Abstract
RNA-binding proteins (RBPs) are pivotal in acute myeloid leukaemia (AML), a lethal disease. Although specific phase separation-competent RBPs are recognized in AML, the effect of their condensate formation on AML leukaemogenesis, and the therapeutic potential of inhibition of phase separation are underexplored. In our in vivo CRISPR RBP screen, fibrillarin (FBL) emerges as a crucial nucleolar protein that regulates AML cell survival, primarily through its phase separation domains rather than methyltransferase or acetylation domains. These phase separation domains, with specific features, coordinately drive nucleoli formation and early processing of pre-rRNA (including efflux, cleavage and methylation), eventually enhancing the translation of oncogenes such as MYC. Targeting the phase separation capability of FBL with CGX-635 leads to elimination of AML cells, suggesting an additional mechanism of action for CGX-635 that complements its established therapeutic effects. We highlight the potential of PS modulation of critical proteins as a possible therapeutic strategy for AML.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The next-generation sequencing data (RiboMeth-seq and polysome fraction mRNA-seq/total mRNA-seq) generated in this study can be accessed in the Gene Expression Omnibus (GEO) under accession code GSE222266. The human reference genome GRCh38 was used for RNA-seq analyses. Previously published proteomic data from ref. 28, reanalysed here, can be found in the Proteomics Identifications Database (PRIDE) under accession codes PXD023201 and PXD028007. Any other potential type of data used to interpret the findings can be provided upon request to the corresponding author. Source data are provided with this paper.
References
Yamashita, M., Dellorusso, P. V., Olson, O. C. & Passegué, E. Dysregulated haematopoietic stem cell behaviour in myeloid leukaemogenesis. Nat. Rev. Cancer 20, 365–382 (2020).
Short, N. J. et al. Advances in the treatment of acute myeloid leukemia: new drugs and new challenges. Cancer Discov. 10, 506–525 (2020).
National Cancer Institute Surveillance, Epidemiology and End Results Program. Cancer Stat Facts: Leukemia—Acute Myeloid Leukemia (AML) (National Cancer Institute); https://seer.cancer.gov/statfacts/html/amyl.html
Cai, D., Liu, Z. & Lippincott-Schwartz, J. Biomolecular condensates and their links to cancer progression. Trends Biochem. Sci. 46, 535–549 (2021).
Boija, A., Klein, I. A. & Young, R. A. Biomolecular condensates and cancer. Cancer Cell 39, 174–192 (2021).
Pappu, R. V., Cohen, S. R., Dar, F., Farag, M. & Kar, M. Phase transitions of associative biomacromolecules. Chem. Rev. 123, 8945–8987 (2023).
Alberti, S. & Hyman, A. A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 22, 196–213 (2021).
Alberti, S., Gladfelter, A. & Mittag, T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019).
Ahn, J. H. et al. Phase separation drives aberrant chromatin looping and cancer development. Nature 595, 591–595 (2021).
Cheng, Y. et al. N6-methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell 39, 958–972 (2021).
Chandra, B. et al. Phase separation mediates NUP98 fusion oncoprotein leukemic transformation. Cancer Discov. 12, 1152–1169 (2022).
Smith, T. et al. Organic phase separation opens up new opportunities to interrogate the RNA-binding proteome. Curr. Opin. Chem. Biol. 54, 70–75 (2020).
Wiedner, H. J. & Giudice, J. It’s not just a phase: function and characteristics of RNA-binding proteins in phase separation. Nat. Struct. Mol. Biol. 28, 465–473 (2021).
Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016).
Frottin, F. et al. The nucleolus functions as a phase-separated protein quality control compartment. Science 365, 342–347 (2019).
Aspesi, A. & Ellis, S. R. Rare ribosomopathies: insights into mechanisms of cancer. Nat. Rev. Cancer 19, 228–238 (2019).
Carotenuto, P., Pecoraro, A., Palma, G., Russo, G. & Russo, A. Therapeutic approaches targeting nucleolus in cancer. Cells 8, 1090 (2019).
Marcel, V. et al. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell 24, 318–330 (2013).
Erales, J. et al. Evidence for rRNA 2′-O-methylation plasticity: control of intrinsic translational capabilities of human ribosomes. Proc. Natl Acad. Sci. USA 114, 12934–12939 (2017).
Yi, Y. et al. A PRC2-independent function for EZH2 in regulating rRNA 2′-O-methylation and IRES-dependent translation. Nat. Cell Biol. 23, 341–354 (2021).
Zhou, F. et al. A dynamic rRNA ribomethylome drives stemness in acute myeloid leukemia. Cancer Discov. 13, 332–347 (2023).
Tessarz, P. et al. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 505, 564–568 (2014).
Iyer-Bierhoff, A. et al. SIRT7-dependent deacetylation of fibrillarin controls histone H2A methylation and rRNA synthesis during the cell cycle. Cell Rep. 25, 2946–2954 (2018).
Yao, R.-W. et al. Nascent pre-rRNA sorting via phase separation drives the assembly of dense fibrillar components in the human nucleolus. Mol. Cell 76, 767–783 (2019).
Platt, R. J. et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
Hentze, M. W., Castello, A., Schwarzl, T. & Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19, 327–341 (2018).
Elcheva, I. A. & Spiegelman, V. S. Targeting RNA-binding proteins in acute and chronic leukemia. Leukemia 35, 360–376 (2021).
Jayavelu, A. K. et al. The proteogenomic subtypes of acute myeloid leukemia. Cancer Cell 40, 301–317 (2022).
Döhner, H. et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129, 424–447 (2017).
Rodriguez-Corona, U., Sobol, M., Rodriguez-Zapata, L. C., Hozak, P. & Castano, E. Fibrillarin from Archaea to human. Biol. Cell 107, 159–174 (2015).
Shubina, M. Y., Musinova, Y. R. & Sheval, E. V. Nucleolar methyltransferase fibrillarin: evolution of structure and functions. Biochemistry (Mosc.) 81, 941–950 (2016).
Aittaleb, M., Visone, T., Fenley, M. O. & Li, H. Structural and thermodynamic evidence for a stabilizing role of Nop5p in S-adenosyl-L-methionine binding to fibrillarin. J. Biol. Chem. 279, 41822–41829 (2004).
Deffrasnes, C. et al. Genome-wide siRNA screening at biosafety level 4 reveals a crucial role for fibrillarin in Henipavirus infection. PLoS Pathog. 12, e1005478 (2016).
Bremer, A. et al. Deciphering how naturally occurring sequence features impact the phase behaviours of disordered prion-like domains. Nat. Chem. 14, 196–207 (2022).
Ruff, K. M., Dar, F. & Pappu, R. V. Polyphasic linkage and the impact of ligand binding on the regulation of biomolecular condensates. Biophys. Rev. 2, 021302 (2021).
Lafontaine, D. L. J., Riback, J. A., Bascetin, R. & Brangwynne, C. P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 22, 165–182 (2021).
Correll, C. C., Bartek, J. & Dundr, M. The nucleolus: a multiphase condensate balancing ribosome synthesis and translational capacity in health, aging and ribosomopathies. Cells 8, 869 (2019).
Elhamamsy, A. R., Metge, B. J., Alsheikh, H. A., Shevde, L. A. & Samant, R. S. Ribosome biogenesis: a central player in cancer metastasis and therapeutic resistance. Cancer Res. 82, 2344–2353 (2022).
Shan, L. et al. Nucleolar URB1 ensures 3′ ETS rRNA removal to prevent exosome surveillance. Nature 615, 526–534 (2023).
Ayadi, L., Motorin, Y. & Marchand, V. Quantification of 2′-O-Me residues in RNA using next-generation sequencing (Illumina RiboMethSeq protocol). Methods Mol. Biol. 1649, 29–48 (2018).
Dong, Z.-W. et al. RTL-P: a sensitive approach for detecting sites of 2′-O-methylation in RNA molecules. Nucleic Acids Res. 40, e157 (2012).
Tafforeau, L. et al. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of pre-rRNA processing factors. Mol. Cell 51, 539–551 (2013).
Liao, H. et al. Human NOP2/NSUN1 regulates ribosome biogenesis through non-catalytic complex formation with box C/D snoRNPs. Nucleic Acids Res. 50, 10695–10716 (2022).
Zhou, F. et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat. Cell Biol. 19, 844–855 (2017).
Pauli, C. et al. Site-specific methylation of 18S ribosomal RNA by SNORD42A is required for acute myeloid leukemia cell proliferation. Blood 135, 2059–2070 (2020).
Ugolini, I. et al. Chromatin localization of nucleophosmin organizes ribosome biogenesis. Mol. Cell 82, 4443–4457 (2022).
Schmidt, E. K., Clavarino, G., Ceppi, M. & Pierre, P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009).
Venugopal, S., Bar-Natan, M. & Mascarenhas, J. O. JAKs to STATs: a tantalizing therapeutic target in acute myeloid leukemia. Blood Rev. 40, 100634 (2020).
Hu, X., Li, J., Fu, M., Zhao, X. & Wang, W. The JAK/STAT signaling pathway: from bench to clinic. Signal Transduct. Target Ther. 6, 402 (2021).
Zhang, L. et al. Myc-Miz1 signaling promotes self-renewal of leukemia stem cells by repressing Cebpα and Cebpδ. Blood 135, 1133–1145 (2020).
Schaub, F. X. et al. Pan-cancer alterations of the MYC oncogene and its proximal network across the Cancer Genome Atlas. Cell Syst 6, 282–300 (2018).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Burger, K. et al. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J. Biol. Chem. 285, 12416–12425 (2010).
Jin, J. et al. Homoharringtonine-based induction regimens for patients with de-novo acute myeloid leukaemia: a multicentre, open-label, randomised, controlled phase 3 trial. Lancet Oncol. 14, 599–608 (2013).
Mi, R. et al. Efficacy and safety of homoharringtonine for the treatment of acute myeloid leukemia: a meta-analysis. Clin. Lymphoma Myeloma Leuk. 21, e752–e767 (2021).
Chen, X.-J. et al. Homoharringtonine deregulates MYC transcriptional expression by directly binding NF-κB repressing factor. Proc. Natl Acad. Sci. USA 116, 2220–2225 (2019).
Ruff, K. M., Dar, F. & Pappu, R. V. Ligand effects on phase separation of multivalent macromolecules. Proc. Natl Acad. Sci. USA 118, e2017184118 (2021).
Leicher, R. et al. Single-stranded nucleic acid binding and coacervation by linker histone H1. Nat. Struct. Mol. Biol. 29, 463–471 (2022).
Mitrea, D. M. et al. Self-interaction of NPM1 modulates multiple mechanisms of liquid–liquid phase separation. Nat. Commun. 9, 842 (2018).
Hein, N., Hannan, K. M., George, A. J., Sanij, E. & Hannan, R. D. The nucleolus: an emerging target for cancer therapy. Trends Mol. Med. 19, 643–654 (2013).
Ide, S., Imai, R., Ochi, H. & Maeshima, K. Transcriptional suppression of ribosomal DNA with phase separation. Sci. Adv. 6, eabb5953 (2020).
Hannan, R. D., Drygin, D. & Pearson, R. B. Targeting RNA polymerase I transcription and the nucleolus for cancer therapy. Expert Opin. Ther. Targets 17, 873–878 (2013).
Bywater, M. J. et al. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 22, 51–65 (2012).
Hein, N. et al. Inhibition of Pol I transcription treats murine and human AML by targeting the leukemia-initiating cell population. Blood 129, 2882–2895 (2017).
Falini, B., Brunetti, L., Sportoletti, P. & Martelli, M. P. NPM1-mutated acute myeloid leukemia: from bench to bedside. Blood 136, 1707–1721 (2020).
Gürel, G., Blaha, G., Moore, P. B. & Steitz, T. A. U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J. Mol. Biol. 389, 146–156 (2009).
Chen, B. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013).
Raj, A. & Tyagi, S. Detection of individual endogenous RNA transcripts in situ using multiple singly labeled probes. Methods Enzymol. 472, 365–386 (2010).
Riback, J. A. et al. Composition-dependent thermodynamics of intracellular phase separation. Nature 581, 209–214 (2020).
Zhang, X. M. et al. A proline-rich motif on VGLUT1 reduces synaptic vesicle super-pool and spontaneous release frequency. eLife 8, e50401 (2019).
Acknowledgements
We thank H. Wang (Zhejiang University) for collecting the primary AML samples and providing the in vitro-transcribed luciferase mRNA. We thank D. Huang (Zhejiang University) for providing HL-60 cells stably expressing luciferase. Cas9-GFP transgenic mice were a gift from Z. Meng (Zhejiang University). This work was supported by grants from the National Key Research and Development Program of China (2022YFA1103500 to P.Q.), the National Natural Science Foundation of China (82222003 and 92268117 to P.Q.; 82200162 to L.Y.), the Zhejiang Provincial Natural Science Foundation of China (Z24H080001 to P.Q.; LQ23H080002 to L.Y.), the Key R&D Program of Zhejiang (2024SSYS0024 to P.Q.), the Department of Science and Technology of Zhejiang Province (2023R01012 to P.Q.), the Fundamental Research Funds for the Central Universities (226-2024-00007 to P.Q.), and the China Postdoctoral Science Foundation (2022M712765 to L.Y). We acknowledge technical support from Y. Wei, Z. Qianbing, S. Shichun and L. Xingguang at the Core Facilities, Zhejiang University School of Medicine, and Liangzhu Laboratory. This manuscript was edited at Life Science Editors. Figures 1a, 6b and 7k were created with BioRender.com.
Author information
Authors and Affiliations
Contributions
L.Y. and P.Q. conceived and designed the study. L.Y. performed the experiments, data analysis and paper writing. Z.Z. performed mouse experiments and cord blood-related experiments. P.J. and Z.Y. analysed sequencing data. D.K. assisted L.Y. with the NSG mouse xenograft studies and data analysis. D.S. and H.Y. performed the SPR assay. Y.H., E.C., W.Z., J. Sun, Y.Z., Y.L., J. Shi and H.H. contributed reagents and resource support. P.Q. supervised the overall project and co-wrote the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
All authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Denis Lafontaine and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Other candidates from CRISPR screen hits do not correlate with the prognosis of AML patients.
a, Recipient mice developed myeloid leukemia after transplantation as determined by flow cytometry analysis of peripheral blood GFP+BFP+ MLL-AF9-transformed donor cells. b, Survival of primary (1st) and secondary (2nd) recipients transplanted with GFP+BFP+ cells. Log rank (Mantel–Cox) test. c and d, Depleted RBPs in the in vivo CRISPR KO screens. Selective known positive regulators of AML are highlighted. FC, fold change. Displayed results derive from MAGeCK’s negative binomial analysis, with one-sided p-values for sgRNA effects. e, Gene ontology analysis of depleted hits in the CRISPR screen. GOBP, gene ontology biological process terms. ClusterProfiler: The one-tailed Fisher’s exact test with FDR. f, Overall survival analysis of other top ranked nucleolar proteins indicate that their expression levels do not correlate with the prognosis in AML patients28. Log rank Mantel–Cox test.
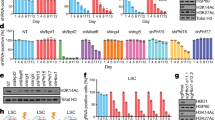
Extended Data Fig. 2 FBL depletion impairs AML cell behaviors in vitro.
a, Immunoblot validation of FBL depletion by two independent shRNAs (shFBL). Scrambled shRNA was used as control (shCT). b, Gating strategy of competition-based assay, cell cyle, differentiation and apoptosis determined by flow cytometry. c, Competition-based assay for the growth of AML cells transduced with shFBL or shCT lentivirus. n = 3 independent cultures. Two-way ANOVA with Dunnett’s multiple comparisons test. d, Flow cytometry analysis of cell cycle stage distribution after shFBL or shCT transduction for 4 days. n = 3 independent cultures. Two-way ANOVA with Dunnett’s test. e, Flow cytometry analysis of the proportion of CD11b-expressing AML cells 4 days after shFBL or shCT lentivirus transduction.n = 4 (HL-60) or 3 (OCI-AML3) independent cultures. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test. f, Flow cytometry analysis for apoptotic cell proportion 6 days post-transduction with shFBL or shCT lentivirus. n = 3 independent cultures. Two-way ANOVA with Dunnett’s test. g-i, representative plots of cell cycle (g), differentiation (h) and apoptosis (i) detection in d-f. Mean ± SEM (c, and d-f).
Extended Data Fig. 3 FBL deficiency impedes LSC self-renewal with minimal impact on normal human HSPCs.
a, Proportion of leukemic stem/progenitor cells (Lin-CD34+CD38−) and leukemic stem cells (Lin−CD34+CD38−TIM3+CD99+) in FBL-knockdown or control cells (GFP+) (4 d post-transfection). n = 3 (OCI-AML3) or 4 (HL-60) independent cultures. One-way ANOVA with Dunnett’s test. b, Gating strategy (upper) and representative plots of LSPC and LSC detection in a. c, Colony formation assay using sorted AML cells transduced with shFBL or shCT lentivirus. n = 4 (OCI-AML3) or 5 (HL-60) independent cultures. One-way ANOVA with Dunnett’s test. d, Colony photos from FBL-depleted or control AML cells, with scale bars shown. e, Colony formation ability of CD34+ HSPCs from human umbilical cord blood (hUCB) expressing shFBL or shCT. Different categories of CFUs are shown. BFU, burst forming unit-erythroid. M, monocyte. G, granulocyte. GM, granulocyte-megakaryocytes. GEMM, granulocyte-erythrocyte-monocyte- megakaryocytes. n = 3 independent hUCB samples. Two tailed unpaired student’s t test. f, Colony formation ability of CD34+ HSPCs from hUCB overexpressing FBL or control. n = 3 independent hUCB samples. Two tailed unpaired student’s t test. Mean ± SEM (a, c, e and f).
Extended Data Fig. 4 The PS domains with non−random sequence features of FBL are required for the growth of AML cells.
a and b, FBL with PS-competent domains rescues growth defects induced by FBL deficiency in HL-60 cells (negative-selection competition assay). n = 3 biological replicates. Two-way ANOVA with Dunnett’s test. c and d, Cell cycle analysis in FBL-depleted HL-60 cells expressing ectopic FBL or mutants. n = 3 biological replicates. Two-way ANOVA with Dunnett’s test. e and f, chimeric PS competent-mutant PLD-MD fails to rescue FBL deficiency-induced growth (e) and colony formation (f) defects. PLD (prion-like domain) is another IDR from hnRNPA1 protein. g, Schematic of FBL domain organization and chimeric mutants (top). Protein disorder prediction of these sequences by IUPred3 (middle) and single amino acid charge distribution analysis along these sequences by EMBOSS (bottom). NCPR represents the net charge per residue. h, Amino acid compositional profile of sequences in g. “Polar” includes Gly, Ser, Thr, Asn, Gln, and Cys. “Aromatic” includes Phe, Tyr, Trp, and His. “Charged” includes Asp, Glu, Arg, and Lys. “Hydrophobic” includes Leu, Iso, Val, and Met. i, Quantitative summary of the area of droplets formed by FBL and its indicated mutants in AML cells (related to Fig. 4i). n = 31 (FBL, MUT-E and GAR), 33 (4KR, 4KQ, MD, H2B-MD and PLD-MD), 35 (MD-GAR) and 36 (RGG-MD) cells from 3 independent replicates. Kruskal-Wallis (Dunnett’s multiple comparisons) test. Mean ± SEM (a, b and d-f).
Extended Data Fig. 5 Phase behavior of FBL and its mutants in vitro and in cells.
a, Purified EGFP-GAR, EGFP-MD, EGFP-FBL proteins were analyzed by SDS–PAGE and detected by Coomassie blue staining. b, Representative fluorescence and DIC images of EGFP-FBL (10 μM) droplets formation in phase separation buffer (see methods) with variable concentrations of NaCl. c, Related to Fig.4j: Fitted recovery curves of the FRAP assay in vitro, as described in the Methods. d, FRAP assay of 4KR, 4KQ and MUT-E mutants shows no significant difference compared with wild type FBL in AML cells. n = 10 (4KQ), 13 (FBL, 4KR) and 14 (MUT-E) cells from 3 independent experiments. e, Related to Fig.4h and Extended Data Fig. 5d: Fitted recovery curves of the FRAP assay in AML cells. f, Representative micrographs of FRAP in AML cells, related to e. g-h, Quantification of the FRAP assay in 293 T cells. n = 5 (GFP), 8 (FBL), 10(RGG-MD, GAR-Helix, 4KQ, MUT-E), 11 (MD, MD-GAR, H2B-MD, 4KR) and 12 (GAR) cells from 2 independent experiments. i, Fitted recovery curves of the FRAP assay in 293 T cells. Mean ± SEM (d-e and g-i). Area within error bands were filled with light color (d, g and h).
Extended Data Fig. 6 The role of FBL on rRNA transcription and rRNA 2’-O-Me modification in AML cells.
a, Immunoblot analysis of H2AQ104me modification using anti-H2AQ104me in FBL-KD HL-60 cells rescued by different FBL mutants as indicated. b and c, Representative images (b) and quantitative summary (c) of nascent rRNA in FBL-KD HL-60 cells expressing WT FBL or the indicated mutants. n = 46 (shCT + EV and shFBL + MD-GAR), 47 (shFBL + EV and shFBL + FBL), 44 (shFBL + 4KR/4KQ), 40 (shFBL + MUT-E), 51 (shFBL + MD), 50 (shFBL + H2B-MD), 39 (shFBL + RGG-MD), 54 (shFBL + GAR) cells from 2 independent replicates. Kruskal-Wallis test. d and e, Quantification of total 2’-O-Me sites (Methscore >0.8) in different rRNA species (d) or of four different nucleotides (adenosine, uridine, guanosine and cytidine). f-h, Methscore values for significant downregulated 2’-O-Me sites in 28 S (f), 18 S (g) and 5.8 S (h) detected by RiboMeth-seq in FBL-KD HL-60 cells, compared to the control group. n = 3 biological replicates. Lines indicate the sites subsequently validated by RTL-P. i, RTL-P assays were conducted to confirm the downregulation of 2’-O-Me sites in 28 S and 5.8 S and 18 S by FBL deficiency as indicated in f-h. n = 3 biological replicates. One-way ANOVA with Dunnett’s test. Mean ± SEM (c-i).
Extended Data Fig. 7 FBL’s effect on rRNA 2’-O-methylation is independent on the regulation of other snoRNPs and snoRNAs in AML cells.
a-b, Densitometry quantification of rRNA intermediates (a) and mature rRNAs (b) (related to Fig. 5l). Intermediates 30 S, 21 S, and 18SE are normalized to 47 S species. Etbr: Ethidium bromide. n = 3 independent experiments. One-way ANOVA with Dunnett’s test. c, Protein levels of other C/D box snoRNPs (NOP58, NOP56, SNU13) in healthy donor CD34+ HSPC (n = 13) and AML patients (n = 177) from the data reported by Oellerich et al.28. Two-tailed unpaired t test with Welch’s correction. d, Normalized mRNA levels of FBL in normal BM (n = 407) and AML patient (n = 151) samples from TCGA datasets. Two-tailed unpaired Mann–Whitney test. e, Kaplan-Meier curve for overall survival analysis of the patients based on the protein level in their AML quantified by mass spectrometry28. Log rank Mantel-Cox test. f and g, QPCR analysis of snoRNPs mRNA levels (f) and western blot analysis of snoRNPs protein levels (g) in HL-60 cells transfected with shFBL lentivirus for 3 days. h, QPCR analysis of C/D box snoRNAs (which have been reported to be highly expressed in AML) in HL-60 cells transfected with shFBL lentivirus for 3 days. n = 3 biological replicates, two-way ANOVA with Dunnett’s test in f and h. Mean ± SEM (a-d, f and h).
Extended Data Fig. 8 FBL enhances the translation of oncogenic genes in AML cells.
a, Gene Ontology analysis of translationally upregulated genes in FBL-KD cells. ClusterProfiler: The one-tailed Fisher’s exact test with FDR. b, Up-(red) and down-(blue) regulated genes at the mRNA level detected by RNA-seq from FBL-KD HL-60 cells, compared to the control. c, Venn diagrams of differentially expressed (ΔmRNA, Log2 | mRNA FC | > 0.7 and P < 0.05, two-sided Wilcoxon test) or differentially translated genes (ΔTE, Log2 | mRNA FC | > 0.7 and P < 0.05, two-sided Wilcoxon test). Red, up-regulated genes in FBL-KD HL-60 cells. Blue, downregulated genes in FBL-KD HL-60 cells. d and e, Gene Ontology analysis of differentially expressed genes in FBL-KD cells, compared to control. ClusterProfiler: The one-tailed Fisher’s exact test with FDR. Down, downregulated by FBL depletion (d). UP, upregulated by FBL depletion (e). f-j, Rescue assay showing that ectopic expression of MYC (f) partially restored the cellular phenotype observed in FBL-deficient cells, such as growth (g), differentiation (h), cell death (i), and colony formation (j). Representative western blot images were shown from 3 independent experiments in n. n = 3 biological replicates, and statistical analysis was performed by two-way ANOVA (g) or One-way ANOVA with Turkey’s multiple comparison test (h-j). k, Rescue effect of IL6 in FBL-KD HL-60 on cell proliferation. n = 3 biological replicates. Two-way ANOVA with Dunnett’s test. Mean ± SEM (g-k).
Extended Data Fig. 9 Specific validation of CGX-635 impact on FBL phase separation.
a and b, SPR assay showing no binding ability of CHX (a) or 5-FU (b) with recombinant FBL protein. c, Immunoblot assay coupled with native PAGE showing no alteration of FBL self-association in AML cells treated with CHX or 5-FU. d, Representative IF staining images show no alteration in nucleolar localization of NPM1(red) and FBL (green) in AML cells treated with CGX-635 (50 nM), CHX (10 μg/ml) or 5-FU (100 μM) for 6 hours. e, FRAP assay shows no influence on the mobility of FBL-GFP puncta in 293 T cells after treated with CHX (10 μg/ml) and 5-FU (100 μM) for 6 h. n = 7 (5-FU) and 8 (Ctrl and CHX) cells from 2 independent experiments. Two-way analysis of ANOVA. f and g, Representative images (f) and quantification (g) of FRAP assay of condensates formed by NPM1 in HL-60 cells. n = 6 (CGX-635, 50 nM) and 7 (Ctrl) cells from 2 independent experiments. Two-way ANOVA test. h and i, Representative images (h) and quantification (i) of FRAP assay of condensates formed by PLD-MD in HL-60 cells. n = 7 (Ctrl) and 8 (CGX-635, 50 nM) cells from 2 independent experiments. Two-way ANOVA test. j-l Representative images and circularity of droplet formation of NPM1 (j, 20 μM), PLD-MD (k, 100 μM) and GAR (l, 5 μM) with or without CGX-635 (50 μM) in vitro. n = 115 (Ctrl) and 117 (CGX-635) droplets in j. n = 144 (CGX-635) and 157 (Ctrl) droplets in h. n = 103 (Ctrl) and 119 (CGX-635) droplets in l. All from 2 independent experiments. Two-tailed unpaired Mann–Whitney test. Mean ± SEM (e, g, i and j-l). Area within error bands were filled with light color (e, g and i).
Supplementary information
Supplementary Table 1
Supplementary Table 1. RBPs CRISPR library and CRISPR score of genes encoding nucleolar proteins. Supplementary Table 2. Oligonucleotides used for this study. Supplementary Table 3. RiboMethSeq data. Supplementary Table 4. List of differentially translated genes upon FBL inhibition. Supplementary Table 5. List of differentially expressed genes upon FBL inhibition.
Source data
Source Data 1
All Statistical data in Figs. 1–8 and Extended Data Fig. 1-9.
Source Data 2
All unprocessed blots and/or gels for each relevant Figure.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, L., Zhang, Z., Jiang, P. et al. Phase separation-competent FBL promotes early pre-rRNA processing and translation in acute myeloid leukaemia. Nat Cell Biol (2024). https://doi.org/10.1038/s41556-024-01420-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41556-024-01420-z