and genetic approaches to the detection of familial hyperaldosteronism type I. J. Hypertens. 13, 1610–1613 (1995).
Chorvátová, A., Gendron, L., Bilodeau, L., Gallo-Payet, N. & Payet, M. D. A Ras-dependent chloride current activated by adrenocorticotropin in rat adrenal zona glomerulosa cells. Endocrinology 141, 684–692 (2000).
Torpy, D. J. et al. Familial hyperaldosteronism type II: description of a large kindred and exclusion of the aldosterone synthase (CYP11B2) gene. J. Clin. Endocrinol. Metab. 83, 3214–3218 (1998).
Krumm, N. et al. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 47, 582–588 (2015).
Verkman, A. S. Development and biological applications of chloride-sensitive fluorescent indicators. Am. J. Physiol. 259, C375–C388 (1990).
Kaneko, H., Putzier, I., Frings, S., Kaupp, U. B. & Gensch, T. Chloride accumulation in mammalian olfactory sensory neurons. J. Neurosci. 24, 7931–7938 (2004).
Bevensee, M. O., Apkon, M. & Boron, W. F. Intracellular pH regulation in cultured astrocytes from rat hippocampus. II. Electrogenic Na/HCO3 cotransport. J. Gen. Physiol. 110, 467–483 (1997).
Chao, A. C., Dix, J. A., Sellers, M. C. & Verkman, A. S. Fluorescence measurement of chloride transport in monolayer cultured cells. Mechanisms of chloride transport in fibroblasts. Biophys. J. 56, 1071–1081 (1989).
Accardi, A. & Pusch, M. Fast and slow gating relaxations in the muscle chloride channel CLC-1. J. Gen. Physiol. 116, 433–444 (2000).
de Santiago, J. A., Nehrke, K. & Arreola, J. Quantitative analysis of the voltage-dependent gating of mouse parotid ClC-2 chloride channel. J. Gen. Physiol. 126, 591–603 (2005).
Rhee, J. S., Ebihara, S. & Akaike, N. Gramicidin perforated patch-clamp technique reveals glycine-gated outward chloride current in dissociated nucleus solitarii neurons of the rat. J. Neurophysiol. 72, 1103–1108 (1994).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Trapnell, C. et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013).
Samocha, K. E. et al. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 46, 944–950 (2014).
Zou, J. et al. Quantifying unobserved protein-coding variants in human populations provides a roadmap for large-scale sequencing projects. Nat. Commun. 7, 13293 (2016).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Acknowledgements
We thank our patients and their families for their invaluable contributions, the Yale Center for Genome Analysis for next-generation sequencing, the Center for Advanced Imaging (CAi) at Heinrich Heine University for providing a confocal microscope, S. Weidtkamp-Peters and S. Hänsch for technical assistance, J. Zhang for helpful discussions, E. Seidel, N. Erlenhardt and N. Klöcker for providing immunoprecipitation protocols and helpful discussions, C. Gomez-Sanchez (University of Mississippi) for providing plasmids, W. Rainey (University of Michigan) for providing HAC15 cells and M. Haase (Heinrich Heine University Düsseldorf) for providing H295R cells. Computational support and infrastructure were in part provided by the Centre for Information and Media Technology (Düsseldorf). This study was supported in part by the Ministerium für Kultur und Wissenschaft des Landes Nordrhein-Westfalen (Rückkehrprogramm and Junges Kolleg) and the Deutsche Forschungsgemeinschaft (SCHO 1386/2-1) (all to U.I.S.) and the NIH Center for Mendelian Genomics (5U54HG006504), NIH P01DK17433 and the Howard Hughes Medical Institute (all to R.P.L.).
Author information
Authors and Affiliations
Contributions
U.I.S., G.S., C.F. and R.P.L. designed the study. M.S. and R.G. recruited and characterized family 3. A.M.O., C.G., D.M., R.W.L., D.P.J., G.C., P.G., E.L., C.N.-W. and R.P.L. ascertained and recruited probands with early-onset primary aldosteronism. A.A.V., E.L. and U.I.S. recruited additional members of selected families. C.N.-W., S.X. and A.W. prepared DNA samples. U.I.S., A.A.V., S.C.J., T.Y., M.C. and R.P.L. analyzed exome sequencing results. U.I.S. identified the disease-associated gene. C.N.-W., A.T. and A.A.V. performed and analyzed the results of targeted DNA sequencing. J.S. performed immunohistochemistry, immunoprecipitation and real-time PCR. J.S. and A.T. made constructs and generated stable cell lines. J.S. and J.C. prepared samples for and analyzed the results of RNA sequencing. J.S. and U.I.S. performed and analyzed the results of splicing assays and confocal microscopy. G.S., H.T., V.U. and C.F. performed and analyzed the results of FLIM and electrophysiology. M.K. and P.M. performed and analyzed the results of mass spectrometry. L.C.R. read and revised the manuscript. U.I.S. wrote the initial draft of the manuscript, with contributions and/or revisions from all authors.
Corresponding author
Ethics declarations
Competing interests
Heinrich Heine University Düsseldorf has filed a patent application: EP17209972, Diagnosis and Therapy of Primary Aldosteronism.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
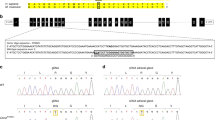
Supplementary Figure 1 Sanger sequences.
a, Sanger sequences of kindreds with CLCN2 variants. M/+ denotes the indicated novel CLCN2 variant in the heterozygous state, and +/+ denotes homozygous wild-type sequence. Mutant bases are indicated by a red frame, and encoded amino acid sequences are shown above. b, Splicing assay for the p.Lys362del mutation. Exons 9–11 of CLCN2 were cloned, and HEK293T cells were transfected with WT DNA and DNA carrying the variant found in kindred 1492. RNA was isolated and cDNA was transcribed. Shown are Sanger sequences of PCRs covering the exon 10/11 splice site affected by the variant. A new donor site on exon 10 is used, resulting in deletion of the last 3 bp of exon 10 (red rectangle in WT sequence, red line in mutant sequence). These results were consistent with in silico prediction.
Supplementary Figure 2 Immunohistochemistry.
a, Immunohistochemistry as in Fig. 2 was performed on the adrenal gland of a second human subject (one of two technical replicates shown). Peptide control, antibody was preincubated with immunogenic peptide. Scale bar, 100 µm. C, capsule; G, glomerulosa; F, fasciculata. b, Immunofluorescent staining of mouse adrenal gland, with DAB2 as a marker of the zona glomerulosa. Scale bar, 10 µm.
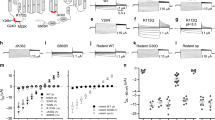
Supplementary Figure 3 Gating analysis, time constants of activation/deactivation and current density–voltage plots of WT and mutant ClC-2 channels.
a, Whole-cell patch–clamp recordings of representative ClC-2MUT and voltage protocol (150 mM Cl– outside, 75 mM Cl– inside; see the Methods for solutions) for the indicated mutations are shown. b, Time constants for WT and mutant ClC-2 channels (see the Methods for details). Mean values ± 95% confidence intervals are shown (WT, p.Arg172Gln and p.Ser865Arg are reproduced from Fig. 3; WT, n = 11; p.Met22Lys, n = 12; p.Tyr26Asn, n = 13; p.Arg172Gln, n = 13; p.Lys362del, n = 12; p.Ser865Arg, n = 11). c, Plot of steady-state current densities. Values are shown as means ± 95% confidence intervals; WT, n = 11; p.Met22Lys, n = 12; p.Tyr26Asn, n = 13; p.Arg172Gln, n = 13; p.Lys362del, n = 12; p.Ser865Arg, n = 11. Median values at –80 mV and the results of Kruskal–Wallis one-way ANOVA (H = 14.823, d.f. = 5) followed by Dunn’s method when appropriate are as follows: WT: –5.75; p.Met22Lys: –10.28, n.s. vs. WT; p.Tyr26Asn: –13.66, P < 0.05 vs. WT; p.Arg172Gln: –9.13, n.s. vs. WT; p.Lys362del: –13.69, P < 0.05 vs. WT; p.Ser865Arg, –12.58, P < 0.05 vs. WT. d–f, Total (d), protopore (e) and common gate (f) open probabilities were determined as described (see the Methods for details; WT, p.Arg172Gln and p.Ser865Arg total and common open probabilities are reproduced from Fig. 3; WT, n = 11; p.Met22Lys, n = 12; p.Tyr26Asn, n = 13; p.Arg172Gln, n =13; p.Lys362del, n = 12; p.Ser865Arg, n = 12). Data points were fit using a Boltzmann function (bold line), and 95% confidence intervals were determined using bootstrap sampling. The overall shift in activation of mutant ClC-2 channels apart from p.Ser865Arg mostly results from a higher common gate open probability, whereas the fast gate open probability of p.Ser865Arg is shifted to more positive potentials. g, Individual data and stable cell line clones used in the analysis shown in Fig. 3 and this figure.
Supplementary Figure 4 Mass spectrometry and confocal microscopy.
a,b, LC–MS/MS-based identification of the phosphorylation site of ClC-2 at Ser865. ClC-2 was identified with 43 peptides and sequence coverage of 52.8% Shown are annotated MS/MS spectra of the unmodified (a) and phosphorylated (b) peptide DSATSSSDTETTEVHALWGPHSR corresponding to amino acids 859–881 of ClC-2. The x axis shows m/z, the left y axis shows relative abundance and the right y axis shows absolute signal intensity. Unmodified peptide: 16 MS/MS counts, posterior error probability (PEP) 1.21 × 10–35, score 160.47; Ser865-phosphorylated peptide: 11 MS/MS counts, PEP 1.61 × 10–36, score 166.49. The phosphorylation site at Ser865 was fully localized, with a localization probability of 0.813811 (mass error of 0.28027 p.p.m.). One of two independent preparations (Methods) is shown; similar results were obtained for another two independent preparations using a different solvent. c,d, Live cell confocal microscopy of YFP-tagged WT and mutant ClC-2 and the surface membrane marker CFP-mem in H295R cells. c, The respective variant is noted on the left. Left, confocal image using the YFP channel; middle, CFP channel; right, overlay. Scale bars, 20 µm. Two independent clones of each plasmid were assessed, and representative images are shown. d, Correlation R between YFP and CFP fluorescence for each construct. WT, 0.29 ± 0.03 (n = 71); p.Met22Lys, 0.15 ± 0.02 (n = 81; P = 0.0001 vs. WT); p.Tyr26Asn, 0.25 ± 0.03 (n = 53; P > 0.9999 vs. WT); p.Arg172Gln, 0.24 ± 0.03 (n = 56; P = 0.5862 vs. WT); p.Lys362del, 0.29 ± 0.03 (n = 65; P > 0.9999 vs. WT); p.Ser865Arg, 0.30 ± 0.02 (n = 59; P > 0.9999 vs. WT); all mean ± s.e.m., Kruskal–Wallis test; Dunn’s multiple-comparisons test; Kruskal–Wallis statistic 28.47, d.f. = 5. Box, interquartile range; whiskers, 1.5 times the interquartile range; line, median; ***P < 0.001; n.s., not significant.
Supplementary Figure 5 CYP11B2 expression in H295R cells after transfection of non-functional CLCN2.
Relative expression levels of CYP11B2 determined by real-time PCR in H295R cells transfected with empty vector control, wild-type CLCN2 (blue) or two constructs with C-terminal deletions that affect ion channel function (yellow, corresponding deletions noted below the graph)30. Values were normalized to empty vector control. CYP11B2 expression is significantly lower after transfection of non-functional channels than after transfection of the wild-type channel (mean ± s.e.m.; WT, 16.35 ± 2.72; p.Arg751Ter, 1.46 ± 0.18, P = 0.0089 vs. WT; p.His573_Leu636del, 2.38 ± 0.50, P = 0.0077 vs. WT; **P < 0.01). n = 5, one-way ANOVA for all constructs, Dunnett’s multiple-comparisons test, F = 31.52, d.f. = 4. Box, interquartile range; whiskers, 1.5 times the interquartile range; line, median.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1–7 and 10, and Supplementary Note
Supplementary Table 8
Details of statistical analysis shown in Figs. 3 and 4, and Supplementary Fig. 3
Supplementary Table 9
Whole-transcriptome analysis of CLCN2-transfected H295R cells
Rights and permissions
About this article
Cite this article
Scholl, U.I., Stölting, G., Schewe, J. et al. CLCN2 chloride channel mutations in familial hyperaldosteronism type II. Nat Genet 50, 349–354 (2018). https://doi.org/10.1038/s41588-018-0048-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0048-5
This article is cited by
-
Molecular pathology of endocrine gland tumors: genetic alterations and clinicopathologic relevance
Virchows Archiv (2024)
-
Update in genetic and epigenetic causes of hypertension
Cellular and Molecular Life Sciences (2024)
-
Primary aldosteronism: molecular medicine meets public health
Nature Reviews Nephrology (2023)
-
Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42
Nature Communications (2023)
-
Biallelic CLCN2 mutations cause retinal degeneration by impairing retinal pigment epithelium phagocytosis and chloride channel function
Human Genetics (2023)