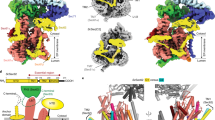
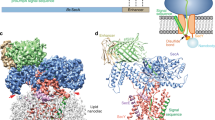
Abstract
Many proteins must translocate through the protein-conducting Sec61 channel in the eukaryotic endoplasmic reticulum membrane or the SecY channel in the prokaryotic plasma membrane1,2. Proteins with highly hydrophobic signal sequences are first recognized by the signal recognition particle (SRP)3,4 and then moved co-translationally through the Sec61 or SecY channel by the associated translating ribosome. Substrates with less hydrophobic signal sequences bypass the SRP and are moved through the channel post-translationally5,6. In eukaryotic cells, post-translational translocation is mediated by the association of the Sec61 channel with another membrane protein complex, the Sec62–Sec63 complex7,8,9, and substrates are moved through the channel by the luminal BiP ATPase9. How the Sec62–Sec63 complex activates the Sec61 channel for post-translational translocation is not known. Here we report the electron cryo-microscopy structure of the Sec complex from Saccharomyces cerevisiae, consisting of the Sec61 channel and the Sec62, Sec63, Sec71 and Sec72 proteins. Sec63 causes wide opening of the lateral gate of the Sec61 channel, priming it for the passage of low-hydrophobicity signal sequences into the lipid phase, without displacing the channel’s plug domain. Lateral channel opening is triggered by Sec63 interacting both with cytosolic loops in the C-terminal half of Sec61 and transmembrane segments in the N-terminal half of the Sec61 channel. The cytosolic Brl domain of Sec63 blocks ribosome binding to the channel and recruits Sec71 and Sec72, positioning them for the capture of polypeptides associated with cytosolic Hsp7010. Our structure shows how the Sec61 channel is activated for post-translational protein translocation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The coordinates of atomic models of the Sec complex without Sec62 were deposited in the Protein Data Bank with accession code 6ND1. The cryo-EM maps of the Sec complex before and after focused refinement with a mask were deposited with accession code EMDB-0440. All other data are available on reasonable request from the corresponding author.
References
Voorhees, R. M. & Hegde, R. S. Toward a structural understanding of co-translational protein translocation. Curr. Opin. Cell Biol. 41, 91–99 (2016).
Rapoport, T. A., Li, L. & Park, E. Structural and mechanistic insights into protein translocation. Annu. Rev. Cell Dev. Biol. 33, 369–390 (2017).
Saraogi, I. & Shan, S.-O. Co-translational protein targeting to the bacterial membrane. Biochim. Biophys. Acta 1843, 1433–1441 (2014).
Wild, K., Halic, M., Sinning, I. & Beckmann, R. SRP meets the ribosome. Nat. Struct. Mol. Biol. 11, 1049–1053 (2004).
Ng, D. T., Brown, J. D. & Walter, P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 134, 269–278 (1996).
Ast, T., Cohen, G. & Schuldiner, M. A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell 152, 1134–1145 (2013).
Deshaies, R. J., Sanders, S. L., Feldheim, D. A. & Schekman, R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature 349, 806–808 (1991).
Panzner, S., Dreier, L., Hartmann, E., Kostka, S. & Rapoport, T. A. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 81, 561–570 (1995).
Matlack, K. E., Misselwitz, B., Plath, K. & Rapoport, T. A. BiP acts as a molecular ratchet during posttranslational transport of prepro-α factor across the ER membrane. Cell 97, 553–564 (1999).
Deshaies, R. J., Koch, B. D., Werner, W. M., Craig, E. A. & Schekman, R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 332, 800–805 (1988).
van den Berg, B. et al. X-ray structure of a protein-conducting channel. Nature 427, 36–44 (2004).
Zimmer, J., Nam, Y. & Rapoport, T. A. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455, 936–943 (2008).
Li, L. et al. Crystal structure of a substrate-engaged SecY protein-translocation channel. Nature 531, 395–399 (2016).
Egea, P. F. & Stroud, R. M. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc. Natl Acad. Sci. USA 107, 17182–17187 (2010).
Voorhees, R. M., Fernandez, I. S., Scheres, S. H. & Hegde, R. S. Structure of the mammalian ribosome–Sec61 complex to 3.4 Å resolution. Cell 157, 1632–1643 (2014).
Voorhees, R. M. & Hegde, R. S. Structure of the Sec61 channel opened by a signal sequence. Science 351, 88–91 (2016).
Deshaies, R. J. & Schekman, R. Structural and functional dissection of Sec62P, a membrane-bound component of the yeast endoplasmic reticulum protein import machinery. Mol. Cell. Biol. 10, 6024–6035 (1990).
Feldheim, D., Rothblatt, J. & Schekman, R. Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol. Cell. Biol. 12, 3288–3296 (1992).
Feldheim, D., Yoshimura, K., Admon, A. & Schekman, R. Structural and functional characterization of Sec66p—a new subunit of the polypeptide translocation apparatus in the yeast endoplasmic reticulum. Mol. Biol. Cell 4, 931–939 (1993).
Tripathi, A., Mandon, E. C., Gilmore, R. & Rapoport, T. A. Two alternative binding mechanisms connect the protein translocation Sec71/Sec72 complex with heat shock proteins. J. Biol. Chem. 292, 8007–8018 (2017).
Linxweiler, M., Schick, B. & Zimmermann, R. Let’s talk about Secs: Sec61, Sec62 and Sec63 in signal transduction, oncology and personalized medicine. Signal Transduct. Target. Ther. 2, 17002 (2017).
Trueman, S. F., Mandon, E. C. & Gilmore, R. A gating motif in the translocation channel sets the hydrophobicity threshold for signal sequence function. J. Cell Biol. 199, 907–918 (2012).
Ponting, C. P. Proteins of the endoplasmic-reticulum-associated degradation pathway: domain detection and function prediction. Biochem. J. 351, 527–535 (2000).
Jan, C. H., Williams, C. C. & Weissman, J. S. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 346, 1257521 (2014).
Costa, E. A., Subramanian, K., Nunnari, J. & Weissman, J. S. Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science 359, 689–692 (2018).
Wittke, S., Dünnwald, M. & Johnsson, N. Sec62p, a component of the endoplasmic reticulum protein translocation machinery, contains multiple binding sites for the Sec-complex. Mol. Biol. Cell 11, 3859–3871 (2000).
Plath, K., Wilkinson, B. M., Stirling, C. J. & Rapoport, T. A. Interactions between Sec complex and prepro-α-factor during posttranslational protein transport into the endoplasmic reticulum. Mol. Biol. Cell 15, 1–10 (2004).
Plath, K. & Rapoport, T. A. Spontaneous release of cytosolic proteins from posttranslational substrates before their transport into the endoplasmic reticulum. J. Cell Biol. 151, 167–178 (2000).
Shao, S. & Hegde, R. S. A calmodulin-dependent translocation pathway for small secretory proteins. Cell 147, 1576–1588 (2011).
Müller, G. & Zimmermann, R. Import of honeybee prepromelittin into the endoplasmic reticulum: energy requirements for membrane insertion. EMBO J. 7, 639–648 (1988).
Zheng, S., Palovcak, E., Armache, J.-P., Cheng, Y. & Agard, D. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Morin, A. et al. Collaboration gets the most out of software. eLife 2, e01456 (2013).
Källberg, M. et al. Template-based protein structure modeling using the RaptorX web server. Nat. Protocols 7, 1511–1522 (2012).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Acknowledgements
We thank Z. Yu, R. Huang, and H.-T. Chou at the HHMI Janelia Cryo-EM Facility for help with microscope operation and data collection, E. Twomey for help with data analysis, R. Gilmore for advice and materials, and S. Shao and M. Catipovic for comments on the manuscript. This work was supported by a Jane Coffin Child fellowship to X.W., and an NIGMS award (R01GM052586) to T.A.R. T.A.R. is a Howard Hughes Medical Institute Investigator.
Reviewer information
Nature thanks G. von Heijne and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
X.W. designed experiments, purified proteins, collected and analysed electron microscopy data, and built the models. C.C. purified proteins, performed mutagenesis and mutant analysis. T.A.R. supervised the project. T.A.R. and X.W. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Cryo-EM analysis of the Sec complex.
a, The image shows a representative cryo-EM image of Sec-complex particles. Some particles are highlighted with green circles. Four electron microscopy grids were screened and had a similar particle distribution. b, Representative 2D class averages of picked particles collected from images of three grids. c, Image processing workflow for 3D classification and refinement. Shown are views of 3D reconstructions parallel to the membrane, with percentages of the particles in each class indicated. Classes in colour were used for subsequent analysis. d, Euler-angle distribution in two different views. e, Fourier shell correlation (FSC) curve with indicated resolution at FSC = 0.143. f, Local resolution was calculated from the unfiltered half-map and coloured according to the scale on the side. Two different views are shown. g, Map filtered to local resolution using cryoSPARC2. Regions corresponding to the different proteins are coloured as in Fig. 1b.
Extended Data Fig. 2 Examples of the fit of the models into the density map.
a, Density map and model for the indicated segments of Sec63. Interactions 1 and 2 show the interfaces between Sec63 and components of the Sec61 complex. Residues in salmon and pink belong to Sec61 and Sss1, respectively. b, As in a, but for segments of the Sec61 complex. c, As in a, but for the loop of Sec71 interacting with Sec63 and Sec61.
Extended Data Fig. 3 Localization of Sec62 in the Sec complex.
a, Shown is the unsharpened map (grey), low-pass filtered to 8 Å, with models for the components of the Sec complex in ribbon diagram. A homology model for the N-terminal DEP domain of Sec62 (purple) was docked into the map. The acidic C-terminal tail of Sec63 (dotted line on the right panel) wraps around the TM8–TM9 loop of Sec61 and interacts with the cytosolic DEP domain of Sec62. b, Cuts through the middle of the unsharpened map are shown in views from the side and from the cytosol. Density for a transmembrane segment of Sec62 (highlighted by red, dotted ovals) is close to the lateral gate.
Extended Data Fig. 4 The Sec61 channel in the Sec complex has open lateral and closed luminal gates.
Space-filling model of the Sec61 channel in the Sec complex. The left panel shows a side view with the lateral gate in the front. The right panel shows a cut through the space-filling model viewed from the cytosol. Note that the plug domain keeps the luminal gate closed.
Extended Data Fig. 5 Lateral gate changes of the Sec61 channel.
a, The structure of Sec61 in the ribosome-primed state (PDB code 3J7Q) is superimposed on that of the idle channel from M. jannaschii (PDB code 1RH5). Shown are the transmembrane segments as cylinders, viewed from the side (left) and from the cytosol (right). Movements of transmembrane segments are indicated by red arrows. The plug domains are indicated by stars in the right panel. b, As in a, but comparison of the Sec61 channel in the Sec complex with the SecY channel from P. furiosus (PDB code 3MP7). c, The Sec61 channel in the Sec complex is shown together with the signal sequence (SS; in cyan) from a structure of the signal-sequence-opened ribosome-bound Sec61 channel (PDB code 3JC2). The alignment of the Sec61 molecules was done as in Fig. 2c. Note that the lateral gate in the Sec complex is open enough to allow a helix to move through.
Extended Data Fig. 6 Alignment of Sec63 sequences from different species.
Transmembrane segments and the Brl and J domains are shown as black bars above the sequences. The β-sandwich and lasso regions in the Brl domain are indicated as blue lines. Conserved residues are highlighted. Mutated residues involved in the interaction with the Sec61 channel are shown as coloured circles and arrows: interaction 3 at the N terminus in blue; interaction 2 in TM3 in red; interaction 1 in the β-sandwich and lasso segments of the Brl domain in green.
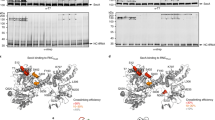
Extended Data Fig. 7 Sec63 expression in wild-type and mutant S. cerevisiae cells.
The strains used in Fig. 3c were analysed for expression of Flag-tagged Sec63. Flag-tagged wild-type or mutant Sec63 was expressed in Sec63Δ S. cerevisiae cells. Equal numbers of cells were analysed by SDS–PAGE, followed by immunoblotting with Flag antibodies. To control for equal loading, the samples were also analysed with antibodies to phosphoglycerate kinase (PGK). The lanes correspond to expression of the following Sec63 proteins: (1) wild-type Sec63; (2) mutations in interface 3 of Sec63 (Y5A/Y7A/D8A); (3) mutations in interface 2 (Y227A/W242A); (4) mutations in interface 2 (Y227A/W242A/W243A); (5) mutations in interface 1 (T444K/S447K/E482A); (6) empty vector. The experiment was repeated twice.
Supplementary information
Rights and permissions
About this article
Cite this article
Wu, X., Cabanos, C. & Rapoport, T.A. Structure of the post-translational protein translocation machinery of the ER membrane. Nature 566, 136–139 (2019). https://doi.org/10.1038/s41586-018-0856-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0856-x
This article is cited by
-
LncRNA WFDC21P interacts with SEC63 to promote gastric cancer malignant behaviors by regulating calcium homeostasis signaling pathway
Cancer Cell International (2024)
-
Metallo-supramolecular branched polymer protects particles from air-water interface in single-particle cryo-electron microscopy
Communications Biology (2024)
-
A common mechanism of Sec61 translocon inhibition by small molecules
Nature Chemical Biology (2023)
-
Signal peptide mimicry primes Sec61 for client-selective inhibition
Nature Chemical Biology (2023)
-
A peroxisomal ubiquitin ligase complex forms a retrotranslocation channel
Nature (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.