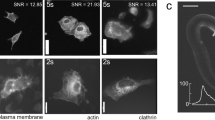
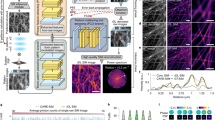
Abstract
Simultaneous imaging of various facets of intact biological systems across multiple spatiotemporal scales is a long-standing goal in biology and medicine, for which progress is hindered by limits of conventional imaging modalities. Here we propose using the refractive index (RI), an intrinsic quantity governing light–matter interaction, as a means for such measurement. We show that major endogenous subcellular structures, which are conventionally accessed via exogenous fluorescence labelling, are encoded in three-dimensional (3D) RI tomograms. We decode this information in a data-driven manner, with a deep learning-based model that infers multiple 3D fluorescence tomograms from RI measurements of the corresponding subcellular targets, thereby achieving multiplexed microtomography. This approach, called RI2FL for refractive index to fluorescence, inherits the advantages of both high-specificity fluorescence imaging and label-free RI imaging. Importantly, full 3D modelling of absolute and unbiased RI improves generalization, such that the approach is applicable to a broad range of new samples without retraining to facilitate immediate applicability. The performance, reliability and scalability of this technology are extensively characterized, and its various applications within single-cell profiling at unprecedented scales (which can generate new experimentally testable hypotheses) are demonstrated.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






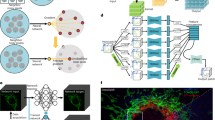
Similar content being viewed by others
Data availability
RI2FL datasets are available at https://github.com/NySunShine/ri2fl. Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
RI2FL source code, example datasets and step-by-step interactive tutorials are available at https://github.com/NySunShine/ri2fl.
References
Mertz, J. Introduction to Optical Microscopy (Cambridge Univ. Press, 2019).
Lichtman, J. W. & Conchello, J.-A. Fluorescence microscopy. Nat. Methods 2, 910–919 (2005).
Christiansen, E. M. et al. In silico labeling: predicting fluorescent labels in unlabeled images. Cell 173, 792–803.e19 (2018).
Ounkomol, C., Seshamani, S., Maleckar, M. M., Collman, F. & Johnson, G. R. Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy. Nat. Methods 15, 917–920 (2018).
Guo, S.-M. et al. Revealing architectural order with quantitative label-free imaging and deep learning. eLife 9, e55502 (2020).
Cheng, S. et al. Single-cell cytometry via multiplexed fluorescence prediction by label-free reflectance microscopy. Sci. Adv. 7, eabe0431 (2021).
Rivenson, Y. et al. PhaseStain: the digital staining of label-free quantitative phase microscopy images using deep learning. Light Sci. Appl. 8, 23 (2019).
Nygate, Y. N. et al. Holographic virtual staining of individual biological cells. Proc. Natl Acad. Sci. USA 117, 9223–9231 (2020).
Kandel, M. E. et al. Phase imaging with computational specificity (PICS) for measuring dry mass changes in sub-cellular compartments. Nat. Commun. 11, 6256 (2020).
Kandel, M. E. et al. Multiscale assay of unlabeled neurite dynamics using phase imaging with computational specificity. ACS Sens. 6, 1864–1874 (2021).
Park, Y., Depeursinge, C. & Popescu, G. Quantitative phase imaging in biomedicine. Nat. Photonics 12, 578–589 (2018).
Kim, K. et al. Correlative three-dimensional fluorescence and refractive index tomography: bridging the gap between molecular specificity and quantitative bioimaging. Biomed. Opt. Express 8, 5688–5697 (2017).
Lee, J. et al. Deep-learning-based label-free segmentation of cell nuclei in time-lapse refractive index tomograms. IEEE Access 7, 83449–83460 (2019).
Park, S. et al. Label-free tomographic imaging of lipid droplets in foam cells for machine-learning-assisted therapeutic evaluation of targeted nanodrugs. ACS Nano 14, 1856–1865 (2020).
Yoon, J. et al. Label-free identification of lymphocyte subtypes using three-dimensional quantitative phase imaging and machine learning. J. Vis. Exp. https://doi.org/10.3791/58305 (2018).
Gal, Y. & Ghahramani, Z. Dropout as a Bayesian approximation: representing model uncertainty in deep learning. Proc. Int. Conf. Mach. Learn. 48, 1050–1059 (2016).
Wang, G. et al. Aleatoric uncertainty estimation with test-time augmentation for medical image segmentation with convolutional neural networks. Neurocomputing 338, 34–45 (2019).
Weigert, M. et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods 15, 1090–1097 (2018).
Xue, Y., Cheng, S., Li, Y. & Tian, L. Reliable deep-learning-based phase imaging with uncertainty quantification. Optica 6, 618–629 (2019).
Kendall, A. & Gal, Y. What uncertainties do we need in Bayesian deep learning for computer vision? In Proc. 31st International Conference on Neural Information Processing Systems (eds von Luxburg, U. et al.) 5574–5584 (Curran Associates, 2017).
Zheng, G., Horstmeyer, R. & Yang, C. Wide-field, high-resolution Fourier ptychographic microscopy. Nat. Photonics 7, 739–745 (2013).
Baek, Y., Lee, K., Shin, S. & Park, Y. Kramers–Kronig holographic imaging for high-space-bandwidth product. Optica 6, 45–51 (2019).
Bray, M.-A. et al. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat. Protoc. 11, 1757–1774 (2016).
Barer, R. Determination of dry mass, thickness, solid and water concentration in living cells. Nature 172, 1097–1098 (1953).
Cooper, K. L. et al. Multiple phases of chondrocyte enlargement underlie differences in skeletal proportions. Nature 495, 375–378 (2013).
Mir, M. et al. Optical measurement of cycle-dependent cell growth. Proc. Natl Acad. Sci. USA 108, 13124–13129 (2011).
McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426 (2018).
Martin, K., Vilela, M., Jeon, N. L., Danuser, G. & Pertz, O. A growth factor-induced, spatially organizing cytoskeletal module enables rapid and persistent fibroblast migration. Dev. Cell 30, 701–716 (2014).
Kim, K., Kim, K. S., Park, H., Ye, J. C. & Park, Y. Real-time visualization of 3-D dynamic microscopic objects using optical diffraction tomography. Opt. Express 21, 32269–32278 (2013).
Inoue, T., Do Heo, W., Grimley, J. S., Wandless, T. J. & Meyer, T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat. Methods 2, 415–418 (2005).
Liu, P. et al. Rab-regulated interaction of early endosomes with lipid droplets. Biochim. Biophys. Acta 1773, 784–793 (2007).
Kounakis, K., Chaniotakis, M., Markaki, M. & Tavernarakis, N. Emerging roles of lipophagy in health and disease. Front. Cell Dev. Biol. 7, 185 (2019).
Pandarinath, C. et al. Inferring single-trial neural population dynamics using sequential auto-encoders. Nat. Methods 15, 805–815 (2018).
Topol, E. J. High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. 25, 44–56 (2019).
Jo, Y. et al. Quantitative phase imaging and artificial intelligence: a review. IEEE J. Sel. Top. Quantum Electron. 25, 1–14 (2019).
Chen, M., Ren, D., Liu, H.-Y., Chowdhury, S. & Waller, L. Multi-layer Born multiple-scattering model for 3D phase microscopy. Optica 7, 394–403 (2020).
Lim, J., Ayoub, A. B., Antoine, E. E. & Psaltis, D. High-fidelity optical diffraction tomography of multiple scattering samples. Light Sci. Appl. 8, 82 (2019).
Doshi-Velez, F. & Kim, B. Towards a rigorous science of interpretable machine learning. Preprint at https://arxiv.org/abs/1702.08608 (2017).
Sussillo, D. & Barak, O. Opening the black box: low-dimensional dynamics in high-dimensional recurrent neural networks. Neural Comput. 25, 626–649 (2013).
Zaritsky, A. et al. Interpretable deep learning uncovers cellular properties in label-free live cell images that are predictive of highly metastatic melanoma. Cell Syst. 12, 733–747.e6 (2021).
Lim, J. et al. Comparative study of iterative reconstruction algorithms for missing cone problems in optical diffraction tomography. Opt. Express 23, 16933–16948 (2015).
Jo, Y., Park, W. S. & Park, Y. Holotomographic imaging of eukaryotic cells. Protoc. Exch. https://doi.org/10.21203/rs.3.pex-1667/v1 (2021).
Kim, S. et al. in International Conference on Medical Image Computing and Computer-Assisted Intervention (eds Shen, D. et al.) 220-228 (Springer, 2019).
Tan, M. & Le, Q. Efficientnet: rethinking model scaling for convolutional neural networks. Proc. Int. Conf. Mach. Learn. 97, 6105–6114 (2019).
Berg, S. et al. Ilastik: interactive machine learning for (bio) image analysis. Nat. Methods 16, 1226–1232 (2019).
Kandel, M. E. et al. Reproductive outcomes predicted by phase imaging with computational specificity of spermatozoon ultrastructure. Proc. Natl Acad. Sci. USA 117, 18302–18309 (2020).
Acknowledgements
We thank the members of KAIST Biomedical Optics Laboratory for helpful discussions. This work was supported by the KAIST Up program, the BK21+ program, Tomocube, the National Research Foundation of Korea (2015R1A3A2066550), an Institute of Information & Communications Technology Planning & Evaluation (IITP; 2021-0-00745) grant and the Commercialization Promotion Agency for R&D Outcomes (COMPA; 055586) funded by the Korean government (MSIT) to Y.P.; the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Korea (HI21C0977) to H.-s.M.; and a KAIST Presidential Fellowship and Asan Foundation Biomedical Science Scholarship to Y.J.
Author information
Authors and Affiliations
Contributions
Y.J. and Y.P. conceived the idea. Y.J. coordinated the project and carried out all analyses. Y.J., H.C. and H.-s.M. designed the deep-learning pipeline, and H.C. implemented and optimized the pipeline. Y.J., W.S.P. and W.D.H. designed the main experiments, and W.S.P. collected the data. W.S.P. and W.D.H. established the molecular biology and imaging protocols. G.K., D.R., Y.S.K. and M.L. contributed to processing the data. H. Joo, H. Jo and Sumin Lee collected and processed the large FOV data. S.P., M.J.L. and Seongsoo Lee contributed new RI tomogram datasets for testing the applicability of the technique. Y.J., H.C., W.S.P. and Y.P. wrote the manuscript with input from all co-authors. Y.P. supervised all aspects of the work.
Corresponding authors
Ethics declarations
Competing interests
H.C., M.L., H. Jo, Sumin Lee, H.-s.M. and Y.P. have financial interests in Tomocube, a company that commercializes ODT and QPI instruments and is one of the sponsors of the work. The remaining authors declare no competing interests.
Additional information
Peer review information Nature Cell Biology thanks Gaudenz Danuser, and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Network architecture and patch-based processing.
(a) We used a single encoder-decoder network, systematically discovered by SCNAS, for all subcellular targets. (b) An example inference from an RI tomogram to the corresponding actin tomogram is illustrated as a flow chart, highlighting the patch-based processing for GPU memory management. All images represent MIPs of 3D data.
Extended Data Fig. 2 Discrete structure-based characterization of RI2FL performance.
Discrete subcellular structures, specifically lipid droplets for acceptable segmentation, were detected and matched in the inferred and ground-truth FL data in the held-out dataset. (a) Schematic summarizing the segmentation procedure (see Methods for details). (b) Matched versus unmatched lipid droplets could be well-defined based on inter-centroid distance and overlap fraction. (c) Distribution of inter-centroid distance in the matched lipid droplets. (d) Correlation between measured and inferred intensity averaged over the lipid droplet segments. The black line represents the least-squares linear regression fit of the data. N = 282 lipid droplets. r, Pearson’s correlation coefficient (calculated using the definition). Numerical source data is provided in source data.
Extended Data Fig. 3 Comparison between pre- and post-staining data.
In order to validate the inference of endogenous subcellular targets, we imaged identical cells before and after staining. The qualitative correspondence between the pre- and post-staining data, despite the irreducible discrepancy due to the temporal difference (from minutes to an hour) and fixation, further validates the successful operation of RI2FL in unlabeled cells.
Extended Data Fig. 4 Iterative tomographic reconstruction and RI2FL performance.
(a) RI reconstruction accuracy over iterations of non-negativity-constrained missing cone recovery algorithm in tomographic reconstruction. Note that the first few iterations are essential for accurate RI reconstruction but around the chosen condition (40 iterations) RI is minimally sensitive to the number of iterations. (b) FL prediction accuracy as a function of RI reconstruction accuracy. Note the strong positive correlation between RI reconstruction accuracy and FL prediction accuracy. Interestingly, this error sensitivity seems to be dependent on the target structure: for example, lipid droplets inference (yellow dots) are least sensitive to RI error. N = 41 tomograms in the test set. Numerical source data is provided in source data.
Extended Data Fig. 5 Uncertainty quantification schemes and visualizations.
(a, b) Quantifications of (a) data uncertainty and (b) model uncertainty were conducted by test-time augmentation and Monte Carlo dropout, respectively. All images in (a) and (b) represent MIPs of 3D data. (c, d) Quantitative visualizations of data uncertainty (c) on a 2D cross-section of Fig. 3a, t = 120 min and (d) along the 1D dashed line shown in (c).
Extended Data Fig. 6 Validation of RI2FL-inferred features.
Total 65 features were extracted with the inferred or ground-truth FL data in the held-out dataset, for one FL channel at a time (but note that most features are defined based on multiple channels; Fig. 3a). (a, b) Example features with inferred or ground-truth actin. (c) Distribution of r values for all features over the six FL channels. r = 0.97 across all features and channels. The least accurate features were actin-mitochondria correlation, mitochondria contrast, RI-lipid droplets correlation, cytoplasmic RI entropy, nuclear sphericity, and RI-nucleoli correlation, for actin, mitochondria, lipid droplets, plasma membranes, nuclei, and nucleoli channels, respectively. N = 102 cells. Numerical source data is provided in source data.
Extended Data Fig. 7 Intracellular distribution of inferred lipid droplets and measured endosomes.
Unexpectedly, the intracellular distribution of the inferred lipid droplets was starkly different from that of the ground-truth endosomes (see main text). While the lipid droplets were strongly correlated with high RI, this was not the case for endosomes.
Extended Data Fig. 8 Deep feature visualization.
In order to facilitate interpreting the operation of the trained networks, feature map activations for a single input tomogram were visualized. (a) Average feature map at each layer. (b, c) Individual feature maps at the last layers of (b) encoder and (c) decoder parts of the network inferring plasma membranes. All images represent MIPs of 3D data.
Supplementary information
Supplementary Information
Supplementary Note 1
Supplementary Tables
Table 1: Comparison of the related cross-modality inference methods. Table 2: Dataset summary. Table 3: Performance metrics. Table 4: Single-cell feature statistics.
Supplementary Video 1
RI2FL across cell types and subcellular targets.
Supplementary Video 2
Dynamics of cell division.
Supplementary Video 3
Large-scale imaging.
Supplementary Video 4
Growth factor stimulation.
Supplementary Video 5
Chemogenetic stimulation.
Rights and permissions
About this article
Cite this article
Jo, Y., Cho, H., Park, W.S. et al. Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning. Nat Cell Biol 23, 1329–1337 (2021). https://doi.org/10.1038/s41556-021-00802-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-021-00802-x
This article is cited by
-
Deep-learning based 3D birefringence image generation using 2D multi-view holographic images
Scientific Reports (2024)
-
On the use of deep learning for phase recovery
Light: Science & Applications (2024)
-
High-resolution assessment of multidimensional cellular mechanics using label-free refractive-index traction force microscopy
Communications Biology (2024)
-
Digital staining in optical microscopy using deep learning - a review
PhotoniX (2023)
-
Machine-learning-based diagnosis of thyroid fine-needle aspiration biopsy synergistically by Papanicolaou staining and refractive index distribution
Scientific Reports (2023)