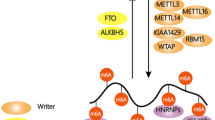
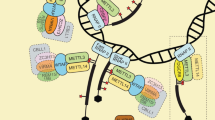
Abstract
RNA methylation to form N 6-methyladenosine (m6A) in mRNA accounts for the most abundant mRNA internal modification and has emerged as a widespread regulatory mechanism that controls gene expression in diverse physiological processes. Transcriptome-wide m6A mapping has revealed the distribution and pattern of m6A in cellular RNAs, referred to as the epitranscriptome. These maps have revealed the specific mRNAs that are regulated by m6A, providing mechanistic links connecting m6A to cellular differentiation, cancer progression and other processes. The effects of m6A on mRNA are mediated by an expanding list of m6A readers and m6A writer-complex components, as well as potential erasers that currently have unclear relevance to m6A prevalence in the transcriptome. Here we review new and emerging methods to characterize and quantify the epitranscriptome, and we discuss new concepts — in some cases, controversies — regarding our understanding of the mechanisms and functions of m6A readers, writers and erasers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
21 August 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41580-023-00654-3
References
Perry, R. P. & Kelley, D. E. Existence of methylated messenger RNA in mouse L cells. Cell 1, 37–42 (1974).
Desrosiers, R., Friderici, K. & Rottman, F. Identification of methylated nucleosides in messenger RNA from novikoff hepatoma cells. Proc. Natl Acad. Sci. USA 71, 3971–3975 (1974).
Sommer, S., Lavi, U. & Darnell, J. E. Jr. The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J. Mol. Biol. 124, 487–499 (1978).
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G. & Rottman, F. M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247 (1997).
Clancy, M. J., Shambaugh, M. E., Timpte, C. S. & Bokar, J. A. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 30, 4509–4518 (2002).
Zhong, S. et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20, 1278–1288 (2008).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012). This study and the study below (ref. 8) provided the first methods to map m 6 A throughout the transcriptome, stimulating the recent renaissance in m 6 A research.
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Schwartz, S. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 8, 284–296 (2014).
Batista, P. J. et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 (2014).
Geula, S. et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347, 1002–1006 (2015). This study documented the role of m 6 A in cellular differentiation by identifying METTL3 as a main regulator for terminating murine naïve pluripotency.
Ke, S. et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29, 2037–2053 (2015).
Ke, S. et al. m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31, 990–1006 (2017). This study demonstrated that m 6 A does not have a major effect on splicing but instead is added in the nucleus to shape the half-life of m 6 A-modified mRNAs in the cytoplasm.
McIntyre, A. B. R. et al. Limits in the detection of m6A changes using MeRIP/m6A-seq. bioXrv https://doi.org/10.1101/657130 (2019).
Yoon, K. J. et al. Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell 171, 877–889 (2017).
Grozhik, A. V. & Jaffrey, S. R. Distinguishing RNA modifications from noise in epitranscriptome maps. Nat. Chem. Biol. 14, 215–225 (2018).
Munns, T. W., Oberst, R. J., Sims, H. F. & Liszewski, M. K. Antibody-nucleic acid complexes: immunospecific recognition of 7-methylguanine- and N6-methyladenine-containing 5′-terminal oligonucleotides of mRNA. J. Biol. Chem. 254, 4327–4330 (1979).
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Meyer, K. D. & Jaffrey, S. R. Rethinking m(6)a readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342 (2017).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Huang, H. et al. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 567, 414–419 (2019).
Bertero, A. et al. The SMAD2/3 interactome reveals that TGFbeta controls m(6)A mRNA methylation in pluripotency. Nature 555, 256–259 (2018).
Barbieri, I. et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552, 126–131 (2017).
Salditt-Georgieff, M. et al. Methyl labeling of HeLa cell hnRNA: a comparison with mRNA. Cell 7, 227–237 (1976).
Darnell, R. B., Ke, S. & Darnell, J. E. Pre-mRNA processing includes N 6 methylation of adenosine residues that are retained in mRNA exons and the fallacy of ‘RNA epigenetics’. RNA 24, 262–267 (2018).
Wang, X. et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015).
Jackson, R. J., Hellen, C. U. T. & Pestova, T. V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 (2010).
Lee, A. S. Y., Kranzusch, P. J. & Cate, J. H. D. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 522, 111–114 (2015).
Meyer, K. D. et al. 5′ UTR m6A promotes cap-independent translation. Cell 163, 999–1010 (2015).
Zhou, J. et al. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594 (2015).
Tatomer, D. C. & Wilusz, J. E. An unchartered journey for ribosomes: circumnavigating circular RNAs to produce proteins. Mol. Cell 66, 1–2 (2017).
Lin, S., Choe, J., Du, P. & Triboulet, R. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345 (2016).
Choe, J. et al. mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature 561, 556–560 (2018).
Kan, L. et al. The m(6)A pathway facilitates sex determination in Drosophila. Nat. Commun. 8, 15737 (2017).
Haussmann, I. U. et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540, 301–304 (2016).
Lence, T. et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature 540, 242–247 (2016).
Liu, N. et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564 (2015).
Louloupi, A., Ntini, E., Conrad, T. & Orom, U. A. V. Transient N-6-methyladenosine transcriptome sequencing reveals a regulatory role of m6A in splicing efficiency. Cell Rep. 23, 3429–3437 (2018).
Wilkinson, F. L. et al. Emerin interacts in vitro with the splicing-associated factor, YT521-B. Eur. J. Biochem. 270, 2459–2466 (2003).
Hartmann, A. M., Nayler, O., Schwaiger, F. W., Obermeier, A. & Stamm, S. The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59fyn. Mol. Biol. Cell 10, 3909–3926 (1999).
Imai, Y., Matsuo, N., Ogawa, S., Tohyama, M. & Takagi, T. Cloning of a gene, YT521, for a novel RNA splicing-related protein induced by hypoxia/reoxygenation. Brain Res. Mol. Brain Res. 53, 33–40 (1998).
Xiao, W. et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016).
Lu, X., Ng, H.-H. & Bubulya, P. A. The role of SON in splicing, development, and disease. Wiley Interdiscip. Rev. RNA 5, 637–646 (2014).
Min, H., Chan, R. C. & Black, D. L. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 9, 2659–2671 (1995).
Ries, R. J. et al. m6A enhances the phase separation potential of mRNA. Nature 571, 424–428 (2019). Ries et al. demonstrate that all YTHDF proteins share the same ability to undergo liquid–liquid phase separation and thus to recruit m 6 A mRNAs in different cytoplasmic compartments, such as P-bodies and stress granules.
Roundtree, I. A. et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. eLife 6, e31311 (2017).
Bahar Halpern, K. et al. Nuclear retention of mRNA in mammalian tissues. Cell Rep. 13, 2653–2662 (2015).
Sledz, P. & Jinek, M. Structural insights into the molecular mechanism of the m(6)A writer complex. eLife 5, e18434 (2016).
Wang, P., Doxtader, K. A. & Nam, Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317 (2016).
Wang, X. et al. Structural basis of N(6)-adenosine methylation by the METTL3–METTL14 complex. Nature 534, 575–578 (2016). The studies by Sledz et al. (ref. 49), P. Wang et al. (ref. 50) and X. Wang et al. (ref. 51) report the crystal structure of the METTL3–METTL14 complex. These studies were essential to finally determining that Mettl3 is the only catalytically active subunit in the complex.
Ma, H. et al. N(6)-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 15, 88–94 (2019).
van Tran, N. et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz619 (2019).
Pendleton, K. E. et al. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169, 824–835 (2017).
Shima, H. et al. S-Adenosylmethionine synthesis is regulated by selective N 6-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep. 21, 3354–3363 (2017).
Warda, A. S. et al. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 18, 2004–2014 (2017).
Bokar, J. A., Rath-Shambaugh, M. E., Ludwiczak, R., Narayan, P. & Rottman, F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 269, 17697–17704 (1994).
Liu, J. et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N(6)-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Wang, Y. et al. N(6)-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198 (2014).
Iyer, L. M., Zhang, D. & Aravind, L. Adenine methylation in eukaryotes: apprehending the complex evolutionary history and functional potential of an epigenetic modification. BioEssays 38, 27–40 (2016).
Lin, Z. et al. Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis. Cell Res. 27, 1216–1230 (2017).
Sharpe, J. J. & Cooper, T. A. Unexpected consequences: exon skipping caused by CRISPR-generated mutations. Genome Biol. 18, 109 (2017).
Slobodin, B. et al. Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell 169, 326–337.e312 (2017).
Spies, N., Nielsen, C. B., Padgett, R. A. & Burge, C. B. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell 36, 245–254 (2009).
Schwartz, S., Meshorer, E. & Ast, G. Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 16, 990–995 (2009).
Tilgner, H. et al. Nucleosome positioning as a determinant of exon recognition. Nat. Struct. Mol. Biol. 16, 996–1001 (2009).
Kolasinska-Zwierz, P. et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 41, 376–381 (2009).
Le Hir, H. & Seraphin, B. EJCs at the heart of translational control. Cell 133, 213–216 (2008).
Patil, D. P. et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 (2016). This suggests that YTHDC1 may influence the function of long noncoding RNAs.
Lichinchi, G. et al. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 1, 16011 (2016).
Gokhale, N. S. et al. N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20, 654–665 (2016).
Alarcon, C. R., Lee, H., Goodarzi, H., Halberg, N. & Tavazoie, S. F. N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485 (2015).
Choe, J. et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 561, 556–560 (2018).
Stoilov, P., Rafalska, I. & Stamm, S. YTH: a new domain in nuclear proteins. Trends Biochem. Sci. 27, 495–497 (2002).
Zhu, T. et al. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 24, 1493–1496 (2014).
Li, F., Zhao, D., Wu, J. & Shi, Y. Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. 24, 1490–1492 (2014).
Wang, X. et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015).
Li, A. et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 27, 444–447 (2017).
Zhou, J. et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594 (2015).
Shi, H. et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27, 315–328 (2017).
Wojtas, M. N. et al. Regulation of m6A transcripts by the 3′→5′ RNA helicase ythdc2 is essential for a successful meiotic program in the mammalian germline. Mol. Cell 68, 374–387 (2017).
Luo, S. & Tong, L. Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl Acad. Sci. USA 111, 13834–13839 (2014).
Theler, D., Dominguez, C., Blatter, M., Boudet, J. & Allain, F. H. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 42, 13911–13919 (2014).
Xu, C. et al. Structural basis for the discriminative recognition of N 6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem. 290, 24902–24913 (2015).
Xu, C. et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10, 927–929 (2014).
Nayler, O., Hartmann, A. M. & Stamm, S. The ER repeat protein YT521-B localizes to a novel subnuclear compartment. J. Cell Biol. 150, 949–962 (2000).
Saitoh, N. et al. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell 15, 3876–3890 (2004).
Patil, D. P., Pickering, B. F. & Jaffrey, S. R. Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol. 28, 113–117 (2017).
Kennedy, E. M. et al. Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 19, 675–685 (2016).
Du, H. et al. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 7, 12626 (2016).
Park, O. H. et al. Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol. Cell 74, 494–507 (2019).
Bailey, A. S. et al. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. eLife 6, e26116 (2017).
Hsu, P. J. et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127 (2017).
Jain, D. et al. ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. eLife 7, e30919 (2018).
Dhote, V., Sweeney, T. R., Kim, N., Hellen, C. U. & Pestova, T. V. Roles of individual domains in the function of DHX29, an essential factor required for translation of structured mammalian mRNAs. Proc. Natl Acad. Sci. USA 109, E3150–E3159 (2012).
Kretschmer, J. et al. The m(6)A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′–3′ exoribonuclease XRN1. RNA 24, 1339–1350 (2018).
Roost, C. et al. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J. Am. Chem. Soc. 137, 2107–2115 (2015).
Spitale, R. C. et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519, 486–490 (2015).
Liu, N. et al. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45, 6051–6063 (2017).
Wu, B. et al. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 9, 420 (2018).
Alarcon, C. R. et al. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 162, 1299–1308 (2015).
Edupuganti, R. R. et al. N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878 (2017).
Choi, J. et al. N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat. Struct. Mol. Biol. 23, 110–115 (2016).
Huang, H. et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 (2018).
Hafner, M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010).
Zhang, F. et al. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 27, 3936–3950 (2018).
Youn, J. Y. et al. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell 69, 517–532 (2018).
Sun, L. et al. RNA structure maps across mammalian cellular compartments. Nat. Struct. Mol. Biol. 26, 322–330 (2019).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Zhao, B. S., Roundtree, I. A. & He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42 (2017).
Gerken, T. et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318, 1469–1472 (2007).
Fedeles, B. I., Singh, V., Delaney, J. C., Li, D. & Essigmann, J. M. The AlkB family of fe(ii)/alpha-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 290, 20734–20742 (2015).
Ougland, R. et al. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol. Cell 16, 107–116 (2004).
Jia, G. et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 582, 3313–3319 (2008).
Hess, M. E. et al. The fat mass and obesity associated gene (FTO) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 16, 1042–1048 (2013).
Mauer, J. et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375 (2017).
Garcia-Campos, M. A. et al. Deciphering the “m6A Code” via antibody-independent quantitative profiling. Cell https://doi.org/10.1016/j.cell.2019.06.013 (2019). Garcia-Campos et al. are the first to develop a quantitative profiling of a subset of m 6 A sites in the transcriptome at single-nucleotide resolution.
Zou, S. et al. N(6)-Methyladenosine: a conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5. Sci. Rep. 6, 25677 (2016).
Wei, C. M. & Moss, B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 16, 1672–1676 (1977).
Wei, C., Gershowitz, A. & Moss, B. N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 257, 251–253 (1975).
Mauer, J. et al. FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 15, 340–347 (2019). Mauer et al. demonstrate that m 6 A m in snRNAs are the main targets of FTO, and that FTO's biological function is to regulate snRNA processing.
Bartosovic, M. et al. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 45, 11356–11370 (2017).
Li, Z. et al. FTO plays an oncogenic role in acute myeloid leukemia as a N 6-methyladenosine RNA demethylase. Cancer Cell 31, 127–141 (2017).
Zhang, S. et al. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31, 591–606 (2017).
Dixit, D., Xie, Q., Rich, J. N. & Zhao, J. C. Messenger RNA methylation regulates glioblastoma tumorigenesis. Cancer Cell 31, 474–475 (2017).
Thalhammer, A. et al. Human AlkB homologue 5 is a nuclear 2-oxoglutarate dependent oxygenase and a direct target of hypoxia-inducible factor 1α (HIF-1α). PLOS ONE 6, e16210 (2011).
Zhang, C. et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc. Natl Acad. Sci. USA 113, E2047–E2056 (2016).
Rubio, R. M., Depledge, D. P., Bianco, C., Thompson, L. & Mohr, I. RNA m(6)A modification enzymes shape innate responses to DNA by regulating interferon beta. Genes Dev. 32, 1472–1484 (2018).
Agarwala, S. D., Blitzblau, H. G., Hochwagen, A. & Fink, G. R. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLOS Genet. 8, e1002732 (2012).
Ping, X. L. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189 (2014).
Schwartz, S. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 8, 284–296 (2014).
Scholler, E. et al. Interactions, localization, and phosphorylation of the m(6)A generating METTL3–METTL14–WTAP complex. RNA 24, 499–512 (2018).
Horiuchi, K. et al. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 288, 33292–33302 (2013).
Yue, Y. et al. VIRMA mediates preferential m(6)A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4, 10 (2018).
Knuckles, P. et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 32, 415–429 (2018).
Guo, J., Tang, H. W., Li, J., Perrimon, N. & Yan, D. Xio is a component of the Drosophila sex determination pathway and RNA N(6)-methyladenosine methyltransferase complex. Proc. Natl Acad. Sci. USA 115, 3674–3679 (2018).
Wen, J. et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69, 1028–1038 (2018).
Fujita, Y. et al. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat. Cell Biol. 4, 222–231 (2002).
Ruzicka, K. et al. Identification of factors required for m(6) A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 215, 157–172 (2017).
Molinie, B. et al. (2016) m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat. Methods 13, 692–698 (2016).
Liu, N. & Pan, T. Probing RNA modification status at single-nucleotide resolution in total RNA. Methods Enzymol. 560, 149–159 (2015).
Wang, Y. et al. N 6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198 (2014).
Sednev, M. V. et al. N 6-methyladenosine-sensitive RNA-cleaving deoxyribosomes. Angew. Chem. 130, 15337–15341 (2018).
Imanishi, M., Tsuji, S., Suda, A. & Futaki, S. Detection of N 6-methyladenosine based on the methyl-sensitivity of MazF RNA endonuclease. Chem. Commun. 53, 12930–12933 (2017).
Harcourt, E. M., Ehrenschwender, T., Batista, P. J., Chang, H. Y. & Kool, E. T. Identification of a selective polymerase enables detection of N 6-methyladenosine in RNA. J. Am. Chem. Soc. 135, 19079–19082 (2013).
Aschenbrenner, J. et al. Engineering of a DNA polymerase for direct m6A sequencing. Angew. Chem. Int. Ed. Engl. 57, 417–421 (2018).
Hong, T. et al. Precise antibody-independent m6A identification via 4SedTTP-involved and FTO-assisted strategy at single-nucleotide resolution. J. Am. Chem. Soc. 140, 5886–5889 (2018).
Xiao, Y. et al. An elongation- and ligation-based qPCR amplification method for the radiolabeling-free detection of locus-specific N 6-methyladenosine modification. Angew. Chem. Int. Ed. Engl. 57, 15995–16000 (2018).
Liu, W. et al. Identification of a selective DNA ligase for accurate recognition and ultrasensitive quantification of -methyladenosine in RNA at one-nucleotide resolution. Chem. Sci. 9, 3354–3359 (2018).
Garalde, D. R. et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206 (2018).
Castellanos-Rubio, A. et al. A novel RT-QPCR-based assay for the relative quantification of residue specific m6A RNA methylation. Sci. Rep. 9, 4220 (2019).
Chen, Y. et al. Study of the whole genome, methylome and transcriptome of Cordyceps militaris. Sci. Rep. 9, 898 (2019).
Bringmann, P. & Lührmann, R. Antibodies specific for N 6-methyladenosine react with intact snRNPs U2 and U4/U6. FEBS Lett. 213, 309–315 (1987).
Thüring, K., Schmid, K., Keller, P. & Helm, M. LC-MS analysis of methylated RNA. Methods Mol. Biol. 1562, 3–18 (2017).
Bodi, Z. & Fray, R. G. Detection and quantification of N 6-methyladenosine in messenger RNA by TLC. Methods Mol. Biol. 1562, 79–87 (2017).
Acknowledgements
The authors thank the members of the Jaffrey laboratory for comments and suggestions.
Author information
Authors and Affiliations
Contributions
S.Z., R.J.R. and S.R.J. researched data for the review; R.J.R. and S.R.J. made substantial contributions to the discussion of the m6A writer complex; S.Z. and S.R.J. made substantial contributions to the rest of the review; S.Z., R.J.R. and S.R.J. wrote, reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
S.R.J. is scientific founder and adviser to Gotham Therapeutics, in which he also owns equity.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Transcriptome turnover
-
Term used to indicate the rate at which mRNAs are produced and degraded in order to control the final mRNA quantity and quality in a cell.
- mRNA looping
-
A mechanism of interaction between the 3′ UTR of an mRNA and the 5′ UTR of the same mRNA to favour ribosome reloading and a high translation rate.
- Liquid–liquid phase separation
-
(LLPS). Condensation into a dense phase of RNA and proteins that often resembles a liquid droplet and is strictly related to microenvironment conditions, such as pH, temperature and salt concentration.
- Low-complexity domain
-
Regions of a protein in which specific amino acids are over-represented compared with their amino acid proportions found in the proteome.
- Stress granules
-
Membraneless cytoplasmic liquid–liquid phase compartments where RNA and RNA-bound proteins are found under conditions of cellular stress.
- P-bodies
-
Membraneless liquid–liquid phase compartments where RNA and RNA-bound proteins are assembled in order to store and/or degrade the RNA.
- RNA granules
-
RNA- and protein-containing liquid–liquid phase compartments assembled in order to transport RNA to dendrites in neurons.
- Allosteric adaptor
-
A protein that indirectly influences the effects of a second protein on the bound target. In this case, the second protein may directly influence the function of the target.
- Exon skipping
-
A specific mechanism of mRNA splicing used to ‘skip over’ an exon normally included in the processed mRNA. Thus, in this case, the final processed mRNA will not include this exon.
- Nuclear speckles
-
Membraneless compartments enriched in pre-mRNA splicing factors, located in the interchromatin regions of the nucleoplasm and implicated in different aspects of RNA metabolism.
- UV crosslinking immunoprecipitation
-
(CLIP). An antibody-based method used to identify the RNA sites directly bound by RNA-binding proteins.
- Ribosome profiling
-
A method used to determine the ribosome footprints on mRNAs and thus to identify the translating mRNAs.
Rights and permissions
About this article
Cite this article
Zaccara, S., Ries, R.J. & Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol 20, 608–624 (2019). https://doi.org/10.1038/s41580-019-0168-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41580-019-0168-5
This article is cited by
-
m6A methylation modification and immune infiltration analysis in osteonecrosis of the femoral head
Journal of Orthopaedic Surgery and Research (2024)
-
Genome-wide identification, characterization, and expression analysis of m6A readers-YTH domain-containing genes in alfalfa
BMC Genomics (2024)
-
Fat mass and obesity-associated protein (FTO) mediated m6A modification of circFAM192A promoted gastric cancer proliferation by suppressing SLC7A5 decay
Molecular Biomedicine (2024)
-
N6-methyladenosine-modified circPLPP4 sustains cisplatin resistance in ovarian cancer cells via PIK3R1 upregulation
Molecular Cancer (2024)
-
FBW7/GSK3β mediated degradation of IGF2BP2 inhibits IGF2BP2-SLC7A5 positive feedback loop and radioresistance in lung cancer
Journal of Experimental & Clinical Cancer Research (2024)