Abstract
In the past decade, membraneless assemblies known as biomolecular condensates have been reported to play key roles in many cellular functions by compartmentalizing specific proteins and nucleic acids in subcellular environments with distinct properties. Furthermore, growing evidence supports the view that biomolecular condensates often form by phase separation, in which a single-phase system demixes into a two-phase system consisting of a condensed phase and a dilute phase of particular biomolecules. Emerging understanding of condensate function in normal and aberrant cellular states, and of the mechanisms of condensate formation, is providing new insights into human disease and revealing novel therapeutic opportunities. In this Perspective, we propose that such insights could enable a previously unexplored drug discovery approach based on identifying condensate-modifying therapeutics (c-mods), and we discuss the strategies, techniques and challenges involved.
Similar content being viewed by others
Introduction
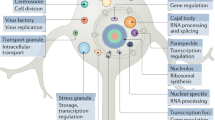
For more than a century, scientists have speculated on the structure and organization of the protoplasm1,2,3,4. In addition to membrane-bound organelles such as the nucleus and mitochondria, microscopists also observed organelles lacking membranes. For instance, the nucleolus was first described in the 1830s5. Additional membraneless organelles were identified at the turn of the twentieth century6,7,8,9,10, and many others have since been reported. Although the functions of these assemblies — now known as biomolecular condensates — have been described in some cases (Table 1), the mechanisms that control their formation, structure, dynamics, composition and activity are only now being studied intensively (Fig. 1).
The molecular community defines the identity of a biomolecular condensate. Examples of three biomolecular condensates and selected components. The centrosome is the central organizer of microtubules and is involved in regulation of mitosis; the image shows a mitotic SiHa cell, with the centrosome structural protein CDK5RAP2 stained green, the nucleus blue and microtubules red. The nucleolus is the site of ribosome biogenesis; the image shows U2OS cells with the nucleolar scaffold protein NPM1 stained green and microtubules red. Stress granules; the image shows stress granules in HeLa cells, visualized via G3BP1 immunofluorescence. lncRNA, long non-coding RNA; rRNA, ribosomal RNA; snoRNA, small nucleolar RNA. The centrosome image is reproduced from https://www.proteinatlas.org/ENSG00000136861-CDK5RAP2/cell#img, and the nucleolus image is reproduced from https://www.proteinatlas.org/ENSG00000181163-NPM1/cell#img.
Experimental evidence to support the hypothesis that biomolecular condensates form by aqueous phase separation was first generated by Cliff Brangwynne, Tony Hyman, Frank Jülicher and colleagues. They demonstrated that P granules — protein–RNA assemblies found in Caenorhabditis elegans — exhibit liquid-like behaviour in cells, including dripping, wetting and fusion, implicating phase separation in their formation11. Subsequent work from Brangwynne and Hyman showed that nucleoli in Xenopus oocytes also behave as liquids, exhibiting rapid ATP-dependent dynamics12.
Publications from other laboratories soon provided additional support for the concept of biological phase separation. The Rosen laboratory was first to recognize the role and importance of weak multivalent interactions in driving phase separation and speculated that cellular organization and regulation across all of biology might be critically dependent upon such phase transitions13. Work from the McKnight laboratory showed that proteins containing low-complexity, intrinsically disordered regions (IDRs) phase separate into hydrogels capable of partitioning ribonucleoprotein (RNP) granule components14,15. Hanazawa, Yonetani and Sugimoto revealed that just two condensate components can reconstitute P granules in cells, supporting the idea that some proteins are necessary and sufficient to promote condensate assembly16. This early work was captured in a series of reviews17,18,19,20,21. The biomolecular condensates field reached an inflexion point in 2015, with multiple publications reporting breakthrough findings (reviewed in ref.22); since then, research in the field has grown rapidly.
Several lines of evidence emerging from such research support the relevance of biomolecular condensates for drug discovery. There are a growing number of examples of ‘aberrant behaviours’ of condensates that are associated with disease states, including neurodegeneration23, cancer (for example, prostate cancer)24, viral infections (for example, respiratory syncytial virus (RSV))25 and cardiac disease26,27 (Table 2). Proteins of high therapeutic interest in neurodegenerative disease such as TDP43 and FUS have been identified inside condensates28,29. The anticancer drugs cisplatin and tamoxifen can partition into transcriptional condensates, altering their composition in cultured cells and in vitro reconstituted model condensates30; initial reports show that small molecules can alter condensate behaviours with functional consequences in cell-based studies31,32. Finally, the tools to study condensates are rapidly maturing, increasing their applicability to efforts to identify condensate-modifying therapeutics (c-mods).
Several components are crucial to the basis of a c-mod discovery campaign. First, observed associations between condensate characteristics and diseases should be rigorously validated, with the aim of identifying associations that are causal. Furthermore, it should be established that molecular and mechanistic aspects of biomolecular condensates identified through in vitro studies are relevant in vivo. Second, assays that reliably reflect disease-relevant aspects of the biology of condensates need to be developed. In contrast to classical drugs that typically target unique macromolecules, the target for c-mods is a community of molecules engaged in an extended network. Major challenges include identifying the biomolecule(s) required for condensate assembly, as well as understanding the thermodynamics of the extended network and the kinetics of processes that disrupt the equilibrium. As discussed in detail in refs.33,34, caution needs to be exercised to not over-interpret qualitative data and results obtained from simplified (for example, in vitro reconstitution) or artificial (for example, overexpression) systems. Nevertheless, these model systems can be leveraged to obtain insights into the structural ensemble and mesoscale organization of a subset of macromolecules inside a condensate-like milieu (reviewed in ref.35) and the effects of putative c-mods on the represented interactions, as we discuss later in the Perspective.
Given the infancy of the field, the aspects of the rationale and strategies for pursuing c-mod discovery discussed are built on disparate pieces of evidence from studies that were not necessarily focused on drug discovery, or from drug discovery studies that were not searching for c-mods. However, we believe that there is a substantial amount of data from such studies that support the feasibility of targeting biomolecular condensates and provide a foundation for a guide to future c-mod discovery and assay development when interpreted with a condensate perspective.
With the goal of contributing to such a guide, in this Perspective we first discuss how understanding the properties and functions of condensates may enable a novel approach to drug discovery. After briefly describing the physics and structural basis for the formation of biomolecular condensates, we outline the diverse roles that condensates play in cellular function and some of the evidence for the associations of aberrant condensates with disease. We then describe approaches and technologies for the identification and characterization of drug candidates that can modulate or otherwise exploit disease-relevant condensates and consider the challenges that need to be addressed for them to be effective.
Overview of condensate biology
Biomolecular condensates have been linked to many cellular processes, including sensing and responding to stress, compartmentalization of biochemical reactions, mechanical regulation and signalling (reviewed in refs.34,36). Their composition is typically complex, consisting of hundreds of different proteins and nucleic acids, which form an extensive intermolecular network spanning length scales of nanometres to micrometres. The underlying mechanisms for biomolecular condensate assembly depend on their composition and architecture (reviewed in refs.33,36,37,38). As a common denominator, this assembly is mediated by multivalent interactions leading to increased local concentration of a select molecular community, which creates a microenvironment with unique properties (reviewed in ref.36). Here, we focus on the widely used model in which biomolecular condensates assemble through phase separation. We propose that targeting the emergent properties of the molecular community within condensates provides an untapped source of therapeutic agents. Notably, however, the c-mod design and discovery strategies discussed later in this Perspective are agnostic to the mechanisms underlying condensate assembly.
Principles of condensate assembly
Many biomolecular condensates are thought to assemble in a concentration-dependent manner to form non-stoichiometric macromolecular assemblies, via spontaneous or nucleated phase separation of a select set of proteins and/or nucleic acids. Phase separation occurs when the concentration of biomolecules exceeds the saturation concentration (Csat). This threshold defines the phase boundary, above which thermodynamics favours self-solvation of these biomolecules rather than solvation by the surrounding environment, driving formation of mesoscale assemblies rather than discrete biomolecular complexes. Minor changes in biomolecule concentration that cross the phase boundary trigger a sharp, switch-like response, leading to either condensate formation or dissolution, effectively changing the local concentration, sometimes over several orders of magnitude (Fig. 2a). Importantly, this process is fast and reversible, making it an ideal element in sensing stress and other environmental changes.
a | Phase separation enables a sharp, switch-like response as the concentration of phase-separating biomolecule(s) exceeds the saturation concentration (Csat). In this case, a small change in bulk concentration can lead to sudden change in a molecule’s local concentration, and may lead to large changes in activity/signalling (top). By contrast, without phase separation, a molecule’s local concentration scales linearly with the bulk concentration, resulting in subtle effects (bottom). b | At atomic and molecular levels (angstrom to nanometre), various types of interactions (top left) and their valency (top right) define condensates. These interactions, in turn, define condensate composition. Examples of biomolecular topology that promote condensation include intrinsically disordered proteins with multivalency encoded in short linear motifs, modular proteins with multivalency encoded in repeat folded binding domains, modular proteins with a condensation-prone intrinsically disordered region (IDR) and one or more folded domains that drive specific localization (for example, transcription factors), and nucleic acids (DNA and RNA). Scaffolds (orange) are characterized by higher partition coefficients and lower Csat values compared with clients (blue). Together, all smaller-scale interactions modulate the biomolecular network of the molecular community (bottom left) and the emergent material properties (bottom right) at the mesoscale (nanometre to micrometre), optimized for condensate function (fit for purpose). Solid-like condensates such as the Balbiani body are reversible under physiological conditions, in contrast to solid-like pathological amyloids. Part b is adapted with permission from ref.124, Elsevier.
Assembly of biomolecular condensates is facilitated by multivalency encoded as multiple copies of folded domains or of structural motifs, and/or low-complexity IDRs13,22,39. These interactions are typically weak and contribute to fine-tuning the material properties of biomolecular condensates40,41 (Fig. 2b).
Physical properties of condensates
Biomolecular condensates exhibit a range of material properties (reviewed in ref.21). Stress granules42,43, P granules11 and nucleoli12 exhibit liquid-like properties. Gel-like assemblies (that is, the centrosome44, RNA expansion repeats45 and the nuclear pore46) and solid-like functional amyloids (that is, the Balbiani body47 and A-bodies48) have reduced internal dynamics. These material properties are correlated with the composition and biological functions of the condensates and can be dynamically modulated through changes in the environment or active biological processes (discussed below). For example, the liquid-like properties allow stress granules to rapidly assemble and disassemble as conditions vary. The gel-like centrosome can withstand the microtubule pulling forces when the mitotic spindle is formed49. The Balbiani body is proposed to promote dormancy during oocyte storage by shutting down all biochemical reactions (reviewed in ref.50).
The complex network of interactions between the various members of a molecular community determines the material properties of a condensate. The biomolecules within a community share features that contribute to their compatibility and co-localization (reviewed in ref.51). These features include similar amino acid bias in IDRs52, certain families of folded domains13,53,54 and similar classes of nucleic acids28,55,56. These molecules can be classified as scaffolds or clients (Fig. 2b) based on whether they are essential for the formation of the underlying network of a condensate53. Scaffolds are multivalent biomolecules required for condensate formation; they typically exhibit the lowest Csat among components, initiating the condensation process, and are characterized by a high partition coefficient (the ratio between concentrations inside versus outside the condensate)13,52,53,57,58. Clients are molecules that partition into condensates via interactions with the scaffolds; they are typically characterized by lower partition coefficients compared with scaffolds59. Typically, multiple macromolecules can function as co-scaffolds (for example, G3BP1/2 in stress granules60 and PGL1/3 in P granules16,61,62). The composition of condensates dynamically adjusts based on changes in bulk concentration of co-scaffolds and clients53,63,64, as well as in response to non-equilibrium processes (for example, activity of energy-consuming enzymes).
Functions of condensates
Condensates provide a distinct environment optimized for function. The intra-condensate milieu regulates enzymatic reactions by compartmentalizing components involved in related biological processes. This unique environment can modulate one or more of the following parameters: diffusion of components, enrichment in substrates and/or depletion of inhibitors65,66,67,68,69,70.
Biomolecular condensates can respond rapidly, with a low energy threshold, to sudden environmental changes, such as temperature, stress, starvation, detection of foreign material or other cellular stimuli71. Phase separation affords a reversible mechanism for increasing the local concentration of a particular component within the condensate, while reducing it in the outside environment. Such processes are involved in mitigating cellular toxicity, by sequestering excess materials in response to stress. For example, stress signals sensed in the cytoplasm trigger assembly of stress granules, which compartmentalize untranslated RNA and RNA-binding proteins from the cytoplasm and nucleus28,72 (Fig. 1). Similarly, stress sensed in the nucleus leads to dynamic changes in the nucleolus73 and formation of nuclear stress condensates74,75,76. Biomolecular condensates also play roles in minimizing cellular noise77, control of genome packaging56,78,79,80, transcription81,82, cell-cycle control and DNA double-strand breaks83, viral assembly84 and immune responses66 (Tables 1,2).
Regulation of condensates
The composition of biomolecular condensates is complex, dynamic and varies with cell type and the type of signal that induces condensate formation. Studies indicate that biological systems are often optimized to reside close to the phase boundary. Thus, small changes in the environment (for example, metabolite or biomolecule concentrations, pH or temperature) tip the equilibrium to either dissolve or assemble the condensate, generating a rapid switch-like signal (reviewed in ref.71) (Fig. 2a). Condensates exist in a non-equilibrium state via the action of energy-consuming enzymes12,85, competitive interactions with ligands63,64,86, hydrotropes87 and other perturbations, which regulate their function, composition and dynamics. Protein quality control, including chaperones, autophagy and proteasome degradation85,88,89,90,91, and the post-translational machinery are intimately involved in the regulation of condensates and their emergent properties.
Post-translational modifications
Assembly and disassembly of condensates is modulated by covalent post-translational modifications (PTMs) of protein components, such as phosphorylation, acetylation, methylation, SUMOylation, ubiquitination, PARylation (poly-ADP-ribosylation) and glycosylation (reviewed in refs.92,93). These modifications can have a dramatic effect on the conformational ensemble and dynamics of IDRs involved in condensate scaffolding (reviewed in refs.94,95). For example, epigenetic modifications were shown to induce changes in material properties of chromatin, modulating access of the transcriptional machinery to the genetic information56. Additionally, epitranscriptomic and post-transcriptional modifications on RNA contribute similarly to modulating phase behaviour of condensates (reviewed in ref.39).
Competitive interactions
Proteins that serve as scaffolds for condensates can interact with chaperones (reviewed in ref.85) and nucleocytoplasmic transporters (for example, karyopherins)96, thereby modulating the Csat, preventing and reversing aberrant phase transitions. Similarly, helicase action controls the partitioning of RNA within biomolecular condensates97. For example, HSP70 is required for the dissolution of aged stress granules containing misfolded SOD1 (ref.88). Karyopherins prevent aberrant phase transitions of prion-like domains and reverse phase separation of amyotrophic lateral sclerosis (ALS)-associated proteins (for example, FUS, TDP43 and hnRNPA1) by directly binding to cognate nuclear localization signals within the target protein, aided by weak, non-specific interactions that compete for the condensation-driving interactions98,99.
Compositional change
As described above, condensate composition, dynamics, material properties and function are interrelated. Partitioning of RNA inside protein condensates tunes the viscosity in vitro and in vivo100,101,102,103. The level of RNA can either promote or dissolve condensates scaffolded by RNA-binding proteins104,105,106,107. Such a regulatory mechanism has significant implications in biology; a notable example is regulatory feedback in transcription, where incipient amounts of RNA synthesis promote stabilization of transcriptional condensates, whereas accumulation of transcripts promotes their dissolution108. In addition to RNA, other nucleotide polymers can tune the stability of biomolecular condensates, including DNA56,66,80 and PAR109,110. Modulation of the protein composition of a condensate can affect the dynamics of individual components. For example, the centrosome nucleator SPD2 diffuses faster when its binding partners PLK1 and TPXL1 are present in the reconstituted condensates44.
In a recent report, An et al.111 demonstrated that aberrant, persistent pathological stress granules formed by an ALS-associated FUS mutant exhibit different proteomics compared with normal stress granules. These stress granules are characterized by enriched physical interactions between components, consistent with earlier observations that pathological stress granules are less dynamic.
Conformation-encoded regulation
Condensate-associated proteins exhibit a modular topology that allows them to function as interaction hubs by engaging with multiple types of macromolecules. They encode structural switches that promote transitions between freely diffusing discreet monomers or oligomers to a ligand-bound scaffold of a large macromolecular network within a condensate, as described for the nucleolar and stress granule scaffolds, NPM1 (ref.86) and G3BP1 (refs.112,113), respectively. In these examples, ligand binding alters the conformation of a protein at atomic scale, triggering remodelling of the nanometre-to-micrometre scale molecular network of the condensate.
Within the condensate microenvironment, IDRs can remain disordered, as seen in DDX4 (ref.114) and FUS107, or can undergo folding upon binding. FUS and TDP43 were shown to form cross-β structures within their LCD domains in hydrogels to stabilize intermolecular interactions115,116, and the carboxy-terminal LCD of TDP43 stabilizes a helix structure upon dimerization in liquid-like droplets117. Modulation of the helical propensity by mutations, including those associated with ALS, affect not only the Csat for phase separation but also the material properties of the resulting condensates and the splicing function of TDP43 (ref.117).
Spatial positioning
Biomolecular condensates provide a means for cells to spatially regulate important processes. For example, at homeostasis, FUS and TDP43 fulfil roles in RNA splicing and metabolism in the nucleus but are sequestered into cytoplasmic stress granules under stress conditions. The function of HSF1, a transcription factor for heat shock chaperones, is modulated under stress conditions via sequestration into nuclear condensates74. Organization of the cytoplasm, such as spatial patterning of specific transcripts in polar cells, is achieved by encapsulation of the target RNA molecules in biomolecular condensates11,101,102.
Multi-compartment organization of the condensate interior (reviewed in ref.37) arises via coexisting, non-miscible phases40,103,118,119,120. The nucleolus exhibits a three-layered architecture, determined by the surface tension with respect to the nucleoplasm40. It was proposed that the material properties of the different nucleolar layers are optimized to promote the correct sequence of steps and the vectorial flux in ribosome biogenesis40,64.
Cellular surfaces, such as chromatin, membranes and the cytoskeleton, can serve as regulators of spatial positioning of biomolecular condensates. Membranes serve as nucleators to drive phase separation by restricting molecular diffusion and promoting local crowding effects (reviewed in ref.95). For example, T cell signalling condensates and TIS granules form at the plasma membrane65 and on the endoplasmic reticulum membrane121, respectively. Su et al. showed that phosphorylation of the T cell receptor triggers clustering of LAT (linker for activation of T cells) into mesoscale condensates at the plasma membrane, and that these condensates recruit components of T cell signalling, which subsequently trigger actin polymerization as a functional output65.
Taken together, these mechanisms governing their behaviour, function and regulation play an important role in the normal function of condensates in cells and provide valuable insights for designing strategies to correct their malfunction in disease.
Condensates in disease
Here, we define a ‘condensatopathy’ as an aberration of a condensate that drives a specific disease phenotype, which has been observed in in vitro model systems and in vivo cellular and animal models of neurodegenerative diseases, dilated cardiomyopathy, certain types of cancer (reviewed in refs.23,27,122,123,124) and viral infections. Intriguingly, there are multiple examples of genetic mutations showing strong clinical association with diseases that affect proteins that have been identified in biomolecular condensates. This suggests the possibility that these mutations might dysregulate condensate function, and thereby drive disease. We discuss a few better-understood examples below. Although correlations between condensate malfunction and disease in these model systems have been well documented, strong support for causation is still under development.
Neurodegeneration
ALS and frontotemporal dementia (FTD) pathology have been linked to environmental factors and a diversity of genetic alterations, including numerous point mutations in the low-complexity regions of RNP-granule localized proteins, as well as repeat expansions. Point mutations in proteins such as FUS42 and hnRNPA1 (ref.43) accelerate the kinetics of phase transition and promote amyloid-like fibril formation within the condensate environment in in vitro studies. Altered kinetics of clearance of RNP granules, namely prolonged persistence of condensates, was associated with the ALS-hallmark phenotype of cytoplasmic inclusions in cultured cells and neurons. For example, repetitive cycling of G3BP1-positive condensate formation and increased persistence, modulated via an optogenetics model, evolved towards cytoplasmic proteinaceous inclusions and caused cellular toxicity125.
A recurring pathological observation in ALS/FTD is the presence of TDP43-rich cytoplasmic granules, irrespective of whether the TDP43 gene harbours mutations126. In vitro and cellular data suggest that these granules arise as condensates and undergo ageing (reviewed in ref.23). This spatial re-localization of nuclear TDP43 into cytoplasmic condensates in cultured neurons was associated with increased condensate viscosity127 and splicing defects in several motor neuron-specific mRNAs, including that encoding stathmin 2 (STMN2)128, a neuron-specific regulator of microtubule stability. The TDP43 condensatopathy causes a loss of function of STMN2 (ref.128) and impaired axonal growth and regeneration127,128. Furthermore, optogenetic formation of TDP43-positive condensates via blue light illumination was sufficient to recapitulate the progressive motor dysfunction observed in patients with ALS in a Drosophila model129.
Repeat expansions of a short nucleotide segment are another specific type of genetic alteration associated with diseases such as ALS/FTD, myotonic dystrophy, spinocerebellar ataxias and Huntington disease. The severity of these diseases scales with the length of the repeat (multivalency) of the transcript and/or translated polypeptide. Furthermore, the diagnosed clinical cases exhibit a minimum threshold length of the repeat. The resulting polyvalent RNAs and polypeptides have the hallmarks of biomolecules that will localize to condensates as scaffolds, and result in condensatopathies that sequester other important biomolecules. C9orf72 G4C2, CAG and CUG repeats are found in ALS/FTD, Huntington disease and myotonic dystrophy, respectively; transcripts containing variable lengths of these repeats form RNA foci in live cells, and dynamically arrested condensates in vitro45. polyGA and polyGR peptides, resulting from an ATG-independent translation of the G4C2 repeats, were shown to sequester proteasomal and nucleolar proteins, respectively (reviewed in ref.130). Furthermore, overexpressed or exogenously added arginine-rich polypeptides of G4C2 repeats insinuate in pre-existing cellular condensates, such as the nucleolus, RNP granules and the nuclear pore complex. As a result, the condensates change their composition, material properties and function due to competition between the G4C2 peptides with the native interactions131,132. Correcting the altered material properties and/or sequestration of biomolecules due to the underlying condensatopathies may prevent or reverse these neurodegenerative diseases.
Cardiomyopathy
Condensatopathies associated with dysregulation of RNP granules are not limited to neurodegenerative diseases. A mutation in the gene coding for the tissue-specific alternative splicing factor RBM20 that is found in patients with congenital dilated cardiomyopathy is characterized by a RNP condensate defect, coupled with contractile dysfunction and aberrant heart anatomy in a heterozygous pig model26. The R636S point mutation localized in the low-complexity disordered RSRS region causes aberrant sarcoplasmic accumulation of RBM20. At the cellular level, the dominant effect of the mutant leads to RBM20 re-localization from nuclear splicing speckles to cytoplasmic condensates that fuse with other cellular condensates harbouring stress granule markers26. This condensatopathy causes sequestration of mRNA, polysomes and cardiac cytoskeleton proteins (for example, ACTC1). Interestingly, mice harbouring the congenital dilated cardiomyopathy mutation exhibited a more severe cardiac dysfunction phenotype than mice lacking RBM20 (ref.133).
Collectively, these observations suggest that the pathological mechanism attributed to the condensate phenotype is complex, involving loss of nuclear splicing function for RBM20, loss of function of proteins that partition into aberrant cytoplasmic RBM20 condensates and a gain of function of these condensates. Therefore, the RBM20 condensatopathy serves as a hub for misregulation of multiple pathways in congenital dilated cardiomyopathy and is an attractive node to target this disease therapeutically.
Cancer
Recent progress has shown a link between condensatopathies and several types of cancer. These condensatopathies deregulate many processes, including, but not limited to, genomic stability, signalling, protein quality control and transcription (reviewed in refs.134,135). Transcription of key developmental genes is often under the control of super-enhancers. Super-enhancers are classically defined by chromatin immunoprecipitation sequencing as clusters of enhancers bearing large amounts of transcriptional machinery (transcription factors, coactivators and RNA Polymerase II (Pol II)). This high-density assembly of proteins at super-enhancers is now understood to constitute transcriptional condensates that drive gene expression81,82,136. These insights challenge the stoichiometric model of transcription, suggesting novel properties and functions of transcription factors and coactivators in a concentrated condensate of protein and DNA. For example, the function of transcription factor activation domains was poorly understood because they contain IDRs not amenable to crystallography. Now it is becoming clear that they may activate genes, in part, by their capacity to condense with coactivators on genomic regulatory elements. Transcription of oncogenes is a general feature of cancer cells. This often occurs through condensatopathies, such as acquisition of aberrant super-enhancers137.
Condensatopathies resulting in aberrant gene expression are also associated with cancers. Several chromosomal translocations have been identified, where a condensation-prone IDR fused to a chromatin-associating folded domain creates aberrant condensates. Two examples are EWS–FLI in Ewing sarcoma138 and NUP98–KDM5A in leukaemia139. NUP98–KDM5A is one of many variations of genetic translocations that fuse the condensation-prone amino-terminal FG-rich IDR of a nucleoporin (for example, NUP98 and NUP124) with a folded domain that anchors it at a specific location on chromatin, such as a DNA-binding domain (for example, HOXA9, HOXA13 and PHF23), a helicase domain (for example, DDX10) or a histone binding domain (for example, KDM5A, NSD1)140. These genetic translocations result in condensatopathies that share a common expression reprogramming phenotype, with upregulation of the developmentally silenced Hox genes. These cancers with diverse genetic aetiology may be treatable by similar drug strategies aimed at the underlying condensatopathies.
Viral infections
Biomolecular condensates are also leveraged by pathogens such as viruses to more effectively hijack the host cell and evade the host innate immunity self-defence mechanisms (reviewed in refs.10,84,141). Literature reports link the roles of biomolecular condensates to multiple steps within the viral replication cycle, including viral entry and egress, transcription, protein synthesis, and genome and virion assembly (reviewed in ref.84).
Certain viral infections (for example, rabies and mammalian orthoreovirus)10 induce formation of stress granules. Interestingly, despite the fact that Negri bodies in cells infected with rabies virus share some protein and RNA components with stress granules, the two biomolecular condensates behave similarly to immiscible liquids142, highlighting the importance of the whole molecular community in determining the identity, function and material properties of a condensate. This concept of a molecular community-imposed selectivity becomes important when designing compounds that target specific biomolecular condensates.
Viruses have evolved to evade the host’s innate immune response via multiple mechanisms. The host senses foreign cytoplasmic genomic material via pathogen receptors such as RIGI and MDA5 and induces PML body assembly in the nucleus, as a part of the interferon-dependent innate immune response. Partitioning the viral RNA within viral factory condensates provides a shielding mechanism, preventing its detection by the cytoplasmic pathogen-sensing machinery. Additionally, DNA and RNA viruses disrupt PML bodies as part of their nuclear replication (reviewed in ref.10).
Viral latency is one of the primary challenges that prevent the development of cures for patients suffering from viral infections such as HIV-1. The histone chaperone CAF1 condenses with the viral HIV-1 LTR to form nuclear bodies that recruit other histone chaperones and epigenetic modifiers, and these condensates maintain the integrated viral genome during latency143. These observations could provide a novel intervention point to reactivate latent HIV-1-infected cells, which has been a long-standing focus of efforts to develop a potential cure for HIV-1 infection.
Insights from in vitro and in cell overexpression model systems into the molecular mechanisms of replication and host evasion of SARS-CoV-2 indicate that the dimerization of the nucleocapsid protein144 promotes phase separation with specific viral RNA elements, primarily located at the 5ʹ and 3ʹ UTRs145, as well as with host heterogeneous nuclear RNPs, such as stress granule proteins146. Phase separation inhibits PTMs such as Lys63-linked polyubiquitination of a host antiviral signalling protein, MAVS, thereby suppressing activation of the innate immune system144.
Drug discovery strategies
The roles of condensates in normal and aberrant cellular functions are becoming clearer, and a range of tools are now available to study these cellular phenomena, including protein proximity labelling, advanced microscopy techniques and computational methods, as discussed further below. Accordingly, there is a growing opportunity to explore condensate-informed approaches to drug discovery.
We introduce the term condensate-modifying therapeutics (c-mods) to describe drugs that modulate the physical properties, macromolecular network, composition, dynamics and/or function of specific biomolecular condensates to prevent or reverse disease. A c-mod discovery programme may have one of the three following objectives: repairing a condensatopathy; disrupting the normal functioning of a condensate implicated in disease; or preventing a target from functioning either by disabling it within its native condensate or by de-partitioning the target from its native condensate (Fig. 3a). In each case, the drug discovery strategy will be based on a screening and validation model where a condensate optical phenotype is reliably correlated with one or more functional, disease-relevant outputs.
a | Condensate-modifying therapeutics (c-mods) are developed to achieve one or more of the following objectives: to repair or eliminate a condensatopathy (left); to prevent a specific target from functioning by either delocalizing it from its native condensate (centre) or rendering it inactive within the condensate; or to disrupt the function of a normal condensate (right). b | Strategies to modulate the emerging properties of condensates with c-mods, described in detail in the text. These strategies can be used individually or in combination, and any one strategy can influence multiple characteristics of a condensate; for example, modulating the scaffold will probably result in changes in composition and material properties.
First, for condensatopathy repair, when an aberrant condensate has clearly been implicated in causing a disease, the objective would be to restore normal condensate behaviour or remove aberrant condensates, either by preventing their formation or eliminating them once formed. This could be considered a phenotypic screening strategy, with condensate behaviour in model systems being assessed in the initial screen, and the hits further validated in a disease-relevant secondary assay, as discussed below. There need not be a specific target or pathway, nor any presumed molecular mechanism by which the effect on the condensate is achieved, although such information may be available at the pathway or target level.
Second, in cases where the normal functioning of a condensate is implicated in a biological process central to a disease, the objective would be to develop a c-mod that interferes with the condensate behaviour, ideally only in the disease-relevant cells. The screening strategy would be similar to that described above.
In the third type of case, the objective would be to render a specific target inactive either by ‘disabling’ its ability to function within its native condensate environment or by removing it from that environment. This is especially relevant for targets of high therapeutic interest that are often described as ‘undruggable’ due to selectivity issues or the intrinsic difficulty of finding chemical matter that interferes with their function. If new condensate knowledge indicates that such targets function within a condensate environment, novel strategies could be adopted to disable them. Programmes focused on condensatopathy repair or the disruption of the normal functioning of a condensate implicated in disease may be entirely driven by phenotype. By contrast, the focus of programmes that seek to block target function is to track the behaviour of that specific target. Those targets might be de-partitioned out of the condensate, thereby rendering them inactive. Alternatively, the targets might remain in the condensate but be prevented from engaging in the interactions necessary for function.
C-mod discovery strategies
A wide variety of strategies may be envisioned to identify c-mods that achieve these three objectives. The preferred strategies in any situation will depend on the desired pharmacological outcomes and the detailed knowledge of components, structure and function of the given condensate.
Modulating the condensate scaffold
Modulating a condensate scaffold is expected to lead to drastic effects on the stability of condensates, such as persistence, Csat (that is, formation or dissolution) (Fig. 3b), material properties and/or composition. If a c-mod intercalates between two or more condensate components, or changes the interaction valency or the interaction strength between (co)-scaffolds (within folded and/or disordered domains), it could also change the material properties. The goal is not full inhibition of a particular protein but, rather, disruption of the composition or stability of a biomolecular condensate, which can be achieved via modest changes in the weak networking interactions.
Scaffold modulation could be achieved in various ways. One approach is tuning valency. For example, a low-valency poly-PR peptide dissolved in vitro heterotypic condensates consisting of NPM1 and a multivalent poly-PR peptide132, suggesting that replacing multivalent interactions that mediate network-stabilizing interactions with monovalent, terminal ones is a feasible c-mod mechanism.
A second approach is directly blocking or stabilizing protein–protein144, protein–nucleic acid or nucleic acid–nucleic acid interactions that contribute to scaffolding147. For example, short bait RNAs prevented formation of TDP43 inclusions in an optogenetic cellular model148, probably via a mechanism that outcompetes TDP43–TDP43 self-interaction. A second example is the topoisomerase inhibitor and nucleic acid intercalator mitoxantrone (Table 3), which inhibited stress granule formation in a phenotypic high-content screen using two different cell lines and multiple types of stress, and was shown to block the RNA-dependent recruitment of RNA-binding proteins, including TDP43. These compounds reduced persistence of TDP43 puncta in induced pluripotent stem cell-derived motor neurons31; the exact mechanism of action and how it relates to the annotated activity of this compound need to be further investigated.
Several encouraging proofs of concept for condensate-targeted antiviral drug discovery have been reported, although the exact mechanisms of action are not fully elucidated. Small molecules such as kanamycin (Table 3) are able to destabilize nucleocapsid-containing biomolecular condensates, both in vitro and in cultured cells143. Additionally, a peptide that inhibits nucleocapsid dimerization prevented condensation and viral replication, and rescued the innate immune response in live cells and mouse models142. Cyclopamine (Table 3) analogues modulated the material properties of RSV viral factories in infected cells by reducing the dynamics of the M2-1 protein recovery upon photobleaching, translating into reduced viral replication in the lungs of living mice25.
A c-mod could stabilize non-productive conformations (for example, in folded domains or disordered regions), thereby preventing scaffolding contacts; alternatively, it can trap an aberrant or excess protein in inactive condensates (for example, depots). Sulforaphane (Table 3) treatment of colorectal cancer cells induces formation of β-catenin nuclear depots that partially co-localize with the transcriptional repressor PRMT5; the appearance of the nuclear depots is associated with a reduction in β-catenin-dependent transcriptional activity149.
Modulating condensate composition
C-mods can be envisioned that inhibit or promote the client–scaffold interactions to drive target exclusion or inclusion into the condensate, respectively. For example, an aberrantly de-partitioned protein could be helped to return to its ‘home’ condensate. Nucleolar protein NPM1 is aberrantly localized to the cytoplasm in acute myeloid leukaemia (AML). The natural product avrainvillamide covalently binds mutant NPM1, returning the protein to the nucleoplasm and nucleolus in cell lines from patients with AML150. As discussed in previous sections, changes in condensate composition can affect numerous features, from material properties (for example, viscosity and surface tension), to dynamics and ability to respond to environmental stimuli (for example, persistence and ageing), to enzymatic activity of individual components (for example, cGAS66, UBC9 (ref.68) and Dcp1/2 (ref.67)).
Modulating the conformational and interaction landscape
C-mods that interact with the IDRs of a protein may alter the ability of that protein to partition into a condensate or prevent that protein from forming various intermolecular interactions with other biomolecules within the condensate. Because IDRs are conformationally highly dynamic, it is challenging to use traditional structure-based methods to screen for c-mods that interact with them. However, c-mods could work by engaging with IDRs to either decrease the population of functionally active states or increase the population of inactive or inhibitory states.
Some drug targets, including transcription factors (for example, MYC), hormone receptors (for example, the androgen receptor) and nucleotide-binding proteins (for example, TDP43), contain IDRs and are known to localize to biomolecular condensates. Although small molecules have been identified that bind to these IDRs, they generally do so at micromolar affinities; covalent binders were reported for MYC151 and the androgen receptor152 IDRs (Table 3), and non-covalent IDR binders were reported for p27Kip1 (refs.153,154). The ability to produce a high local drug concentration within a condensate might allow for the development of lower affinity, but highly partitioned, drugs that are effective in targeting proteins in these families that have so far been highly challenging.
Differences in protein conformation inside versus outside a condensate might also be leveraged to develop c-mods that are selective for one of the conformations, potentially minimizing off-target effects.
Degraders
One approach to effectively remove a specific protein from a condensate is to degrade it using proteolysis-targeting chimera (PROTAC) or molecular glue strategies155. There are now several reports suggesting that E3 ligases involved in protein degradation function within condensates156,157,158,159. Several PROTAC design strategies for neurodegeneration targets, including TDP43, α-synuclein, tau and huntingtin, are discussed in ref.160. Similar in concept, RIBOTACs are bifunctional molecules that target specific RNA molecules for ribonucleolytic degradation161.
It has been shown that nuclear p62-containing condensates are essential to efficient proteasomal function, serving as a hub for efficient nuclear protein turnover162. Autophagy is also critically dependent upon phase separation. For example, autophagosome-tethering compounds are molecular glues that selectively target the mutant huntingtin to degradation via autophagy, by selectively binding to the expanded polyQ and LC3 (ref.163).
Using similar approaches, one can envision degrading a scaffold to reduce the effective concentration below Csat, thereby preventing or reversing condensate assembly.
Modulating condensate regulatory processes
Enzymes such as chaperones and helicases can play critical roles in regulating the condensate environment95. Such regulation could affect the condensate environment generally, or may more selectively affect the behaviour of particular proteins or nucleic acids, either by preventing them from interacting with their usual partner molecules or by dramatically affecting their properties (for example, conformation, solubility and valency), causing them to de-partition out of the condensate. Affecting turnover kinetics may be another mechanism to regulate condensate composition.
Another option could be to change the post-translational state of a protein, or the epigenetic or epitranscriptomic state of DNA or RNA, respectively. As discussed earlier, this may either modify the ability of a biomolecule to nucleate the formation of the condensate, change its residence time in the condensate or alter the function of that biomolecule inside the condensate.
Phosphorylation and methylation are some of the most studied PTMs that modulate protein condensation93. RNA post-transcriptional modifications are essential regulators of RNA function and may affect the ability of those RNA to phase separate39,164. Furthermore, epigenetic regulation via histone methylation and acetylation status tunes the phase separation of chromatin56.
Optimize partitioning of drug into condensates
Condensates contain key drug targets such as enzymes, transcription factors, DNA and coactivators. This creates a unique local microenvironment that may selectively increase or decrease the concentration of small molecules, thereby having an impact on their target engagement and therapeutic efficacy (Fig. 3b). For example, cisplatin and JQ1 (Table 3) are antineoplastic compounds that act by intercalating DNA and inhibiting transcription, respectively. Both have recently been shown to specifically partition in transcriptional condensates30. Transformed cells often acquire transcriptional condensates at oncogenes, and high concentrations of intercalating agents or inhibitors at these key genes might account for the heightened sensitivity of cancer cells to agents that target universal cellular processes82. This partitioning behaviour might explain their ability to preferentially kill cancer cells, but it is not yet clear whether these compounds function inside the condensates, and a systematic comparison of efficacy versus condensate partitioning within a drug analogue series is yet to be reported.
Considerations and challenges
The compositional complexity, size and dynamics of biomolecular condensates pose several challenges for drug discovery. First, reliable models reflecting relevant biology and well-defined metrics for characterization of condensates are imperative. Challenges in quantitative characterization of condensates and development of model systems are discussed in refs.33,34. Condensates are exquisitely sensitive to variations in expression levels of their components and regulators, and changes in environment. For example, an overexpressed protein or a protein engineered to undergo phase separation more readily58,81,136,138,148,165,166,167 in a model cell line could induce formation of condensates, whereas under endogenous expression levels in the disease-relevant cell line it exists below Csat, raising the question of relevance of the screening outcome. Such artificial model systems have been used extensively in the field of transcription, where the small size and transient nature of the condensates or hubs make their quantification via conventional microscopy methods challenging.
Second, c-mod discovery will depend on cellular phenotypic assays (as described below), and so the identification of the target(s) driving the observed phenotypes will seldom be straightforward. C-mods may affect condensates via a wide range of mechanisms, from direct interactions with one or more biomolecules within the condensate, to general effects on the emergent properties of the condensate, to altered PTM of proteins, thereby preventing them from entering the condensate. For some c-mod discovery efforts, the target(s) will not be known, and the compound optimization effort will be driven solely by phenotypic cellular assays. In cases where the targets of interest are known, a different challenge will emerge: correlating often-subtle measures of condensate phenotypic behaviours with more traditional measures used in drug discovery programmes, such as biochemical read-outs, intracellular target engagement, measures of gene expression, disease-relevant functional cellular read-outs or pharmacological effects.
A third challenge is how to optimize selective partitioning of the c-mod into the disease-relevant condensate. The properties of condensates vary widely and are difficult to quantify. To improve the therapeutic index, properties such as polarity, lipophilicity, hydrogen bond donor/acceptor count, charge, overall shape, flexibility, aromaticity and the presence of specific functional groups may influence partitioning and be optimized for a specific condensate environment.
An additional selectivity challenge results from the fact that many biomolecular components are found in multiple condensates. We hypothesize that a c-mod which partitions non-selectively into many condensates may lead to unacceptable off-target effects. However, if a c-mod concentrates in the condensate of interest, as each condensate contains in the order of a hundred other kinds of gene products, the functional selectivity is likely to be very high because many other potential binding partners for that c-mod are not present in the condensate, leading to a high therapeutic index. In addition, if a c-mod can bind, even weakly, to multiple related proteins within the target condensate, this may yield a pharmacologically relevant effect.
Building a c-mod discovery platform
Converting our knowledge of biomolecular condensates and their involvement in disease into c-mods requires a novel drug discovery platform, which can be divided into four main parts (Fig. 4): identification of a disease-relevant target condensate, and formulation of a hypothesis on how modification of the target condensate could have desired functional effects in cellular models of disease (the condensate hypothesis); characterization of the target condensate and development of assays to measure such effects that enable validation of the hypothesis; high-throughput screening (HTS) to identify potential c-mods that reverse or prevent the aberrant condensate phenotype; and hit-to-lead optimization based on the desired, disease-relevant functional outcome. Such a discovery platform will be built on existing (reviewed in ref.35) as well as novel interdisciplinary assays.
a | The first step is validation of the condensate hypothesis by testing the correlation between the genetic alteration (NUP98 and HOX9 fusion), aberrant condensate phenotype and aberrant transcription of HOX genes. b | A proof-of-concept drug discovery pipeline for the NUP98–HOXA9 condensatopathy. A primary phenotypic high-throughput screen (HTS) in a cell line expressing NUP98–HOXA9 could identify compounds that change the morphology of aberrant condensates. Hit compounds with various chemotypes could be filtered and prioritized (for example, with the aid of machine learning/artificial intelligence or through traditional methods) based on various characteristics (two are shown in the graph). Selected hits would then move into secondary validation assays, where one or more functional outcomes are monitored, in disease-relevant cell lines (for example, genome occupancy by ChIP-seq, and in vitro pharmacology by proliferation kinetics) and in vivo activity (for example, tumour growth and survival rates in animal models). Lead compound characteristics would then be optimized in a panel of assays, ranging from in vitro binding studies to the target, biophysical characterization of the lead compound effects on composition and material properties of in vitro reconstituted and endogenous condensates, partitioning measurements, toxicity and off-target measurements, in addition to secondary functional assays. Parts a and b are adapted with permission from refs.168,169, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/), Elsevier.
A foundational piece in the development of a c-mod discovery pipeline is establishing a reliable connection between the target condensate phenotype and a disease-relevant functional read-out. For example, a correlation between the decapping activity of Dcp1/2 (ref.67) and condensate formation was determined by tracking the fluorescence of a dual-labelled RNA probe. MED1-IDR-induced condensation was correlated with the transcriptional output measured in an in vitro transcription assay81. Fluorescence microscopy was used to monitor and quantify actin polymerization in response to signalling cluster formation65. Splicing26 and cardiac defects in transgenic pigs26 were linked to the phenotypic observation of RBM20 condensatopathy. Behavioural changes (for example, crawling ability) in a Drosophila ALS model129 and survival curves in a cardiomyopathy pig model26 have been successfully used to correlate the condensatopathy with clinical presentations of the disease in vivo.
These functional assays should also be utilized again later in the pipeline as secondary hit validation assays and to optimize c-mod efficacy. To illustrate how such a c-mod discovery pipeline could look in practice, we use the example of repairing the NUP98–HOXA9 condensatopathy in AML, based on a range of complementary assays described by Chandra et al.168 and Xu et al.169 (Fig. 4).
Formulate a condensate hypothesis
The initial step of condensate-centric drug discovery is to select a target condensate and formulate a condensate hypothesis. Targets could originate from several sources, including pre-existing data on the disease relevance of a condensate, de novo identification of a condensatopathy or de novo identification of a condensate-association of a conventional target. Curated databases of genetic variants with strong disease association170,171, when combined with predictors of condensation-prone features172 of the mutated proteins, provide a rich source to draw upon for hypothesis generation and can help prioritize proteins that probably play central roles in condensate assembly52.
Additionally, computational methods (Box 1) have the potential to decode, a priori, how biomolecules cooperate to form condensates or how putative c-mods affect condensates, and to aid de novo discovery of c-mods. However, the complexity of condensates creates challenges for computational methods. Computational analysis can take the form of data mining the existing knowledge of condensate composition and the properties of simplified systems in vitro to generate predictions. In this vein, several databases of phase-separating proteins or condensate components have begun to be curated (reviewed in ref.173). Such databases can be an excellent first source when exploring a condensate hypothesis.
A hypothesis that a NUP98–HOXA9 condensatopathy is responsible for cellular transformation in AML cells can be formulated based on the following findings (Fig. 4a). First, human genetics data show a strong correlation between expression of NUP98 fusion oncogenes and AML clinical manifestation174. Second, expression of NUP98–HOXA9 in cultured cells induces formation of nuclear puncta, driven by the FG-repeat IDR of NUP98 (refs.168,175,176).
Condensate characterization
To understand the condensate, one must characterize, to the greatest extent possible, the community of biomolecules which comprise it. This information can be leveraged to characterize the condensatopathy, to select a suitable HTS phenotypic assay to identify c-mods and to interrogate the mechanism of action of c-mods. The compositional analysis can be performed via subcellular proteomic and transcriptomic analysis177 (Box 2) or multiplexed imaging methods178.
Due to the compositional complexity of condensates and the various parallel modes of regulation of phase behaviour, disentangling the contributions of specific components to the phenotype and behaviour of a condensate in living cells remains challenging.
Typically, live-cell fluorescence confocal microscopy is used to characterize the localization and emergent properties of condensates (reviewed in ref.179). Condensates with sizes below the limit of detection of conventional confocal microscopes136,180,181 may be visualized, albeit at the expense of speed and throughput, with the advanced techniques182 described in Box 3. In-cell phase boundaries of biomolecular condensates can be quantified by correlating the variable levels of expression of a fluorescently tagged marker protein with the formation of condensates58. This analysis can measure the effects of disease-associated mutations183, or identify co-scaffold interdependencies64,184. Furthermore, it can be readily implemented into the HTS pipeline to determine the identity of c-mods and obtain mechanistic insights.
Complementary to cellular assays, in vitro reconstituted condensates that recapitulate a subset of relevant features of the biological condensate can be used to address more specific questions related to the nature of interactions that drive condensation or are affected by c-mods. For example, monitoring the shift in the phase boundary and changes in emergent biophysical properties (such as number, size, morphology, material properties, dynamics and composition) as a function of various parameters (such as ionic strength, pH, ligand concentration and temperature) could identify the most promising points for therapeutic intervention inside the macromolecular network (reviewed in ref.179). This informs strategies for c-mod design (for example, a protein–ligand interaction, a hydrophobic-driven interaction or an electrostatically driven interaction) and hit optimization.
Methods for probing material properties, such as viscosity, surface tension and component dynamics (for example, diffusion and mobile fraction), can be applied in vitro and in live cells40,44,185 (Box 4); this information can be leveraged as a read-out to detect a change in condensate milieu, and/or to gain insights into the mechanisms driving a condensatopathy or a c-mod.
For example, the condensate hypothesis for the NUP98–HOXA9 condensatopathy is supported by the correlation between the phenotypic observation of aberrant nuclear puncta and transcriptional reprogramming of the HOX cluster and p53 that leads to leukaemogenesis in primary cells and mouse models168,169. The composition of these aberrant condensates has been characterized by proximity labelling proteomic assays186 (Box 2), as well as Co-IP and ChIP-seq169 assays showing an expansive network of interactions with chromatin remodelling factors168,169. NUP98–HOXA9 within nuclear condensates recovers from photobleaching in the order of seconds168, indicating dynamic on/off binding kinetics and rapid diffusion with and within the extended condensate network. Importantly, these disease-relevant functions depend on the ability of NUP98–HOXA9 to form condensates via FG motif multivalency, and the ability to nucleate condensates by binding DNA through the HOXA9 folded domain168.
Primary screens
At the onset of the HTS campaign for potential c-mods, one should have the following: a validated condensate hypothesis; a robust and scalable set of phenotypic and disease-relevant functional assays; and an assay that enables investigation of structure–activity relationships for hit-to-lead optimization. C-mods are selected based on phenotypic HTS, which monitors a combination of emergent properties (for example, size, number and morphology) and/or co-localization of selected markers. Considerations for the selection of the appropriate HTS set-up that balances speed, throughput and resolution/sensitivity as appropriate for the target condensate are discussed below.
High-content imaging assays can be optimized for screening of large libraries (~106 compounds), while monitoring the optical phenotype of condensates in live or fixed cells31,32,187, and in vitro reconstituted condensates188 with sizes above the diffraction limit. This approach has been used to identify hits that inhibit stress-induced aggregation of TDP43 (ref.187), stress granule formation31,32 and p53–Mdm2 interaction188.
The imaging technique depends on the size of the condensate. Generally, increasing the optical resolution and signal sensitivity comes at the expense of throughput. Initial target assessment and hit follow-up studies of 1,000 compounds might be achievable with the advanced techniques presented in Box 3. Alternatively, the model system can be altered to artificially increase the size of the condensate, by using optogenetics58,189,190 and repeat operon arrays30,136,138,191,192. These engineered systems and practical considerations for their selection are reviewed in ref.179.
Monitoring the material properties of condensates can also identify c-mods (Box 4). These methods are amenable to multiplexing and HTS applications, with libraries of up to 104–105 compounds, depending on the technique.
For the example of repairing the NUP98–HOXA9 condensatopathy, a phenotypic primary HTS would identify compounds that change the optical phenotype of the NUP98–HOXA9 nuclear puncta (Fig. 4b). A change from punctate to diffuse staining of NUP98–HOXA9 could indicate c-mods that dissolve the condensates, whereas a change to fewer, larger condensates could indicate c-mods that inhibit binding to chromatin168. Similar phenotypic screens have been performed to identify small molecules that prevent oxidative stress-induced TDP43 and G3BP1 cytoplasmic puncta formation in PC12 (ref.187), as well as HEK293xT and neural precursor cells31, respectively; these serve as model systems for ALS.
Secondary screens and hit optimization
The primary hits from the HTS are filtered based on cytotoxicity and condensate selectivity (for example, using a phenotypic screen against a panel of unrelated condensates), validated based on disease-relevant functional assays (for example, induced pluripotent stem cell-derived or patient-derived cells) and their mechanism of action characterized via biophysical measurements. Optimization of drug partitioning inside a target condensate (Box 5) provides the opportunity to improve the therapeutic index by increasing exposure of a drug to its target and minimizing off-target effects.
In our NUP98–HOXA9 condensatopathy example (Fig. 4b), the primary screen hits would be evaluated and further optimized in cell proliferation/transformation (for example, proliferation rates and colony formation) and/or transcriptional reprogramming (for example, qRT-PCR and ChIP-seq) assays, followed by validation in animal models (for example, tumour growth and survival)169. Leptomycin B (Table 3), a well-characterized inhibitor of the nucleocytoplasmic transporter CRM1, exhibited c-mod properties when it inhibited formation of NUP98–HOXA9 aberrant condensates, and transcriptional reprogramming193. We hypothesize that the c-mod acts by inhibiting CRM1-dependent nucleation of NUP98–HOXA9 condensates on chromatin168,193,194.
Outlook
Biomolecular condensates are emerging as attractive novel targets for drug discovery. Many proteins and nucleic acids of high therapeutic interest, including numerous targets previously considered ‘undruggable’, operate within condensates. Importantly, there is emerging evidence that condensates are ‘druggable’. First, some approved drugs have been shown to partition into condensates30. Second, high-content cellular screening has identified drug-like molecules that modulate condensate behaviours in a selective manner31,148,187. Third, it is now understood that PTMs can strongly regulate the formation, behaviour and dissolution of condensates. Taken together, it is tempting to speculate that many approved drugs may, in part, be acting as c-mods, for example by exerting a portion of their pharmacological benefit through the modification of disease-relevant condensates.
We hypothesize that condensates could represent nodes of misregulation in polygenic diseases. For example, mutations in binding motifs within IDRs (such as degrons, nuclear export and import signals) can alter regions of transient structure120 and/or the interactome of the affected protein126. Consequently, changes in the IDR interactome can lead to alterations in the condensate scaffolding, composition, dynamics, material properties and functional output. Alternatively, the mutations can lie outside canonical binding regions, where they might affect condensation by changing the physicochemical properties and valency. This paradigm might explain the pathophysiology of certain diseases that exhibit stereotyped phenotypes but complex genetic and environmental causes. Each individual type of ALS/FTD-associated genetic mutation accounts for a relatively small number of patients. However, despite differences in the cause of onset, all ALS subtypes share condensate dysfunction as the common denominator, namely formation and persistence of cytoplasmic TDP43 granules in affected neurons23. Targeting the condensate rather than individual mutations within that molecular community might provide an avenue to deliver broader therapies to a larger patient population. It is also possible that the dysfunctional cellular processes observed in some cancers are driven by mutations in diverse genes, all of which form a single condensate. This condensate may integrate oncogenic signals into a single output, such as a high proliferative capacity or sustained signalling.
The high complexity and dynamic nature of condensates raise several unique challenges and opportunities for c-mod discovery. For example, c-mods can exhibit unusual dose–response behaviour, which can vary depending on the experimental conditions. This behaviour, however, could provide insights into the mechanism of action of the c-mod, such as preferential engagement with one of the phases195 or engagements of multiple targets196. For this reason, a range of biologically relevant assay time frames, windows of drug treatment and phenotypic responses must be measured. In addition, tight control over assay conditions must be maintained to achieve the necessary assay reproducibility required for HTS and medicinal chemistry. Appropriate biochemical and disease-relevant functional read-outs are required to demonstrate clear correlations with the observed condensate phenotypes.
Because of the complexity of condensates, and the nature of the forces that lead to condensate formation, combinations of drugs that engage multiple components of the molecular community may be of particular importance. Furthermore, a c-mod may be envisioned that binds weakly to multiple sites on one protein or to many related proteins; such monovalent compounds, binding in a super-stoichiometric fashion, are expected to reduce the valency on the scaffolds, thereby destabilizing the condensate. To stabilize a conformational state that promotes interaction, one could adopt a molecular glue strategy to force biomolecules to remain in an associated or proximal state.
Many questions and challenges are topics of active investigation by the community. For example, how do we demonstrate causality between disease and condensatopathies? How do we identify ‘hub’ condensatopathies for polygenic diseases? What are the different signalling and regulatory pathways that are dysregulated via any one target condensate? What are the most informative components for understanding the function of the condensate and the effects of c-mods? To address each of these challenges, the drug-hunter must understand the individual components of a target condensate as well as the collective behaviour of the molecular community. However, this remains challenging, both due to technological limitations of spatial and temporal resolution as well as biological complexity (for example, fluctuations in composition due to stochastic variability in protein expression or differences in cell-cycle state).
The more complete the condensate map, the more opportunities for a successful drug discovery programme. The complexity of the condensate environment requires creative medicinal chemistry approaches to develop c-mods. For example, knowledge about drugging individual targets that localize to or regulate a condensate can be leveraged to create combination therapies or multifunctional drugs. This information may, in turn, address other challenges, including how to mitigate toxicity by, for example, avoiding inhibition of components that do not exclusively function within the target condensate, or overcoming drug resistance. A promising result in this direction has been reported for multiple myeloma, where patients with high expression of the protein SRC3 experience poor outcomes. Liu et al.197 discovered that resistance to the proteasome inhibitor bortezomib results from interactions between steroid receptor coactivator SRC3 and the histone methyltransferase NSD2, leading to the stabilization and phase separation of SRC3. A small-molecule compound, SI-2 (Table 3), disrupts the interaction between SRC3 and NSD2, eliminating the condensate and restoring the activity of bortezomib197.
Incorporating a ‘condensate perspective’ into the drug discovery process holds significant potential to create medicines that operate through fundamentally different mechanisms. However, to capitalize on the insights that are emerging in the condensate field, it is clear that a novel approach is required. We suggest that successful discovery of c-mods will result from integrating deep understanding of condensate properties and function, pragmatic drug discovery expertise, and robust commitment to the development and application of suitable technologies to measure emergent properties of condensates, to characterize the broad effects of c-mods on condensate behaviour and function, and to further understand the thermodynamics and kinetics of these interactions at a molecular level. Synergy between efforts in the biotechnology and pharmaceutical industries and academia, and expertise from disparate fields, has been and will continue to be the key for success in developing new medicines by targeting biomolecular condensates.
References
Wilson, E. B. The structure of protoplasm. Science 10, 33–45 (1899).
Hopkins, F. G. The dynamic side of biochemistry. Br. Med. J. 2, 713–717 (1913).
Heilbrunn, L. V. The colloid chemistry of protoplasm. Am. J. Physiol. 63, 481–498 (1923).
Oparin, A. I. The Origin of Life (Foreign Language Publishing House, 1936).
Pederson, T. The nucleolus. Cold Spring Harb. Perspect. Biol. 3, a000638 (2011).
Cajal, S. R. Un sencillo metodo de coloracion selectiva del reticulo protoplasmico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados [Spanish]. Trab. Lab. Investig. Biol. Univ. Madr. 2, 129–221 (1903).
Negri, A. Contributo allo studio dell’eziologia della rabbia [Italian]. Boll. Della Soc. Med. chirurgica di Pavia 2, 88–115 (1904).
Hegner, R. W. Effects of removing the germ-cell determinants from the eggs of some chrysomelid beetles. Biol. Bull. 16, 19–26 (1908).
Gall, J. G. The centennial of the Cajal body. Nat. Rev. Mol. Cell Biol. 4, 975–980 (2003).
Nevers, Q., Albertini, A. A., Lagaudrière-Gesbert, C. & Gaudin, Y. Negri bodies and other virus membrane-less replication compartments. Biochim. Biophys. Acta Mol. Cell Res. 1867, 118831 (2020).
Brangwynne, C. P. et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009).
Brangwynne, C. P., Mitchison, T. J. & Hyman, A. A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl Acad. Sci. USA 108, 4334–4339 (2011).
Li, P. et al. Phase transitions in the assembly of multi-valent signaling proteins. Nature 483, 336–340 (2012).
Han, T. W. et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779 (2012).
Kato, M. et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012).
Hanazawa, M., Yonetani, M. & Sugimoto, A. PGL proteins self associate and bind RNPs to mediate germ granule assembly in C. elegans. J. Cell Biol. 192, 929–937 (2011).
Hyman, A. A. & Brangwynne, C. P. Beyond stereospecificity: liquids and mesoscale organization of cytoplasm. Dev. Cell 21, 14–16 (2011).
Hyman, A. A. & Simons, K. Beyond oil and water — phase transitions in cells. Science 337, 1047–1049 (2012).
Keating, C. D. Aqueous phase separation as a possible route to compartmentalization of biological molecules. Acc. Chem. Res. 45, 2114–2124 (2012).
Brangwynne, C. P. Phase transitions and size scaling of membrane-less organelles. J. Cell Biol. 203, 875–881 (2013).
Hyman, A. A., Weber, C. A. & Jülicher, F. Liquid–liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014).
Mitrea, D. M. & Kriwacki, R. W. Phase separation in biology; functional organization of a higher order short linear motifs — the unexplored frontier of the eukaryotic proteome. Cell Commun. Signal. 14, 1–20 (2016).
Portz, B., Lee, B. L. & Shorter, J. FUS and TDP-43 phases in health and disease. Trends Biochem. Sci. 46, 550–563 (2021).
Zhang, F. et al. Dynamic phase separation of the androgen receptor and its coactivators to regulate gene expression. Preprint at bioRxiv https://doi.org/10.1101/2021.03.27.437301 (2021).
Risso-Ballester, J. et al. A condensate-hardening drug blocks RSV replication in vivo. Nature 595, 596–599 (2021).
Schneider, J. W. et al. Dysregulated ribonucleoprotein granules promote cardiomyopathy in RBM20 gene-edited pigs. Nat. Med. 26, 1788–1800 (2020).
Alberti, S. & Dormann, D. Liquid–liquid phase separation in disease. Annu. Rev. Genet. 53, 171–194 (2019).
Markmiller, S. et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604.e13 (2018).
Li, Y. R., King, O. D., Shorter, J. & Gitler, A. D. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372 (2013).
Klein, I. A. et al. Partitioning of cancer therapeutics in nuclear condensates. Science 368, 1386–1392 (2020).
Fang, M. Y. et al. Small-molecule modulation of TDP-43 recruitment to stress granules prevents persistent TDP-43 accumulation in ALS/FTD. Neuron 103, 802–819.e11 (2019).
Wheeler, R. J. et al. Small molecules for modulating protein driven liquid–liquid phase separation in treating neurodegenerative disease. Preprint at bioRxiv https://doi.org/10.1101/721001 (2019).
McSwiggen, D. T., Mir, M., Darzacq, X. & Tjian, R. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes. Dev. 33, 1619–1634 (2019).
Alberti, S., Gladfelter, A. & Mittag, T. Considerations and challenges in studying liquid–liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019).
Mitrea, D. M. et al. Methods for physical characterization of phase-separated bodies and membrane-less organelles. J. Mol. Biol. 430, 4773–4805 (2018).
Holehouse, A. S. & Pappu, R. V. Functional implications of intracellular phase transitions. Biochemistry 57, 2415–2423 (2018).
Fare, C. M., Villani, A., Drake, L. E. & Shorter, J. Higher-order organization of biomolecular condensates. Open. Biol. 11, 210137 (2021).
Choi, J. M., Holehouse, A. S. & Pappu, R. V. Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 49, 107–133 (2020).
Roden, C. & Gladfelter, A. S. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 22, 183–195 (2021).
Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016).
Weber, S. C. Sequence-encoded material properties dictate the structure and function of nuclear bodies. Curr. Opin. Cell Biol. 46, 62–71 (2017).
Patel, A. et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015).
Molliex, A. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015).
Woodruff, J. B. et al. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169, 1066–1077.e10 (2017).
Jain, A. & Vale, R. D. RNA phase transitions in repeat expansion disorders. Nature 546, 243–247 (2017).
Frey, S., Richter, R. P. & Görlich, D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817 (2006).
Boke, E. et al. Amyloid-like self-assembly of a cellular compartment. Cell 166, 637–650 (2016).
Audas, T. E. et al. Adaptation to stressors by systemic protein amyloidogenesis. Dev. Cell 39, 155–168 (2016).
Mittasch, M. et al. Regulated changes in material properties underlie centrosome disassembly during mitotic exit. J. Cell Biol. 219, e201912036 (2020).
Woodruff, J. B., Hyman, A. A. & Boke, E. Organization and function of non-dynamic biomolecular condensates. Trends Biochem. Sci. 43, 81–94 (2018).
Ditlev, J. A., Case, L. B. & Rosen, M. K. Who’s in and who’s out — compositional control of biomolecular condensates. J. Mol. Biol. 430, 4666–4684 (2018).
Wang, J. et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16 (2018).
Banani, S. F. et al. Compositional control of phase-separated cellular bodies. Cell 166, 651–663 (2016).
Mitrea, D. M. et al. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife 5, e13571 (2016).
Hubstenberger, A. et al. P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell 68, 144–157.e5 (2017).
Gibson, B. A. et al. Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470–484.e21 (2019).
Nott, T. J. et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 (2015).
Bracha, D. et al. Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell 175, 1467–1480.e13 (2018).
Xing, W., Muhlrad, D., Parker, R. & Rosen, M. K. A quantitative inventory of yeast P body proteins reveals principles of composition and specificity. eLife 9, e56525 (2020).
Matsuki, H. et al. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells 18, 135–146 (2013).
Kawasaki, I. et al. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94, 635–645 (1998).
Kawasaki, I. et al. The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics 167, 645–661 (2004).
Ferrolino, M. C., Mitrea, D. M., Michael, J. R. & Kriwacki, R. W. Compositional adaptability in NPM1–SURF6 scaffolding networks enabled by dynamic switching of phase separation mechanisms. Nat. Commun. 9, 5064 (2018).
Riback, J. A. et al. Composition-dependent thermodynamics of intracellular phase separation. Nature 581, 209–214 (2020).
Su, X. et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–600 (2016).
Du, M. & Chen, Z. J. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709 (2018).
Tibble, R. W., Depaix, A., Kowalska, J., Jemielity, J. & Gross, J. D. Biomolecular condensates amplify mRNA decapping by biasing enzyme conformation. Nat. Chem. Biol. 17, 615–623 (2021).
Peeples, W. & Rosen, M. K. Mechanistic dissection of increased enzymatic rate in a phase-separated compartment. Nat. Chem. Biol. 17, 693–702 (2021).
O’Flynn, B. G. & Mittag, T. The role of liquid–liquid phase separation in regulating enzyme activity. Curr. Opin. Cell Biol. 69, 70–79 (2021).
Prouteau, M. & Loewith, R. Regulation of cellular metabolism through phase separation of enzymes. Biomolecules 8, 160 (2018).
Yoo, H., Triandafillou, C. & Drummond, D. A. Cellular sensing by phase separation: using the process, not just the products. J. Biol. Chem. 294, 7151–7159 (2019).
Khong, A. et al. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 68, 808–820.e5 (2017).
Andersen, J. S. et al. Nucleolar proteome dynamics. Nature 433, 77–83 (2005).
Gaglia, G. et al. HSF1 phase transition mediates stress adaptation and cell fate decisions. Nat. Cell Biol. 22, 151–158 (2020).
Ninomiya, K. et al. lncRNA-dependent nuclear stress bodies promote intron retention through SR protein phosphorylation. EMBO J. 39, e102729 (2020).
Biamonti, G. & Vourc’h, C. Nuclear stress bodies. Cold Spring Harb. Perspect. Biol. 2, a000695 (2010).
Klosin, A. et al. Phase separation provides a mechanism to reduce noise in cells. Science 367, 464–468 (2020).
Larson, A. G. et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 (2017).
Strom, A. R. et al. Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017).
Falk, M. et al. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 570, 395–399 (2019).
Sabari, B. R. et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018).
Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K. & Sharp, P. A. A phase separation model for transcriptional control. Cell 169, 13–23 (2017).
Pessina, F. et al. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat. Cell Biol. 21, 1286–1299 (2019).
Etibor, T. A., Yamauchi, Y. & Amorim, M. J. Liquid biomolecular condensates and viral lifecycles: review and perspectives. Viruses 13, 9–14 (2021).
Alberti, S. & Hyman, A. A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 22, 196–213 (2021).
Mitrea, D. M. et al. Self-interaction of NPM1 modulates multiple mechanisms of liquid–liquid phase separation. Nat. Commun. 9, 842 (2018).
Patel, A. et al. ATP as a biological hydrotrope. Science 356, 753–756 (2017).
Mateju, D. et al. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 36, 1669–1687 (2017).
Dao, T. P. et al. Ubiquitin modulates liquid–liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell 69, 965–978.e6 (2018).
Kroschwald, S. et al. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife 4, e06807 (2015).
Frottin, F. et al. The nucleolus functions as a phase-separated protein quality control compartment. Science 365, 342–347 (2019).
Bah, A. & Forman-Kay, J. D. Modulation of intrinsically disordered protein function by post-translational modifications. J. Biol. Chem. 291, 6696–6705 (2016).
Hofweber, M. & Dormann, D. Friend or foe-post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 294, 7137–7150 (2019).
Spannl, S., Tereshchenko, M., Mastromarco, G. J., Ihn, S. J. & Lee, H. O. Biomolecular condensates in neurodegeneration and cancer. Traffic 20, 890–911 (2019).
Snead, W. T. & Gladfelter, A. S. The control centers of biomolecular phase separation: how membrane surfaces, PTMs, and active processes regulate condensation. Mol. Cell 76, 295–305 (2019).
Springhower, C. E., Rosen, M. K. & Chook, Y. M. Karyopherins and condensates. Curr. Opin. Cell Biol. 64, 112–123 (2020).
Hondele, M. et al. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148 (2019).
Yoshizawa, T. et al. Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell 173, 693–705 (2018).
Guo, L. et al. Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173, 677–692 (2018).
Elbaum-Garfinkle, S. et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl Acad. Sci. USA 112, 7189–7194 (2015).
Zhang, H. et al. RNA controls PolyQ protein phase transitions. Mol. Cell 60, 220–230 (2015).
Langdon, E. M. et al. mRNA structure determines specificity ofa polyQ-driven phase separation. Science 360, 922–927 (2018).
Boeynaems, S. et al. Spontaneous driving forces give rise to protein−RNA condensates with coexisting phases and complex material properties. Proc. Natl Acad. Sci. USA 116, 7889–7898 (2019).
Maharana, S. et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921 (2018).
Banerjee, P. R., Milin, A. N., Moosa, M., Onuchic, P. L. & Deniz, A. A. Reentrant phase transition drives dynamic substructure formation in ribonucleoprotein droplets. Angew. Chem. Int. Ed. 56, 11354–11359 (2017).
Krainer, G. et al. Reentrant liquid condensate phase of proteins is stabilized by hydrophobic and non-ionic interactions. Nat. Commun. 12, 1085 (2021).
Burke, K. A., Janke, A. M., Rhine, C. L. & Fawzi, N. L. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell 60, 231–241 (2015).
Henninger, J. E. et al. RNA-mediated feedback control of transcriptional condensates. Cell 184, 207–225.e24 (2021).
McGurk, L. et al. Poly(ADP-ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell 71, 703–717.e9 (2018).
Altmeyer, M. et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 6, 8088 (2015).
An, H. et al. ALS-linked cytoplasmic FUS assemblies are compositionally different from physiological stress granules and sequester hnRNPA3, a novel modifier of FUS toxicity. Neurobiol. Dis. 162, 105585 (2021).
Guillén-Boixet, J. et al. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361.e17 (2020).
Yang, P. et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345.e28 (2020).
Brady, J. P. et al. Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc. Natl Acad. Sci. USA 114, E8194–E8203 (2017).
Murray, D. T. et al. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell 171, 615–627.e16 (2017).
Lin, Y. et al. Redox-mediated regulation of an evolutionarily conserved cross-β structure formed by the TDP43 low complexity domain. Proc. Natl Acad. Sci. USA 117, 28727–28734 (2020).
Conicella, A. E. et al. TDP-43 α-helical structure tunes liquid–liquid phase separation and function. Proc. Natl Acad. Sci. USA 117, 5883–5894 (2020).
Jain, S. et al. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 (2016).
Wheeler, J. R., Matheny, T., Jain, S., Abrisch, R. & Parker, R. Distinct stages in stress granule assembly and disassembly. eLife 5, e18413 (2016).
Fei, J. et al. Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J. Cell Sci. 130, 4180–4192 (2017).
Ma, W. & Mayr, C. A membraneless organelle associated with the endoplasmic reticulum enables 3′UTR-mediated protein–protein interactions. Cell 175, 1492–1506.e19 (2018).
Bradner, J. E., Hnisz, D. & Young, R. A. Transcriptional addiction in cancer. Cell 168, 629–643 (2016).
Sabari, B. R., Dall’Agnese, A. & Young, R. A. Biomolecular condensates in the nucleus. Trends Biochem. Sci. 45, 961–977 (2020).
Boija, A., Klein, I. A. & Young, R. A. Biomolecular condensates and cancer. Cancer Cell 39, 174–192 (2021).
Zhang, P. et al. Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS–FTD pathology. eLife 8, e39578 (2019).
Taylor, J. P., Brown, R. H. J. & Cleaveland, D. W. Decoding ALS: from genes to mechanism. Nature 539, 197–206 (2016).
Gopal, P. P., Nirschl, J. J., Klinman, E. & Holzbaurb, E. L. F. Amyotrophic lateral sclerosis-linked mutations increase the viscosity of liquid-like TDP-43 RNP granules in neurons. Proc. Natl Acad. Sci. USA 114, E2466–E2475 (2017).
Klim, J. R. et al. ALS implicated protein TDP-43 sustains levels of STMN2 a mediator of motor neuron growth and repair. Nat. Neurosci. 22, 167–179 (2019).
Otte, C. G. et al. Optogenetic TDP-43 nucleation induces persistent insoluble species and progressive motor dysfunction in vivo. Neurobiol. Dis. 146, 105078 (2020).
Freibaum, B. D. & Taylor, J. P. The role of dipeptide repeats in C9ORF72-related ALS-FTD. Front. Mol. Neurosci. 10, 35 (2017).
Lee, K. H. et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774–788.e17 (2016).
White, M. R. et al. C9orf72 poly(PR) dipeptide repeats disturb biomolecular phase separation and disrupt nucleolar function. Mol. Cell 74, 713–728.e6 (2019).
Ihara, K. et al. A missense mutation in the RSRSP stretch of Rbm20 causes dilated cardiomyopathy and atrial fibrillation in mice. Sci. Rep. 10, 17894 (2020).
Jiang, S., Fagman, J. B., Chen, C., Alberti, S. & Liu, B. Protein phase separation and its role in tumorigenesis. eLife 9, e60264 (2020).
Lu, J. et al. Emerging roles of liquid–liquid phase separation in cancer: from protein aggregation to immune-associated signaling. Front. Cell Dev. Biol. 9, 631486 (2021).
Boija, A. et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16 (2018).
Lovén, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Chong, S. et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361, eaar2555 (2018).
Terlecki-Zaniewicz, S. et al. Biomolecular condensation of NUP98 fusion proteins drives leukemogenic gene expression. Nat. Struct. Mol. Biol. 28, 190–201 (2021).
Michmerhuizen, N. L., Klco, J. M. & Mullighan, C. G. Mechanistic insights and potential therapeutic approaches for NUP98-rearranged hematologic malignancies. Blood 136, 2275–2289 (2020).
Su, J. M., Wilson, M. Z., Samuel, C. E. & Ma, D. Formation and function of liquid-like viral factories in negative-sense single-stranded rna virus infections. Viruses 13, 126 (2021).
Nikolic, J. et al. Negri bodies are viral factories with properties of liquid organelles. Nat. Commun. 8, 58 (2017).
Ma, X. et al. Histone chaperone CAF-1 promotes HIV-1 latency by leading the formation of phase-separated suppressive nuclear bodies. EMBO J. 40, e106632 (2021).
Wang, S. et al. Targeting liquid–liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nat. Cell Biol. 23, 718–732 (2021).
Iserman, C. et al. Genomic RNA elements drive phase separation of the SARS-CoV-2 nucleocapsid. Mol. Cell 80, 1078–1091.e6 (2020).
Perdikari, T. M. et al. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. 39, e106478 (2020).
Warner, K. D., Hajdin, C. E. & Weeks, K. M. Principles for targeting RNA with drug-like small molecules. Nat. Rev. Drug. Discov. 17, 547–558 (2018).
Mann, J. R. et al. RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron 102, 321–338.e8 (2019).
Bernkopf, D. B., Daum, G., Brückner, M. & Behrens, J. Sulforaphane inhibits growth and blocks Wnt/β-catenin signaling of colorectal cancer cells. Oncotarget 9, 33982–33994 (2018).
Mukherjee, H. et al. Interactions of the natural product (+)-avrainvillamide with nucleophosmin and exportin-1 mediate the cellular localization of nucleophosmin and its aml-associated mutants. ACS Chem. Biol. 10, 855–863 (2015).
Boike, L. et al. Discovery of a functional covalent ligand targeting an intrinsically disordered cysteine within MYC. Cell Chem. Biol. 28, 4–13.e17 (2021).
De Mol, E. et al. EPI-001, a compound active against castration-resistant prostate cancer, targets transactivation unit 5 of the androgen receptor. ACS Chem. Biol. 11, 2499–2505 (2016).
Iconaru, L. I. et al. Discovery of small molecules that inhibit the disordered protein, p27 Kip1. Sci. Rep. 5, 15686 (2015).
Iconaru, L. I. et al. Small molecule sequestration of the intrinsically disordered protein, p27Kip1, within soluble oligomers. J. Mol. Biol. 433, 167120 (2021).
Dale, B. et al. Advancing targeted protein degradation for cancer therapy. Nat. Rev. Cancer 21, 638–654 (2021).
Gallego, L. D. et al. Phase separation directs ubiquitination of gene body nucleosomes. Nature 579, 592–597 (2020).
Zhang, L., Cao, J., Dong, L. & Lin, H. TiPARP forms nuclear condensates to degrade HIF-1α and suppress tumorigenesis. Proc. Natl Acad. Sci. USA 117, 13447–13456 (2020).
Yasuda, S. et al. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 578, 296–300 (2020).
Albert, S. et al. Direct visualization of degradation microcompartments at the ER membrane. Proc. Natl Acad. Sci. USA 117, 1069–1080 (2020).
Hyun, S. & Shin, D. Chemical-mediated targeted protein degradation in neurodegenerative diseases. Life 11, 607 (2021).
Costales, M. G., Suresh, B., Vishnu, K. & Disney, M. D. Targeted degradation of a hypoxia-associated non-coding RNA enhances the selectivity of a small molecule interacting with RNA. Cell Chem. Biol. 26, 1180–1186.e5 (2019).
Fu, A., Cohen-Kaplan, V., Avni, N., Livneh, I. & Ciechanover, A. p62-containing, proteolytically active nuclear condensates, increase the efficiency of the ubiquitin–proteasome system. Proc. Natl Acad. Sci. USA 118, e2107321118 (2021).
Li, Z. et al. Allele-selective lowering of mutant HTT protein by HTT–LC3 linker compounds. Nature 575, 203–209 (2019).
Ries, R. J. et al. m6A enhances the phase separation potential of mRNA. Nature 571, 424–428 (2019).
Erdel, F. et al. Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid–liquid phase separation. Mol. Cell 78, 236–249.e7 (2020).
Reed, E. H., Schuster, B. S., Good, M. C. & Hammer, D. A. SPLIT: stable protein coacervation using a light Induced transition. ACS Synth. Biol. 9, 500–507 (2020).
Wei, M. T. et al. Nucleated transcriptional condensates amplify gene expression. Nat. Cell Biol. 22, 1187–1196 (2020).
Chandra, B. et al. Phase separation mediates NUP98 fusion oncoprotein leukemic transformation. Cancer Discov. 12, 1152–1169 (2022).
Xu, H. et al. NUP98 fusion proteins interact with the NSL and MLL1 complexes to drive leukemogenesis. Cancer Cell 30, 863–878 (2016).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Plenge, R. M., Scolnick, E. M. & Altshuler, D. Validating therapeutic targets through human genetics. Nat. Rev. Drug. Discov. 12, 581–594 (2013).
Vernon, R. M. C. et al. Pi–Pi contacts are an overlooked protein feature relevant to phase separation. eLife 7, e31486 (2018).
Li, Q. et al. Protein databases related to liquid–liquid phase separation. Int. J. Mol. Sci. 21, 6796 (2020).
Gough, S. M., Slape, C. I. & Aplan, P. D. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood 118, 6247–6257 (2011).
Schmidt, H. B. & Görlich, D. Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. eLife 4, e04251 (2015).
Ahn, J. H. et al. Phase separation drives aberrant chromatin looping and cancer development. Nature 595, 591–595 (2021).
Christopher, J. A. et al. Subcellular proteomics. Nat. Rev. Methods Primers 1, 32 (2021).
Chandrasekaran, S. N., Ceulemans, H., Boyd, J. D. & Carpenter, A. E. Image-based profiling for drug discovery: due for a machine-learning upgrade? Nat. Rev. Drug. Discov. 20, 145–159 (2021).
Levin, B. et al. Harnessing the power of fluorescence to characterize biomolecular condensates. Methods Microbiol. 48, 1–47 (2021).
Favard, C. et al. HIV-1 Gag specifically restricts PI(4,5)P2 and cholesterol mobility in living cells creating a nanodomain platform for virus assembly. Sci. Adv. 5, eaaw8651 (2019).
Cho, W. K. et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 (2018).
Shakya, A. & King, J. T. Modern optical microscopy methods to study biomolecular condensates. Curr. Opin. Colloid Interface Sci. 52, 101421 (2021).
Heidenreich, M. et al. Designer protein assemblies with tunable phase diagrams in living cells. Nat. Chem. Biol. 16, 939–945 (2020).
Sanders, D. W. et al. Competing protein–RNA interaction networks control multiphase intracellular organization. Cell 181, 306–324.e28 (2020).
Hobson, C. M., O’Brien, E. T., Falvo, M. R. & Superfine, R. Combined selective plane illumination microscopy and FRAP maps intranuclear diffusion of NLS-GFP. Biophys. J. 119, 514–524 (2020).
Mendes, A., Jühlen, R., Bousbata, S. & Fahrenkrog, B. Disclosing the interactome of leukemogenic NUP98–HOXA9 and SET–NUP214 fusion proteins using a proteomic approach. Cells 9, 1666 (2020).
Boyd, J. D. et al. A high content screen identifies novel compounds that inhibit stress-induced TDP-43 cellular aggregation and associated cytotoxicity. J. Biomol. Screen. 19, 44–56 (2014).
Zhou, M. et al. Phase-separated condensate-aided enrichment of biomolecular interactions for high-throughput drug screening in test tubes. J. Biol. Chem. 295, 11420–11434 (2020).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Shin, Y. et al. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell 168, 159–171 (2017).
Janicki, S. M. et al. From silencing to gene expression: real-time analysis in single cells. Cell 116, 683–698 (2004).
Zamudio, A. V. et al. Mediator condensates localize signaling factors to key cell identity genes. Mol. Cell 76, 753–766.e6 (2019).
Oka, M. et al. Chromatin-prebound Crm1 recruits Nup98–HoxA9 fusion to induce aberrant expression of Hox cluster genes. eLife 5, e09540 (2016).
Oka, M. et al. Chromatin-bound CRM1 recruits SET-Nup214 and NPM1c onto HOX clusters causing aberrant HOX expression in leukemia cells. eLife 8, e46667 (2019).
Ruff, K. M., Dar, F. & Pappu, R. V. Ligand effects on phase separation of multivalent macromolecules. Proc. Natl Acad. Sci. USA 118, e2017184118 (2021).
Douglass, E. F., Miller, C. J., Sparer, G., Shapiro, H. & Spiegel, D. A. A comprehensive mathematical model for three-body binding equilibria. J. Am. Chem. Soc. 135, 6092–6099 (2013).
Liu, J. et al. Targeting NSD2-mediated SRC-3 liquid–liquid phase separation sensitizes bortezomib treatment in multiple myeloma. Nat. Commun. 12, 1022 (2021).
Theodoridis, P. R. et al. Local translation in nuclear condensate amyloid bodies. Proc. Natl Acad. Sci. USA 118, e2014457118 (2021).
Wang, M., Bokros, M., Theodoridis, P. R. & Lee, S. Nucleolar sequestration: remodeling nucleoli into amyloid bodies. Front. Genet. 10, 1179 (2019).
Machyna, M., Heyn, P. & Neugebauer, K. M. Cajal bodies: where form meets function. Wiley Interdiscip. Rev. RNA 4, 17–34 (2013).
Li, L. et al. Dynamic nature of cleavage bodies and their spatial relationship to DDX1 bodies, Cajal bodies, and gems. Mol. Biol. Cell 17, 1126–1140 (2006).
Fijen, C. & Rothenberg, E. The evolving complexity of DNA damage foci: RNA, condensates and chromatin in DNA double-strand break repair. DNA Repair 105, 103170 (2021).
Bekker-Jensen, S. & Mailand, N. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair 9, 1219–1228 (2010).
Morimoto, M. & Boerkoel, C. F. The role of nuclear bodies in gene expression and disease. Biology 2, 976–1033 (2013).
Larson, A. G. & Narlikar, G. J. The role of phase separation in heterochromatin formation, function, and regulation. Biochemistry 57, 2540–2548 (2018).
Duronio, R. J. & Marzluff, W. F. Coordinating cell cycle-regulated histone gene expression through assembly and function of the Histone locus body. RNA Biol. 14, 726–738 (2017).
Chen, Y. & Belmont, A. S. Genome organization around nuclear speckles. Curr. Opin. Genet. Dev. 55, 91–99 (2019).
Galganski, L., Urbanek, M. O. & Krzyzosiak, W. J. Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res. 45, 10350–10368 (2017).
Spector, D. L. & Lamond, A. I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 3, a000646 (2011).
Rawat, P. et al. Stress-induced nuclear condensation of NELF drives transcriptional downregulation. Mol. Cell 81, 1013–1026.e11 (2021).
Lafontaine, D. L. J., Riback, J. A., Bascetin, R. & Brangwynne, C. P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 22, 165–182 (2021).
Berkamp, S., Mostafavi, S. & Sachse, C. Structure and function of p62/SQSTM1 in the emerging framework of phase separation. FEBS J. 288, 6927–6941 (2021).
Hsu, K. S. & Kao, H. Y. PML: regulation and multifaceted function beyond tumor suppression. Cell Biosci. 8, 5 (2018).
Everett, R. D. & Chelbi-Alix, M. K. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89, 819–830 (2007).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934 (2013).
Schmidt, H. B. & Görlich, D. Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem. Sci. 41, 46–61 (2016).
Vasquez-Limeta, A. & Loncarek, J. Human centrosome organization and function in interphase and mitosis. Semin. Cell Dev. Biol. 117, 30–41 (2021).
Fox, A. H., Nakagawa, S., Hirose, T. & Bond, C. S. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem. Sci. 43, 124–135 (2018).
Pisani, G. & Baron, B. Nuclear paraspeckles function in mediating gene regulatory and apoptotic pathways. Noncoding RNA Res. 4, 128–134 (2019).
Yamazaki, T. et al. Paraspeckles are constructed as block copolymer micelles. EMBO J. 40, e107270 (2021).
Luo, Y., Na, Z. & Slavoff, S. A. P-bodies: composition, properties, and functions. Biochemistry 57, 2424–2431 (2018).
Standart, N. & Weil, D. P-bodies: cytosolic droplets for coordinated mRNA storage. Trends Genet. 34, 612–626 (2018).
Updike, D. & Strome, S. P granule assembly and function in Caenorhabditis elegans germ cells. J. Androl. 31, 53–60 (2010).
Costa Dalla, I. et al. The functional organization of axonal mRNA transport and translation. Nat. Rev. Neurosci. 22, 77–91 (2021).
Campos-Melo, D., Hawley, Z. C. E., Droppelmann, C. A. & Strong, M. J. The integral role of RNA in stress granule formation and function. Front. Cell Dev. Biol. 9, 621779 (2021).
Van Treeck, B. & Parker, R. Principles of stress granules revealed by imaging approaches. Cold Spring Harb. Perspect. Biol. 11, a033068 (2019).
Liu, J. L. & Gall, J. G. U bodies are cytoplasmic structures that contain uridine-rich small nuclear ribonucleoproteins and associate with P bodies. Proc. Natl Acad. Sci. USA 104, 11655–11659 (2007).
Chen, X., Wu, X., Wu, H. & Zhang, M. Phase separation at the synapse. Nat. Neurosci. 23, 301–310 (2020).
McDonald, N. A. & Shen, K. Finding functions of phase separation in the presynapse. Curr. Opin. Neurobiol. 69, 178–184 (2021).
Al-Husini, N. et al. BR-bodies provide selectively permeable condensates that stimulate mRNA decay and prevent release of decay intermediates. Mol. Cell 78, 670–682.e8 (2020).
Vladimirova, O. et al. Phase separation and DAXX redistribution contribute to LANA nuclear body and KSHV genome dynamics during latency and reactivation. PLoS Pathog. 17, e1009231 (2021).
López, D. J., Rodríguez, J. A. & Bañuelos, S. Nucleophosmin, a multifunctional nucleolar organizer with a role in DNA repair. Biochim. Biophys. Acta Proteins Proteom. 1868, 140532 (2020).
Di Matteo, A. et al. Molecules that target nucleophosmin for cancer treatment: an update. Oncotarget 7, 44821–44840 (2016).
Cheng, K. et al. The leukemia-associated cytoplasmic nucleophosmin mutant is an oncogene with paradoxical functions: Arf inactivation and induction of cellular senescence. Oncogene 26, 7391–7400 (2007).
Hiscox, J. A. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5, 119–127 (2007).
Hofmann, S., Stubbe, M., Mai, J. & Schreiner, S. Double-edged role of PML nuclear bodies during human adenovirus infection. Virus Res. 295, 198280 (2021).
Advani, V. M. & Ivanov, P. Stress granule subtypes: an emerging link to neurodegeneration. Cell. Mol. Life Sci. 77, 4827–4845 (2020).
Zhang, Q., Sharma, N. R., Zheng, Z. M. & Chen, M. Viral regulation of RNA granules in infected. Cell Virol. Sin. 34, 175–191 (2019).
White, J. P. & Lloyd, R. E. Regulation of stress granules in virus systems. Trends Microbiol. 20, 175–183 (2012).
Andresen, V. et al. Anti-proliferative activity of the NPM1 interacting natural product avrainvillamide in acute myeloid leukemia. Cell Death Dis. 7, e2497 (2016).
Lemos, C. et al. Identification of small molecules that modulate mutant p53 condensation. iScience 23, 101517 (2020).
Gallo, R., Rai, A. & Pelkmans, L. DYRK3-controlled phase separation organizes the early secretory pathway. Preprint at bioRxiv https://doi.org/10.1101/2020.02.10.941757 (2020).
Hoerr, V. et al. Characterization and prediction of the mechanism of action of antibiotics through NMR metabolomics. BMC Microbiol. 16, 82 (2016).
Schmidt, H. B. et al. Oxaliplatin kills cells via liquid–liquid demixing of nucleoli. Preprint at bioRxiv https://doi.org/10.1101/2021.06.10.447918 (2021).
Leone, A. et al. Sulforaphane for the chemoprevention of bladder cancer: molecular mechanism targeted approach. Oncotarget 8, 35412–35424 (2017).
Bolognesi, B. et al. A concentration-dependent liquid phase separation can cause toxicity upon increased protein expression. Cell Rep. 16, 222–231 (2016).
Lancaster, A. K., Nutter-Upham, A., Lindquist, S. & King, O. D. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30, 2501–2502 (2014).
Shen, B. et al. Computational screening of phase-separating proteins. Genom. Proteom. Bioinform. 19, 13–24 (2021).
Oates, M. E. et al. D2P2: database of disordered protein predictions. Nucleic Acids Res. 41, 508–516 (2013).
Piovesan, D. et al. MobiDB 3.0: more annotations for intrinsic disorder, conformational diversity and interactions in proteins. Nucleic Acids Res. 46, D471–D476 (2018).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Nguyen Ba, A. N., Pogoutse, A., Provart, N. & Moses, A. M. NLStradamus: a simple hidden Markov model for nuclear localization signal prediction. BMC Bioinformatics 10, 202 (2009).
Ba, A. N. N. et al. Proteome-wide discovery of evolutionary conserved sequences in disordered regions. Sci. Signal. 5, rs1 (2016).
Davey, N. E., Shields, D. C. & Edwards, R. J. SLiMDisc: short, linear motif discovery, correcting for common evolutionary descent. Nucleic Acids Res. 34, 3546–3554 (2006).
Zhu, J., Salvatella, X. & Robustelli, P. Small molecules targeting the disordered transactivation domain of the androgen receptor induce the formation of collapsed helical states. Preprint at bioiRxiv https://doi.org/10.1101/2021.12.23.474012 (2021).
Qin, W., Cho, K. F., Cavanagh, P. E. & Ting, A. Y. Deciphering molecular interactions by proximity labeling. Nat. Methods 18, 133–143 (2021).
Dunham, W. H., Mullin, M. & Gingras, A. C. Affinity-purification coupled to mass spectrometry: basic principles and strategies. Proteomics 12, 1576–1590 (2012).
Choi-Rhee, E., Schulman, H. & Cronan, J. E. Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci. 13, 3043–3050 (2004).
Lam, S. S. et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54 (2014).
Kotani, N. et al. Biochemical visualization of cell surface molecular clustering in living cells. Proc. Natl Acad. Sci. USA 105, 7405–7409 (2008).
Kim, D. I. et al. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell 27, 1188–1196 (2016).
Ramanathan, M. et al. RN A–protein interaction detection in living cells. Nat. Methods 15, 207–212 (2018).
Branon, T. C. et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–898 (2018).
Yap, K., Chung, T. H. & Makeyev, E. V. Hybridization-proximity labeling reveals spatially ordered interactions of nuclear RNA compartments. Mol. Cell 82, 463–478.e11 (2022).
Chou, C. C. et al. TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 21, 228–239 (2018).
Dingar, D. et al. BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors. J. Proteom. 118, 95–111 (2015).
Gingras, A. C., Abe, K. T. & Raught, B. Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles. Curr. Opin. Chem. Biol. 48, 44–54 (2019).
Garcia, D. A. et al. An intrinsically disordered region-mediated confinement state contributes to the dynamics and function of transcription factors. Mol. Cell 81, 1484–1498.e6 (2021).
Rego, E. H. et al. Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50-nm resolution. Proc. Natl Acad. Sci. USA 109, E135–E143 (2012).
Chen, B.-C. et al. Lattice light sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 (2014).
Wang, J. T. et al. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife 3, e04591 (2014).
Narayanan, A. et al. A first order phase transition mechanism underlies protein aggregation in mammalian cells. eLife 8, e39695 (2019).
Yang, B. et al. Epi-illumination SPIM for volumetric imaging with high spatial-temporal resolution. Nat. Methods 16, 501–504 (2019).
Azuma, T. & Kei, T. Super-resolution spinning-disk confocal microscopy using optical photon reassignment. Opt. Express 23, 15003 (2015).
Huff, J. The Airyscan detector from ZEISS: confocal imaging with improved signal-to-noise ratio and super-resolution. Nat. Methods 12, i–ii (2015).
Zakrzewski, F. et al. Automated detection of the HER2 gene amplification status in fluorescence in situ hybridization images for the diagnostics of cancer tissues. Sci. Rep. 9, 8231 (2019).
Weigert, M. et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods 15, 1090–1097 (2018).
Wang, H. et al. Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat. Methods 16, 103–110 (2019).
Taylor, N. O., Wei, M. T., Stone, H. A. & Brangwynne, C. P. Quantifying dynamics in phase-separated condensates using fluorescence recovery after photobleaching. Biophys. J. 117, 1285–1300 (2019).
Wolstenholme, C. H. et al. AggFluor: fluorogenic toolbox enables direct visualization of the multi-step protein aggregation process in live cells. J. Am. Chem. Soc. 142, 17515–17523 (2020).
Laine, R. F. et al. Fast fluorescence lifetime imaging reveals the aggregation processes of α-synuclein and polyglutamine in aging Caenorhabditis elegans. ACS Chem. Biol. 14, 1628–1636 (2019).
Tian, F. et al. Monitoring peripheral nerve degeneration in ALS by label-free stimulated Raman scattering imaging. Nat. Commun. 7, 13283 (2016).
Picardi, G. et al. Tissue degeneration in ALS affected spinal cord evaluated by Raman spectroscopy. Sci. Rep. 8, 4–10 (2018).
Miao, K. & Wei, L. Live-cell imaging and quantification of PolyQ aggregates by stimulated Raman scattering of selective deuterium labeling. ACS Cent. Sci. 6, 478–486 (2020).
Putnam, A., Cassani, M., Smith, J. & Seydoux, G. A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol. 26, 220–226 (2019).
Fritsch, A. W. et al. Local thermodynamics govern formation and dissolution of Caenorhabditis elegans P granule condensates. Proc. Natl Acad. Sci. USA 118, e2102772118 (2021).
Mittasch, M. et al. Non-invasive perturbations of intracellular flow reveal physical principles of cell organization. Nat. Cell Biol. 20, 344–351 (2018).
Kroschwald, S., Maharana, S. & Simon, A. Hexanediol: a chemical probe to investigate the material properties of membrane-less compartments. Matters https://doi.org/10.19185/matters.201702000010 (2017).
Hernández-Candia, C. N., Pearce, S. & Tucker, C. L. A modular tool to query and inducibly disrupt biomolecular condensates. Nat. Commun. 12, 1809 (2021).
Chu, Y. H. et al. Systemic delivery and biodistribution of cisplatin in vivo. Mol. Pharm. 13, 2677–2682 (2016).
Marrero-Alonso, J. et al. Unique SERM-like properties of the novel fluorescent tamoxifen derivative FLTX1. Eur. J. Pharm. Biopharm. 85, 898–910 (2013).
Freibaum, B. D., Messing, J., Yang, P., Kim, H. J. & Taylor, J. P. High-fidelity reconstitution of stress granules and nucleoli in mammalian cellular lysate. J. Cell Biol. 220, e202009079 (2021).
Tipping, W. J., Lee, M., Serrels, A., Brunton, V. G. & Hulme, A. N. Stimulated Raman scattering microscopy: an emerging tool for drug discovery. Chem. Soc. Rev. 45, 2075–2089 (2016).
Sepp, K. et al. Utilizing stimulated Raman scattering microscopy to study intracellular distribution of label-free ponatinib in live cells. J. Med. Chem. 63, 2028–2034 (2020).
Coskun, A. F. et al. Nanoscopic subcellular imaging enabled by ion beam tomography. Nat. Commun. 12, 789 (2021).
Rovira-Clave, X. et al. Subcellular localization of biomolecules and drug distribution by high-definition ion beam imaging. Nat. Commun. 12, 4628 (2021).
Acknowledgements
The authors thank J. Bouchard for significant contributions to illustrations, literature review and manuscript editing, E. Martin for contributions to the computational methods section and M. Wagner, R. Young, P. Sharp, R. Pappu, A. Chakraborty, S. Mong, A. Gladfelter, S. Alberti, E. Boczek, K. Rizzolo, A. Nathwani and A. Patel for critical review of the manuscript. They thank R. Manoukian for the image of stress granules shown in Fig. 1.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
All authors are employees or board members of Dewpoint Therapeutics, a drug discovery company that studies condensates, and have a financial stake in the company.
Peer review
Peer review information
Nature Reviews Drug Discovery thanks John Lowe, Peter Tompa and the other, anonymous, reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Centrosome
-
The main microtubule organizing centre of all animal cells. It comprises a pair of centrioles on its core that are surrounded by a mass of proteins called pericentriolar material. Centrosomes nucleate microtubules that form the mitotic spindle upon cell division and are also involved in other functions such as cell polarity and cell signalling.
- Clients
-
Molecules that partition into condensates, but are not required for the condensate formation.
- C-mods
-
(Condensate-modifying therapeutics). Compounds that modify the properties of a biomolecular condensate.
- Condensate hypothesis
-
A testable proposition that links the modification of a particular biomolecular condensate (for example, normal or abnormal) to a desirable functional change in diseased cells.
- Condensatopathy
-
An aberration of a condensate that drives a specific disease phenotype.
- C sat
-
(Saturation concentration). The threshold concentration above which condensates form.
- Mesoscale assemblies
-
Biomolecular networks that span the nanometre-to-micrometre length scale.
- Multivalency
-
The presence of repeated, identical or similar interaction sites or domains on the same molecule.
- Nuclear pore
-
A nuclear pore is a protein assembly embedded in the nuclear envelope that provides a selectively permeable barrier via FG-rich intrinsically disordered regions separating the inside of the nucleus from the cytoplasm.
- Nucleolus
-
A ribonucleoprotein condensate that functions as the site of ribosome biogenesis and in stress sensing. Nucleoli exhibit a three-layer architecture, composed of the fibrillar centre (the site of ribosomal DNA transcription), the dense fibrillar component (the site of pre-ribosomal RNA processing) and the granular component (the site of ribososmal RNA assembly with ribosomal proteins into pre-ribosomal particles).
- Partition coefficient
-
The ratio between component concentrations inside versus outside the condensate.
- PARylation
-
(Poly-ADP-ribosylation). A post-translational modification (PTM) where polymers of ADP-ribose (poly(adenosine diphosphate ribose)) are covalently attached to proteins. This process is mediated by PAR polymerase enzymes.
- P granules
-
Perinuclear ribonucleoprotein granules found in Caenorhabditis elegans germline cells.
- RNA expansion repeats
-
Repeats of a short nucleotide sequence of variable length.
- Scaffolds
-
Multivalent biomolecules required for condensate formation.
- Stress granules
-
Membraneless compartments consisting of RNA and proteins that form in the cell cytoplasm upon stress conditions.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mitrea, D.M., Mittasch, M., Gomes, B.F. et al. Modulating biomolecular condensates: a novel approach to drug discovery. Nat Rev Drug Discov 21, 841–862 (2022). https://doi.org/10.1038/s41573-022-00505-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41573-022-00505-4
This article is cited by
-
Misfolded protein oligomers: mechanisms of formation, cytotoxic effects, and pharmacological approaches against protein misfolding diseases
Molecular Neurodegeneration (2024)
-
Phase separation-mediated biomolecular condensates and their relationship to tumor
Cell Communication and Signaling (2024)
-
The E3 ubiquitin ligase MARCH2 protects against myocardial ischemia-reperfusion injury through inhibiting pyroptosis via negative regulation of PGAM5/MAVS/NLRP3 axis
Cell Discovery (2024)
-
Targeting the chromatin binding of exportin-1 disrupts NFAT and T cell activation
Nature Chemical Biology (2024)
-
Pharmacological inhibition of α-synuclein aggregation within liquid condensates
Nature Communications (2024)