Abstract
Reproducibility has long been a tenet of science but has been challenging to achieve—we learned this the hard way when our old approaches proved inadequate to efficiently reproduce our own work. Here we describe how several free software tools have fundamentally upgraded our approach to collaborative research, making our entire workflow more transparent and streamlined. By describing specific tools and how we incrementally began using them for the Ocean Health Index project, we hope to encourage others in the scientific community to do the same—so we can all produce better science in less time.
Similar content being viewed by others
Science, now more than ever, demands reproducibility, collaboration and effective communication to strengthen public trust and effectively inform policy. Recent high-profile difficulties in reproducing and repeating scientific studies have put the spotlight on psychology and cancer biology1–3, but it is widely acknowledged that reproducibility challenges persist across scientific disciplines4–6. Environmental scientists face potentially unique challenges in achieving goals of transparency and reproducibility because they rely on vast amounts of data spanning natural, economic and social sciences that create semantic and synthesis issues exceeding those for most other disciplines7–9. Furthermore, proposed environmental solutions can be complex, controversial and resource intensive, increasing the need for scientists to work transparently and efficiently with data to foster understanding and trust.
Environmental scientists are expected to work effectively with ever-increasing quantities of highly heterogeneous data even though they are seldom formally trained to do so10–14. This was recently highlighted by a survey of 704 US National Science Foundation principal investigators in the biological sciences, which found training in data skills to be the largest unmet need15. Without training, scientists tend to develop their own bespoke workarounds to keep pace, but with this comes wasted time struggling to create their own conventions for managing, wrangling and versioning data. If done haphazardly or without a clear protocol, these efforts are likely to result in work that is not reproducible—by the scientist's own ‘future self’ or by anyone else12. As a team of environmental scientists tasked with reproducing our own science annually, we experienced this struggle first-hand. When we began our project, we worked with data in the same way as we always had, taking extra care to make our methods reproducible for planned future re-use. But when we began to reproduce our workflow a second time and repeat our methods with updated data, we found our approaches to reproducibility were insufficient. However, by borrowing philosophies, tools, and workflows primarily created for software development, we have been able to dramatically improve the ability for ourselves and others to reproduce our science, while also reducing the time involved to do so: the result is better science in less time (Fig. 1).
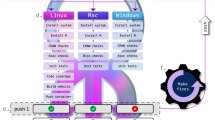
Every year since 2012 we have repeated Ocean Health Index (OHI) methods to track change in global ocean health36,37. Increased reproducibility and collaboration has reduced the amount of time required to repeat methods (size of bubbles) with updated data annually, allowing us to focus on improving methods each year (text labels show the biggest innovations). The original assessment in 2012 focused solely on scientific methods (for example, obtaining and analysing data, developing models, calculating, and presenting results; dark shading). In 2013, by necessity we gave more focus to data science (for example, data organization and wrangling, coding, versioning, and documentation; light shading), using open data science tools. We established R as the main language for all data preparation and modelling (using RStudio), which drastically decreased the time involved to complete the assessment. In 2014, we adopted Git and GitHub for version control, project management, and collaboration. This further decreased the time required to repeat the assessment. We also created the OHI Toolbox, which includes our R package ohicore for core analytical operations used in all OHI assessments. In subsequent years we have continued (and plan to continue) this trajectory towards better science in less time by improving code with principles of tidy data33; standardizing file and data structure; and focusing more on communication, in part by creating websites with the same open data science tools and workflow. See text and Table 1 for more details.
Here we share a tangible narrative of our transformation to better science in less time—meaning more transparent, reproducible, collaborative and openly shared and communicated science—with an aim of inspiring others. Our story is only one potential path because there are many ways to upgrade scientific practices—whether collaborating only with your ‘future self’ or as a team—and they depend on the shared commitment of individuals, institutions and publishers6,16,17. We do not review the important, ongoing work regarding data management architecture and archiving8,18, workflows11,19–21, sharing and publishing data22–25 and code25–27, or how to tackle reproducibility and openness in science28–32. Instead, we focus on our experience, because it required changing the way we had always worked, which was extraordinarily intimidating. We give concrete examples of how we use tools and practices from data science, the discipline of turning raw data into understanding33. It was out of necessity that we began to engage in data science, which we did incrementally by introducing new tools, learning new skills and creating deliberate workflows—all while maintaining annual deadlines. Through our work with academics, governments and non-profit groups around the world, we have seen that the need to improve practices is common if not ubiquitous. In this narrative we describe specific software tools, why we use them, how we use them in our workflow, and how we work openly as a collaborative team. In doing so we underscore two key lessons we learned that we hope encourage others to incorporate these practices into their own research. The first is that powerful tools exist and are freely available to use; the barriers to entry seem to be exposure to relevant tools and building confidence using them. The second is that engagement may best be approached as an evolution rather than as a revolution that may never come.
Improving reproducibility and collaboration
Having found that our homegrown conventions made it difficult to repeat our own data methods, we now use open data science tools that are created specifically to meet modern demands for collaborative data analysis and communication.
From then to now. The Ocean Health Index (OHI) operates at the interface of data-intensive marine science, coastal management and policy, and now, data science34,35. It is a scientific framework to quantify ocean-derived benefits to humans and to help inform sustainable ocean management using the best available information36,37. Assessments using the OHI framework require synthesizing heterogeneous data from nearly one hundred different sources, ranging from categorical tabular data to high-resolution remotely sensed satellite data. Methods must be reproducible, so that others can produce the same results, and also repeatable, so that newly available data can be incorporated in subsequent assessments. Repeated assessments using the same methods enable quantifiable comparison of changes in ocean health through time, which can be used to inform policy and track progress34.
Using the OHI framework, we lead annual global assessments of 220 coastal nations and territories, completing our first assessment in 201236. Despite our best efforts, we struggled to efficiently repeat our own work during the second assessment in 2013 because of our approaches to data preparation37. Data preparation is a critical aspect of making science reproducible but is seldom explicitly reported in research publications; we thought we had documented our methods sufficiently in 130 pages of published supplemental materials36, but we had not.
However, by adopting the data science principles and freely available tools that we describe below, we began building an ‘OHI Toolbox’ and fundamentally changed our approach to science (Fig. 1). The OHI Toolbox provides a file structure, data, code, and instruction, spans computer operating systems, and is shared online for free so that anyone can begin building directly from previous OHI assessments without reinventing the wheel34. While these changes required an investment of our team's time to learn and develop the necessary skills, the pay-off has been substantial. Most significantly we are now able to share and extend our workflow with a growing community of government, non-profit and academic collaborations around the world that use the OHI for science-driven marine management. There are currently two dozen OHI assessments underway, most of which are led by independent groups34, and the OHI Toolbox has helped lower the barriers to entry. Further, our own team has just released the fifth annual global OHI assessment38 and continues to lead assessments at smaller spatial scales, including the northeastern United States, where the OHI is included in President Obama's first Ocean Plan39.
We thought we were doing reproducible science. For the first global OHI assessment in 2012 we employed an approach to reproducibility that is standard to our field, which focused on scientific methods, not data science methods36. Data from nearly one hundred sources were prepared manually—that is, without coding, typically in Microsoft Excel—which included organizing, transforming, rescaling, gap-filling and formatting data. Processing decisions were documented primarily within the Excel files themselves, e-mails, and Microsoft Word documents. We programmatically coded models and meticulously documented their development, (resulting in the 130-page supplemental materials)36, and upon publication we also made the model inputs (that is, prepared data and metadata) freely available to download. This level of documentation and transparency is beyond the norm for environmental science16,40.
We also worked collaboratively in the same ways we always had. Our team included scientists and analysts with diverse skill sets and disciplines, and we had distinct, domain-specific roles assigned to scientists and to a single analytical programmer. Scientists were responsible for developing the models conceptually, preparing data and interpreting modelled results, and the programmer was responsible for coding the models. We communicated and shared files frequently, with long, often-forwarded and vaguely titled e-mail chains (for example, ‘Re: Fwd: data question’) with manually versioned data files (for example, ‘data_final_updated2.xls’). All team members were responsible for organizing those files with their own conventions on their local computers. Final versions of prepared files were stored on the servers and used in models, but records of the data processing itself were scattered.
Upon beginning the second annual assessment in 2013, we realized that our approach was insufficient because it took too much time and relied heavily on individuals’ data organization, e-mail chains and memory—particularly problematic as original team members moved on and new team members joined. We quickly realized we needed a nimble and robust approach to sharing data, methods and results within and outside our team—we needed to completely upgrade our workflow.
Actually doing reproducible science. As we began the second global OHI assessment in 2013 we faced challenges across three main fronts: (1) reproducibility, including transparency and repeatability, particularly in data preparation; (2) collaboration, including team record keeping and internal collaboration; and (3) communication, with scientific and broader communities. We knew that environmental scientists are increasingly using R because it is free, cross-platform, and open source11, and also because of the training and support provided by developers33 and independent groups12,41 alike. We decided to base our work in R and RStudio for coding and visualization42,43, Git for version control44, GitHub for collaboration45, and a combination of GitHub and RStudio for organization, documentation, project management, online publishing, distribution and communication (Table 1). These tools can help scientists organize, document, version and easily share data and methods, thus not only increasing reproducibility but also reducing the amount of time involved to do so14,46,47. Many available tools are free so long as work is shared publicly online, which enables open science, defined by Hampton et al.40 as “the concept of transparency at all stages of the research process, coupled with free and open access to data, code, and papers”. When integrated into the scientific process, data science tools that enable open science—let's call them ‘open data science’ tools—can help realize reproducibility in collaborative scientific research6,16,40,48,49.
Open data science tools helped us upgrade our approach to reproducible, collaborative and transparent science, but they did require a substantial investment to learn, which we did incrementally over time (Fig. 1 and Box 1). Previous to this evolution, most team members with any coding experience—not necessarily in R—had learned just enough to accomplish whatever task had been before them using their own unique conventions. Given the complexity of the OHI project, we needed to learn to code collaboratively and incorporate best50,51 or good-enough practices12,52 into our coding, so that our methods could be co-developed and vetted by multiple team members. Using a version control system not only improved our file and data management, but allowed individuals to feel less inhibited about their coding contributions, since files could always be reverted back to previous versions if there were problems. We built confidence using these tools by sharing our imperfect code, discussing our challenges and learning as a team. These tools quickly became the keystone of how we work, and have overhauled our approach to science, perhaps as much as e-mail did in decades prior. They have changed the way we think about science and about what is possible. The following describes how we have been using open data science practices and tools to overcome the biggest challenges we encountered to reproducibility, collaboration and communication.
Reproducibility
It is paramount that our methods are transparent, reproducible, and also repeatable with additional data for tracking changes through time. We now collaboratively code and use version control for all our work, which not only provides a historical record of what we did, but saves time since it can be rerun when new data are available.
Data preparation: coding and documenting. Our first priority was to code all data preparation, create a standard format for final data layers, and do so using a single programmatic language, R42. Code enables us to reproduce the full process of data preparation, from data download to final model inputs37,53, and a single language makes it more practical for our team to learn and contribute collaboratively. We code in R and use RStudio43 to power our workflow because it has a user-friendly interface and built-in tools useful for coders of all skill levels, and, importantly, it can be configured with Git to directly sync with GitHub online (See ‘Collaboration’). We have succeeded in transitioning to R as our primary coding language for data preparation, including for spatial data, although some operations still require additional languages and tools such as ArcGIS, QGIS, and Python54–56.
All our code is underpinned by the principles of tidy data, the grammar of data manipulation, and the tidyverse R packages developed by Wickham33,57–59. This deliberate philosophy for thinking about data helped bridge our scientific questions with the data processing required to get there, and the readability and conciseness of tidyverse operations makes our data analysis read more as a story arc. Operations require less syntax—which can mean fewer potential errors that are easier to identify—and they can be chained together, minimizing intermediate steps and data objects that can cause clutter and confusion33,60. The tidyverse tools for wrangling data have expedited our transformation as coders and made R less intimidating to learn. We heavily rely on a few packages for data wrangling and visualization that are bundled in the tidyverse package58,59—particularly dplyr, tidyr, and ggplot2—as well as accompanying books, cheatsheets and archived webinars (Box 1).
We keep detailed documentation describing metadata (for example, source, date of access, links) and data processing decisions—trying to capture not only the processing we decided to do, but what we decided against. We started with small plain text files accompanying each R file, but have transitioned to documenting with R Markdown61,62 because it combines plain text and executable chunks of R code within the same file and serves as a living lab notebook. Every time R Markdown output files are regenerated the R code is rerun so the text and figures will also be regenerated and reflect any updates to the code or underlying data. R Markdown files increase our reproducibility and efficiency by streamlining documentation and eliminating the need to constantly paste updated figures into reports as they are developed.
Modelling: R functions and packages. Once the data are prepared, we develop assessment-specific models to calculate OHI scores. Models were originally coded in multiple languages to accommodate disparate data types and formatting. By standardizing our approach to data preparation and final data layer format, we have been able to translate all models into R. In addition to assessment-specific models, the OHI framework includes core analytical operations that are used by all OHI assessments34, and thus we created an R package called ohicore63, which was greatly facilitated by the devtools and roxygen2 packages64–66. The ohicore package is maintained in and installed from a dedicated GitHub repository—using devtools::install_github(‘ohi-science/ohicore’)—from any computer with R and an internet connection, enabling groups leading independent OHI assessments to use it for their own work34.
Version control. We use Git44 as a version control system. Version control systems track changes within files and allow you to examine or rewind to previous versions. This saves time that would otherwise be spent duplicating, renaming and organizing files to preserve past versions. It also makes folders easier to navigate since they are no longer overcrowded with multiple files suffixed with dates or initials (for example, ‘final_JL-2012-02-26.csv’)67–69. Once Git is configured on each team member's machine, they work as before but frequently commit to saving a snapshot of their files, along with a human-readable ‘commit message’67,68. Any line modified in a file tracked by Git will then be attributed to that user.
We interface with Git primarily through RStudio, using the command line for infrequently encountered tasks. Using RStudio to interact with Git was key for our team's uptake of a version control system, since the command line can be an intimidating hurdle or even a barrier for beginners to get on board with using version control. We were less resistant because we could use a familiar interface, and as we gained fluency in Git's operations through RStudio we translated that confidence to the command line.
Organization. Our team developed conventions to standardize the structure and names of files to improve consistency and organization. Along with the GitHub workflow (see ‘Collaboration’), having a structured approach to file organization and naming has helped those within and outside our team navigate our methods more easily. We organize parts of the project in folders that are both RStudio ‘projects’ and GitHub ‘repositories’, which has also helped us collaborate using shared conventions rather than each team member spending time duplicating and organizing files.
Collaboration within our team
Our team collaborates in real-time and also treats our future selves as collaborators, knowing that ‘future us’ can only rely on detailed records. Open data science software streamlines collaboration by weaving together code and text, which lets us capture decision-making right alongside our code.
Coding collaboratively. We transitioned from a team of distinct roles (scientists and programmer) to becoming a team with overlapping skill sets (scientists-as-programmers, or simply, data scientists). Having both environmental expertise and coding skills in the same person increases project efficiency, enables us to vet code as a team, and reduces the bottleneck of relying on a single programmer. We, like Duhigg70, have found that “groups tend to innovate faster, see mistakes more quickly and find better solutions to problems”. Developing these skills and creating the team culture around them requires leadership with the understanding that fostering more-efficient and productive scientists is worth the long-term investment. Our team had the freedom to experiment with available tools and their value was recognized with a commitment that we, as a team, would adopt and pursue these methods further. In addition to supportive leadership, having a ‘champion’ with experience of how tools can be introduced over time and interoperate can expedite the process, but is not the only path (Box 2). Taking the time to experiment and invest in learning data science principles, tools and skills enabled our team to establish a system of best practices for developing, using and teaching the OHI Toolbox.
Our (simplified) GitHub workflow. GitHub is one of many web-based platforms that enables files tracked with Git to be collaboratively shared online so contributors can keep their work synchronized45,68,69, and it is increasingly being adopted by scientific communities for project management71. Versioned files are synced online with GitHub similar to the way Dropbox operates, except syncs require a committed, human-readable message and reflect deliberate snapshots of changes made that are attributed to the user, line-by-line, through time. Built for large, distributed teams of software developers, GitHub provides many features that we as a scientific team, new to data science, do not immediately need, and thus we mostly ignore features such as branching, forking and pull requests. Our team uses a simplified GitHub workflow whereby all members have administrative privileges to the repositories within our ohi-science organization. Each team member is able to sync their local work to GitHub.com, making it easier to attribute contribution, as well as identify to whom questions should be directed.
GitHub is now central to many facets of our collaboration as a team and with other communities—we use it along with screensharing to teach and troubleshoot with groups leading independent OHI assessments, as well as to communicate our ongoing work and final results (see ‘Communication’). Now there are very few files e-mailed back and forth within our team since we all have access to all repositories within the ohi-science organization, and can navigate to and edit whatever we need. Additionally, these organized files are always found with the same file path, whether on GitHub.com or on someone's local computer; this, along with ‘RStudio.Rproj’ files, eases the file path problems that can plague collaborative coding and frustrate new coders.
Internal communication. We use a feature of GitHub called ‘Issues’ in place of e-mail for discussions about data preparation and analysis. We use Issues in a separate private repository to keep our conservations private but our work public. All team members can see and contribute to all conversations, which are a record of all our decisions and discussions across the project and are searchable in a single place. Team members can communicate clearly by linking to specific lines of code in current or past versions of specific files since they are stored on GitHub and thus have a URL. We can also paste images and screenshots, link to other websites, and send an e-mail to specific team members directly by mentioning their GitHub username. In addition to discussing analytical options, we use Issues to track ongoing tasks, tricks we have learned, and future ideas. Issues provide a written reference of institutional memory so new team members can get up to speed more easily. Most importantly, GitHub Issues have helped us move past the never-ending forwarded e-mail chains and instead to conversations available to any current or future team member.
Communication outside the project
Open data science tools have made us re-imagine what communication can mean for science and management. They enable us to not only share our code online, but to create reports, e-books, interactive web applications, and entire websites, which we can share for free to communicate our work.
Sharing data and code. Our code is online in GitHub repositories, publicly available for any researcher or interested person to see and access (http://github.com/ohi-science). As we work, GitHub renders code, text, images and tabular and spatial data, and displays differences between versions, essentially creating webpages that can be easily shared with collaborators, whether or not they use GitHub. Additionally, we create ‘Releases’ for each global assessment36,37 so the code and data we use for peer-reviewed publication are preserved while we continue our work (https://github.com/ohi-Science/ohi-global/releases).
Sharing methods and instruction. We use R Markdown not only for data preparation but also for broader communication. R Markdown files can be generated into a wide variety of formatted outputs, including PDFs, slides, Microsoft Word documents, HTML files, books or full websites61,62. These can all be published online for free through GitHub using the same RStudio–GitHub workflow that we use for our analyses, which has made communication an ongoing part of our work, instead of a final step in completed analyses.
We built a website using GitHub and RStudio publishing tools: http://ohi-science.org. Team members can update content directly, and using the same workflow makes it easier for us to keep it current. Our website is intended for scientists interested in our methods as well as those leading their own assessments34. Thus, the website provides scientific methods, publications, data, and code, as well as instruction, news, blog posts, and a map displaying where all ongoing OHI assessments are taking place so that groups can learn directly from and build off of each other's code. Technical information provided on http://ohi-science.org complements http://oceanhealthindex.org, our overview website intended for more general audiences.
Meeting scientists where they are
We are environmental scientists whose impetus for upgrading approaches to collaborative, data-intensive science was driven by our great difficulty reproducing our own methods. Many researchers do not attempt to reproduce their own work17,72—ourselves included before 2013—and thus may not realize that there could be reproducibility issues in their own approaches. But they can likely identify inefficiencies. Integrating open data science practices and tools into science can save time, while also improving reproducibility for our most important collaborators: our future selves. We have found this as individuals and as a team: we could not be as productive34,35 without open data science practices and tools. We would also not be able to efficiently share and communicate our work while it is ongoing rather than only post-publication, which is particularly important for bridging science and policy. As environmental scientists who are still learning, we hope sharing our experiences will empower other scientists to upgrade their own approaches, helping further shift the scientific culture to value transparency and openness as a benefit to all instead of as a vulnerability16,40,48.
From our own experience and from teaching other academic, non-profit, and government groups through the Ocean Health Index project34, we find that the main barriers to engagement boil down to exposure and confidence: first knowing which tools exist that can be directly useful to one's research, and then having the confidence to develop the skills to use them. These two points are simple but critical. We are among the many environmental scientists who were never formally trained to work deliberately with data. Thus, we were unaware of how significantly open data science tools could directly benefit our research11,73, and upon learning about them we were hesitant, or even resistant, to engage. However, we were able to develop confidence in large part because of the open, inclusive and encouraging online developer community that builds tools and creates tutorials that meet scientists where they are (Boxes 1,2). It takes motivation, patience, diligence, and time to overcome the conceptual and technical challenges involved in developing computing skills but resources are available to help scientists get started11,51,73. Coding is “as important to modern scientific research as telescopes and test tubes”50, but it is critical to “dispel the misconception that these skills are intuitive, obvious, or in any way inherent”41.
There is ongoing and important work by the informatics community on the architecture and systems for data management and archiving7,8,18,74, as well as efforts to enable scientists to publish the code that they do have26,31,52. This work is critical, but comes with the a priori assumption that scientists are already thinking about data and coding in a way that they would seek out further resources. In reality, this is not always the case, and without visible examples of how to use these tools within their scientific fields, common stumbling blocks will be continually combatted with individual workarounds instead of addressed with intention. These workarounds can greatly delay focusing on actual scientific research, particularly when scientific questions that may not yet have answers—for example, how the behavior of X changes with Y—are conflated with data science questions that have many existing answers—for example, how to operate on only criteria X and Y.
Scientific advancement comes from building off the past work of others; scientists can also embrace this principle for using software tools to tackle some of the challenges encountered in modern scientific research. In a recent survey in Nature, 90% of the 1,500 respondents across scientific fields agreed that there was a reproducibility crisis in science, and one third of the respondents reported not having their own “established procedures for reproducibility”4. While reproducibility means distinct things within the protocols of each sub-discipline or specialty, underpinning reproducibility across all disciplines in modern science is working effectively and collaboratively with data, including wrangling, formatting and other tasks that can take 50–80% of a data scientist's time75. While reaching full reproducibility is extremely difficult5,76, incrementally incorporating open data science practices and tools into scientific workflows has the potential to alleviate many of the troubles plaguing science, including collaboration and preserving institutional memory12. Further, sharing openly is fundamental to truly expediting scientific progress because others can build directly off previous work if well-documented, re-usable code is available16,47,48,77. Until quite recently, making research open required a great deal of extra work for researchers and was less likely to be done. Now, with available tools, the benefits of openness can be a by-product of time-saving efficiencies, because tools that reduce data headaches also result in science that is more transparent, reproducible, collaborative and freely accessible to others.
Ecologists and environmental scientists arguably have a heightened responsibility for transparency and openness, as data products provide important snapshots of systems that may be forever altered due to climate change and other human pressures16,18. There is particular urgency for efficiency and transparency, as well as opportunity to democratize science in fields that operate at the interface of science and policy. Individuals play an important part by promoting good practices and creating supportive communities16,41,48. But it is also critical for the broader science community to build a culture where openness and reproducibility are valued, formally taught and practiced, where we all agree that they are worth the investment.
Additional information
How to cite this article: Lowndes, J.S.S. et al. Our path to better science in less time using open data science tools. Nat. Ecol. Evol. 1, 0160 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Baker, M. Over half of psychology studies fail reproducibility test. Nature Newshttp://dx.doi.org/10.1038/nature.2015.18248 (2015).
Baker, M. & Dolgin, E. Cancer reproducibility project releases first results. Nature Newshttp://dx.doi.org/10.1038/541269a (2017).
Open Science Collaboration. Estimating the reproducibility of psychological science. Science 349, aac4716 (2015).
Baker, M. 1,500 scientists lift the lid on reproducibility. Nature 533, 452–454 (2016).
Aschwanden, C. Science isn't broken. FiveThirtyEighthttp://go.nature.com/2qaz0Tz (19 August 2015)
Buck, S. Solving reproducibility. Science 348, 1403–1403 (2015).
Frew, J. & Dozier, J. Environmental informatics. Annu. Rev. Environ. Resources 37, 449–472 (2012).
Jones, M. B., Schildhauer, M. P., Reichman, O. J. & Bowers, S. The new bioinformatics: Integrating ecological data from the gene to the biosphere. Annu. Rev. Ecol. Evol. Syst. 37, 519–544 (2006).
Michener, W. K. & Jones, M. B. Ecoinformatics: Supporting ecology as a data-intensive science. Trends Ecol. Evol. 27, 85–93 (2012).
Check Hayden, E. Mozilla plan seeks to debug scientific code. Nature Newshttp://dx.doi.org/10.1038/501472a (2013).
Boettiger, C., Chamberlain, S., Hart, E. & Ram, K. Building software, building community: Lessons from the rOpenSci project. J. Open Res. Softw. 3, e8 (2015).
Wilson, G. et al. Good enough practices in scientific computing. Preprint at https://arxiv.org/abs/1609.00037 (2016).
Wilson, G. V. Where's the real bottleneck in scientific computing? Am. Sci. 94, 5–6 (2006).
Baker, M. Scientific computing: Code alert. Nature 541, 563–565 (2017).
Barone, L., Williams, J. & Micklos, D. Unmet needs for analyzing biological big data: A survey of 704 NSF principal investigators. Preprint at bioRxivhttps://doi.org/10.1101/108555 (2017).
Wolkovich, E. M., Regetz, J. & O’Connor, M. I. Advances in global change research require open science by individual researchers. Global Change Biol. 18, 2102–2110 (2012).
Nosek, B. A. et al. Promoting an open research culture. Science 348, 1422–1425 (2015).
Reichman, O. J., Jones, M. B. & Schildhauer, M. P. Challenges and opportunities of open data in ecology. Science 331, 703–705 (2011).
Shade, A. & Teal, T. K. Computing workflows for biologists: A roadmap. PLoS Biol. 13, e1002303 (2015).
Goodman, A. et al. Ten simple rules for the care and feeding of scientific data. PLoS Comput. Biol. 10, e1003542 (2014).
Sandve, G. K., Nekrutenko, A., Taylor, J. & Hovig, E. Ten simple rules for reproducible computational research. PLoS Comput. Biol. 9, e1003285 (2013).
White, E. P. et al. Nine simple ways to make it easier to (re)use your data. Ideas Ecol. Evol.http://doi.org/10.4033/iee.2013.6b.6.f (2013).
Kervin, K., Michener, W. & Cook, R. Common errors in ecological data sharing. J. eScience Librarianshiphttp://dx.doi.org/10.7191/jeslib.2013.1024 (2013).
Lewandowsky, S. & Bishop, D. Research integrity: don't let transparency damage science. Nature Newshttp://dx.doi.org/10.1038/529459a (2016).
Michener, W. K. Ten simple rules for creating a good data management plan. PLoS Comput. Biol. 11, e1004525 (2015).
Mislan, K. A. S., Heer, J. M. & White, E. P. Elevating the status of code in ecology. Trends Ecol. Evol. 31, 4–7 (2016).
Kratz, J. & Strasser, C. Data publication consensus and controversies. F1000Researchhttp://dx.doi.org/10.12688/f1000research.3979.3 (2014).
Munafò, M. R. et al. A manifesto for reproducible science. Nat. Hum. Behav. 1, 0021 (2017).
Martinez, C. et al. Reproducibility in Science: A Guide to Enhancing Reproducibility in Scientific Results and Writinghttp://ropensci.github.io/reproducibility-guide/ (2014).
Tuyl, S. V. & Whitmire, A. L. Water, water, everywhere: defining and assessing data sharing in academia. PLoS ONE 11, e0147942 (2016).
Baker, M. Why scientists must share their research code. Nature Newshttp://dx.doi.org/10.1038/nature.2016.20504 (2016).
Kidwell, M. C. et al. Badges to acknowledge open practices: A simple, low-cost, effective method for increasing transparency. PLoS Biol. 14, e1002456 (2016).
Wickham, H. & Grolemund, G. R for Data Science (O’Reilly, 2016); http://r4ds.had.co.nz/
Lowndes, J. S. S. et al. Best practices for assessing ocean health in multiple contexts using tailorable frameworks. PeerJ 3, e1503 (2015).
Lowndes, J. A biography of the ocean health index. ohi-sciencehttp://ohi-science.org/news/Biography-OHI (13 January 2017).
Halpern, B. S. et al. An index to assess the health and benefits of the global ocean. Nature 488, 615–620 (2012).
Halpern, B. S. et al. Patterns and emerging trends in global ocean health. PLoS ONE 10, e0117863 (2015).
Five years of global ocean health index assessments. ohi-sciencehttp://ohi-science.org/ohi-global (2016).
Goldfuss, C. & Holdren, J. P. The nation's first ocean plans. The White Househttps://obamawhitehouse.archives.gov/blog/2016/12/07/nations-first-ocean-plans (7 December 2016).
Hampton, S. E. et al. The tao of open science for ecology. Ecosphere 6, art 120 (2015).
Mills, B. Introducing mozilla science study groups. Mozillahttps://science.mozilla.org/blog/introducing-mozilla-science-study-groups (22 April 2015).
R Core Team R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2016); https://www.R-project.org/
RStudio Team RStudio: Integrated Development for R (RStudio, 2016); www.rstudio.com
Git Team Git Version Control System (Git, 2016); https://git-scm.com/
GitHub: A Collaborative Online Platform To Build Software (GitHub, 2016); https://github.com
Wilson, G. V. Software carpentry: getting scientists to write better code by making them more productive. Comput. Sci. Eng. 8, 66–69 (2006).
Broman, K. Initial steps toward reproducible research. http://kbroman.org/steps2rr/ (2016).
McKiernan, E. C. et al. How open science helps researchers succeed. eLife 5, e16800 (2016).
Seltenrich, N. Scaling the heights of data science. Breakthroughshttps://nature.berkeley.edu/breakthroughs/opensci-data (2016).
Wilson, G. et al. Best practices for scientific computing. PLoS Biol. 12, e1001745 (2014).
Haddock, S. H. & Dunn, C. W. Practical Computing for Biologists (Sinauer Associates, 2011).
Barnes, N. Publish your computer code: it is good enough. Nature 467, 753 (2010).
Frazier, M., Longo, C. & Halpern, B. S. Mapping uncertainty due to missing data in the global ocean health index. PLoS ONE 11, e0160377 (2016).
ESRI ArcGIS Platform (2016); http://www.esri.com/software/arcgis
The QGIS Team QGIS Project (2016); http://www.qgis.org
The Python Team Python (2016); https://www.python.org/
Wickham, H. Tidy data. J. Stat. Softw. 59, 1–23 (2014).
Wickham, H. Tidyverse Tidyweb (accessed 29 January 2017); http://tidyverse.org/
Wickham, H. Tidyverse: Easily Install and Load ‘tidyverse’ Packages (2016); https://CRAN.R-project.org/package=tidyverse
Fischetti, T. How dplyr replaced my most common r idioms. StatsBlogshttp://www.onthelambda.com/2014/02/10/how-dplyr-replaced-my-most-common-r-idioms/ (10 February 2014).
RStudio Team R Markdown (2016); http://rmarkdown.rstudio.com/
Allaire, J. J. et al. R Markdown: Dynamic Documents for R (2016); https://CRAN.R-project.org/package=rmarkdown
Ocean Health Index ohicore Package (Ocean Health Index Team, 2016).
Wickham, H. R Packages (O’Reilly, 2015); http://r-pkgs.had.co.nz/
Wickham, H. & Chang, W. Devtools: Tools to Make Developing R Packages Easier (2016); https://CRAN.R-project.org/package=devtools
Wickham, H., Danenberg, P. & Eugster, M. Roxygen2: In-Source Documentation for R (2015); https://CRAN.R-project.org/package=roxygen2
Ram, K. Git can facilitate greater reproducibility and increased transparency in science. Source Code Biol. Med. 8, 7 (2013).
Blischak, J. D., Davenport, E. R. & Wilson, G. A quick introduction to version control with git and GitHub. PLoS Comput. Biol. 12, e1004668 (2016).
Perez-Riverol, Y. et al. Ten simple rules for taking advantage of Git and GitHub. PLoS Comput. Biol. 12, e1004947 (2016).
Duhigg, C. What Google learned from its quest to build the perfect team. The New York Times (25 February 2016).
Perkel, J. Democratic databases: Science on GitHub. Nature 538, 127–128 (2016).
Casadevall, A. & Fang, F. C. Reproducible science. Infect. Immun. 78, 4972–4975 (2010).
Wilson, G. Software carpentry: lessons learned. F1000Researchhttp://dx.doi.org/10.12688/f1000research.3-62.v2 (2016).
Hampton, S. E. et al. Big data and the future of ecology. Front. Ecol. Environ. 11, 156–162 (2013).
Lohr, S. For big-data scientists, ‘janitor work’ is key hurdle to insights. The New York Times (17 August 2014).
FitzJohn, R., Pennell, M., Zanne, A. & Cornell, W. Reproducible research is still a challenge. ROpenSci https://ropensci.org//blog/2014/06/09/reproducibility/ (9 June 2014).
Boland, M. R., Karczewski, K. J. & Tatonetti, N. P. Ten simple rules to enable multi-site collaborations through data sharing. PLoS Comput. Biol. 13, e1005278 (2017).
Perkel, J. M. Scientific writing: the online cooperative. Nature 514, 127–128 (2014).
How Twitter improved my ecological model. R-bloggershttps://www.r-bloggers.com/how-twitter-improved-my-ecological-model/ (26 February 2015).
Acknowledgements
The Ocean Health Index is a collaboration between Conservation International and the National Center for Ecological Analysis and Synthesis at the University of California at Santa Barbara. We thank J. Polsenberg, S. Katona, E. Pacheco and L. Mosher who are our partners at Conservation International. We thank all past contributors and funders that have supported the Ocean Health Index, including B. Wrigley and H. Wrigley and The Pacific Life Foundation. We also thank all the individuals and groups that openly make their data, tools and tutorials freely available to others. Finally, we thank H. Wickham, K. Ram, K. Woo and M. Schildhauer for friendly review of the developing manuscript. See http://ohi-science.org/betterscienceinlesstime as an example of a website built with RMarkdown and the RStudio–GitHub workflow, and for links and resources referenced in the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Lowndes, J., Best, B., Scarborough, C. et al. Our path to better science in less time using open data science tools. Nat Ecol Evol 1, 0160 (2017). https://doi.org/10.1038/s41559-017-0160
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41559-017-0160