Abstract
Olfactory receptors (ORs) which are mainly known as odor-sensors in the olfactory epithelium are shown to be expressed in several non-sensory tissues. Despite the specified role of some of these receptors in normal physiology of the kidney, little is known about their potential effect in renal disorders. In this study, using the holistic view of systems biology, it was determined that ORs are significantly changed during the progression of kidney fibrosis. For further validation, common differentially expressed ORs resulted from reanalysis of two time-course microarray datasets were selected for experimental evaluation in a validated murine model of unilateral ureteral obstruction (UUO). Transcriptional analysis by real-time quantitative polymerase chain reaction demonstrated considerable changes in the expression pattern of Olfr433, Olfr129, Olfr1393, Olfr161, and Olfr622 during the progression of kidney fibrosis. For localization of these ORs, single-cell RNA-sequencing datasets of normal and UUO mice were reanalyzed. Results showed that Olfr433 is highly expressed in macrophages in day-2 and 7 post-injury in UUO mice and not in normal subgroups. Besides, like previous findings, Olfr1393 was shown to be expressed prominently in the proximal tubular cells of the kidney. In conclusion, our combinatorial temporal approach to the underlying mechanisms of chronic kidney disease highlighted the potential role of ORs in progression of fibrosis. The expression of Olfr433 in the macrophages provides some clue about its relation to molecular mechanisms promoted in the fibrotic kidney. The proposed ORs in this study could be the subject of further functional assessments in the future.
Similar content being viewed by others
Introduction
Olfactory receptors (ORs), belonging to a super-family of G protein-coupled receptors, are well-recognized for their role in odor-sensation in the olfactory epithelium1. This family of receptors was first discovered by Linda Buck and Richard Axel1, leading to a Nobel prize in 20042. More investigations on ORs determined that they are not only expressed in the olfactory epithelium but also non-sensory organs3. The study by Parmentier et al. determined the functionality of these receptors in sperm chemotaxis during fertilization4. Furthermore, it was demonstrated that these receptors have roles in cytoskeletal remodeling, pulmonary hyperplasia5, angiogenesis6, as well as heart metabolism in the cardiovascular system7. Besides, identification of the role of these receptors in other parts of the body, such as skin8,9, gastrointestinal10,11,12, and the immune system13,14 highlights the importance of further investigations on their function in the body.
One of the tissues, in which the presence and function of ORs are investigated, is the kidney15,16,17,18,19. Studies using unsupervised high-throughput techniques have discovered the existence of these receptors in the kidney20,21. Nevertheless, few studies have specifically focused on their role in renal function. The study by Pluznick et al. determined Olfr78 role in blood pressure regulation through interactions with the byproducts of gut microbiota22. Furthermore, Shepard et al. demonstrated that Olfr1393 participates in glucose transportation in both normal23 and pathological24 states of the kidney. Considering the magnitude of this gene family (around 1000 genes in the mouse and 400 genes in the human), as well as their role in chemosensation19,25, more investigations are needed to uncover the role of other OR subtypes in kidney function and hemostasis. Despite the above-mentioned few findings on the role of these receptors in the normal state of the kidney, no remarkable study has focused on their function in the progression of kidney fibrosis. Renal fibrosis is the common pathological manifestation of a variety of disorders leading to chronic kidney disease (CKD)26,27 which causes vast tubular atrophy and glomerulosclerosis27. Unfortunately, dialysis and kidney transplantation are the only treatments in the progressive states of the disease which cause serious complications27. In this regard, basic studies on the molecular mechanisms of the disease are crucial for developing new treatment strategies.
In this study, two time-course microarray datasets from a mouse model of unilateral ureteral obstruction (UUO) were reanalyzed and the gene interaction networks of differentially expressed genes (DEGs) were constructed. Experimental evaluation of common ORs between two datasets determined significant changes in the expression patterns of these genes during the progression of kidney fibrosis. Moreover, by analyzing single-cell RNA-sequencing (scRNA-seq) datasets, we could find some clues about the localization and function of these ORs.
Results
In order to investigate the underlying molecular mechanisms activated during the progression of CKD, two time-course microarray datasets, GSE3649628 and GSE9657129 were reanalyzed. GSE36496 dataset contains transcriptomics data of the UUO and sham-operated C57BL/6 mice at days-1, 2, 5, and 9 postoperation28. The results of this study by Wu et al. suggest CEBPB and HNF4A signaling pathways as important regulators of kidney fibrosis28. The other dataset, GSE96571 which is also deposited by Wu et al., comprises the UUO and sham samples at hours-0.5, 1, 3, 5, 7, 12, as well as days-1, 3, 5, and 7 postoperation29. The single-time point analysis of this dataset by Wu et al. determined overexpression of stress responder genes in the first hours of obstruction besides nephrotoxic damage-related genes at later time points29. This dataset was used for the validation of our findings from the first dataset.
Unsupervised evaluation of microarray dataset determined the quality of datasets
The quality assessment of the GSE36496 dataset by principal component analysis (PCA) demonstrated the separation of sham and UUO groups from each other. Furthermore, the UUO samples were separated based on the times of harvest, which determines the model quality (Fig. 1a). This result was also validated by the hierarchical clustering (Fig. 1b). The DEGs were determined using the linear models for microarray data (LIMMA)30 Package of R software31, which according to our recent study32 is the most reliable tool for time-course analysis of microarray data. A Comparison of the UUO and sham samples determined 2583 DEGs with an adjusted p value < 0.05. The DEGs were used for the construction of a gene interaction network and further topological analysis (Fig. 1c).
Principal component analysis (PCA) and heatmap clustering of GSE36496 dataset demonstrated good quality of samples regarding treatment and times of harvest (a, b). Gene interaction network created from 2583 differentially expressed genes (DEGs) of GSE36496 dataset (c). The first- (d) and the second-ranked (e) modules resulted from topological analysis of the network constitute NDUF and olfactory receptor (OR) gene families, respectively. Gene Interaction network from 3176 DEGs of GSE96571 dataset (f). The top-ranked module from topological analysis of the constructed network is constituted of ORs (g). The overlap between differentially expressed ORs of GSE36496 and GSE96571 datasets (h).
ORs constitute a dense module in the interactome map of kidney fibrosis
In order to discover the functional units of the network, module analysis was performed. Modules are individual units of biological networks that are similar in physical, chemical, or functional aspects and supposed to have a specific function in the networks33,34. Assessment of densely connected network regions, based on the clustering coefficient, revealed 36 significant modules. The first and second modules were mainly related to NDUF and ORs signaling pathways (Fig. 1d, e). NADH:ubiquinone oxidoreductase supernumerary subunits (NDUF) family of genes are expressed in the mitochondria and their relation to kidney fibrosis has previously been demonstrated by Granata et al.35. Despite evidence on the contribution of Olfr1393 to the progression of type 2 diabetes24, no study has shown the relation of ORs to kidney fibrosis. Two non-OR genes, Arrb2 and Grk3, were also observable in the ORs module. Studies revealed the impact of these genes on ORs regulation, as Arrb2 inhibits the activation of ORs36 and Grk3 has a role in ORs desensitization37.
For further investigation on the role of ORs in kidney fibrosis, another time-course UUO dataset was analyzed. LIMMA results determined 3176 DEGs (adjusted p value < 0.05), which were further used for the construction of a gene interaction network (Fig. 1f). Topological analysis of the constructed network revealed 44 significant modules, of which ORs-related module obtained the first rank (Fig. 1g). Overlaying the significantly expressed ORs between two datasets determined Olfr433, Olfr129, Olfr1393, Olfr161, and Olfr622 as common ORs (Fig. 1h and Supplementary Fig. 1). These five common ORs were selected for in vivo expression analysis.
Histopathological analysis of the UUO model validated the robustness of the constructed model
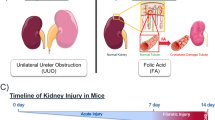
In order to experimentally evaluate the expression of selected ORs, a mouse model of UUO was developed. Both UUO and sham-operated mice were followed over 21 days (Fig. 2a). For validation of the quality of the constructed model, the histopathological analysis was performed. Assessment of the sections revealed significant diffused tubulointerstitial fibrosis along with glomerular injuries, increased mesangial matrix, and diffused glomerulosclerosis in UUO-operated mice compared to the sham group over 21 days of UUO treatment (p value < 0.05), all of which determined the robustness of constructed UUO model (Fig. 2b and c). Furthermore, to show that the developed animal model recapitulates what was reported in both studies by Wu et al., we assessed the expression of some of the genes by real-time quantitative polymerase chain reaction (RT-qPCR). In agreement with the results of study published in 201228, Hnf4a was down-regulated and Cebpb was up-regulated in our model at day-12. Also, Serpina3n and Tgfb1 that were shown to be up-regulated in a more recent study29, were increased in our expression analysis (Supplementary Table 1).
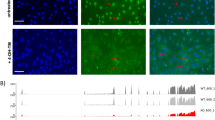
The scheme of experimental design (a). Trichrome and hematoxylin and eosin (H&E) -stained renal sections in normal, sham (day-21), and UUO-operated mice at days-1, 3, 6, 9, 12, 15, 18, and 21 postsurgery. Yellow, red, and black arrows stand for increased mesangial matrix, mesangial cell proliferation, and diffuse glomerulosclerosis, respectively (b). The percentages of glomeruli with increased mesangial matrix (p value = 1.1451e-04) as well as cortical fibrosis (p value = 1.3868e-04) for sham and UUO-operated mice. Data are mean ± SEM (c). * Scale bars: 50 µm.
Gene expression patterns demonstrated significant changes in ORs in the fibrotic kidney
As ORs belong to a highly conserved superfamily which most of them have a similar sequence, the specificity of designed primers was checked and validated by sanger sequencing (Supplementary Data 1).
The expression of Olfr433, Olfr129, Olfr1393, Olfr161, and Olfr622 was evaluated over 21 days in a time-course manner in both UUO and sham-operated mice (Fig. 3 and Supplementary Data 2). Except for Olfr1393, other genes demonstrated upregulation over time in comparison to the sham group. To test whether these changes between the sham and UUO groups were significant during time, Friedman’s two-way ANOVA test was applied. The results of the analysis determined significant changes in patterns of expression in the UUO group compared to sham for all the genes (p value < 0.05). Considering the expression patterns over time revealed that Olfr433, Olfr129, Olfr161, and Olfr622 had a sharp downregulation from day-3 to 6. Although Olfr433 and Olfr161 expression continued with a smoother augmenting response, Olfr129 and Olfr622 had a second sharp peak at day-12 followed by a reduction in the consecutive days. Furthermore, the expression of these four genes in the sham group over time revealed oscillatory patterns, which is an initial clue about the inherent rhythmic pattern of OR genes in the kidney.
Olfr433 is highly expressed by macrophages of the fibrotic kidney
To get insight into the kidney cell types expressing the selected ORs, publicly available scRNA-seq datasets of a normal kidney38 and a mouse model of UUO at day-2 and 7 postinjury39 were reanalyzed. After data quality control and removing unwanted cells, findings demonstrated expression of Olfr1393 by proximal tubular (third segment) cells of the normal kidney. Consistent with our RT-qPCR results, no expression of Olfr1393 was detected in the UUO samples. On the other hand, the expression of Olfr433 was identified in both UUO day-2 and 7 postinjury. Our analysis showed that in day-2, Olfr433 is mainly expressed by macrophages and to some extent proliferating proximal tubules. Likewise, in day-7 postinjury, Olfr433 was shown to be predominantly expressed by macrophages (Fig. 4).
Expression of Olfr1393 in normal kidney (GSE119531) (a). Expression of Olfr433 in UUO samples at day-2 (b) and 7 (c) postinjury (GSE140023). The left uniform manifold approximation and projection (UMAP) plots represent unsupervised clustering of the scRNA-seq datasets. The middle UMAP plots show the normalized average expression of ORs genes (yellow and grey color show high and low expression of genes). The dot plots indicate expression of Olfr1393 and Olfr433 across identified clusters (the circle size denotes percent of cells expressing OR genes and red and blue colors indicate high and low expression of OR genes, respectively).
Discussion
In order to acquire a holistic view of molecular mechanisms of kidney fibrosis, two time-course microarray datasets were reanalyzed. Gene interaction map evaluation determined modular structures of densely connected ORs in both networks. In the next step, to validate in silico results, the common ORs between two datasets were selected for further in vivo analysis in the mouse model of UUO. Additionally, the localization of ORs was inspected by analysis of kidney scRNA-seq datasets.
Although the relationship between some of the ORs and the normal physiology of the kidney has been determined in recent years15,22,23, few studies have focused on the role of ORs in pathological states of the kidney. The study by Shepard et al. focused on the role of Olfr1393 in diabetes24. Additionally, our previous bioinformatic analysis determined a significant change in the expression of ORs in the rat model of ischemia-reperfusion injury40. In this study, based on in silico and in vivo investigations, we identified five ORs which all of them were significantly related to the progression of kidney fibrosis. Analysis of scRNA-seq datasets determined that Olfr1393 is expressed by proximal tubular cells of the normal kidney. Similar to RT-qPCR results, no expression of this gene was detected in the UUO samples. These findings are in line with the results of the study by Shepard et al., which reported expression of Olfr1393 in the proximal tubular cells of normal kidney23. On the other hand, our findings showed that Olfr433 is predominantly expressed by macrophages of the injured kidney and not in normal kidney. The presence of macrophages in the site of injury is correlated with severity of tubulointerstitial damages41. It is shown that macrophages are capable of activating fibrotic pathways through which uncontrolled wound-healing processes and tissue fibrosis are promoted42. Study by Feng et al. demonstrated that transforming growth factor-beta (TGF-β) signaling pathway is activated in M2 macrophages in the injured kidney43. Also, wingless-related integration site (Wnt) signaling pathway in renal macrophages promotes their polarization into M2 phenotype and progression of fibrosis44. Moreover, macrophages are a potential source of extracellular matrix components like collagen45, platelet-derived growth factor (PDGF)46, and matrix metalloproteinase-9 (MMP9)47. This data provides a clue that this receptor has a potential role in inflammatory response and generation of myofibroblasts and is suggestive of the critical role of ORs in the pathogenesis of renal failure. We also appreciate that the localization of these ORs must be validated by other experimental approaches. Activation-inhibition approaches are also necessitated to determine the ORs function in kidney fibrosis.
In this study, we reanalyzed two qualified time-course microarray datasets generated by Wu et al. In both studies, key RNA biomarkers and molecular mechanisms of obstructive nephropathy were introduced28,29. Time-course study designs are worth as they would better demonstrate the dynamism of cellular behavior and also reduce misinterpretations that may occur in single-point studies48,49. Unfortunately, due to cost and difficulty, such experiments are less considered by investigators and most of the gene expression studies are performed statically50. On the other hand, appropriate analysis of time-series data is of great importance and should be performed by suitable mathematical approaches32. Although both studies by Wu et al. were designed temporally, single time-point analysis prevented them from finding significant changes in the expression of OR genes over the course of the disease.
Beside analyzing microarray datasets, we developed a time-course in vivo model of renal obstruction and followed both sham and UUO samples over 21 days. Expression analysis of Olfr433, Olfr129, Olfr1393, Olfr161, and Olfr622 determined differential expression patterns for these genes during 21 days. For distinguishing real changes from noisy fluctuations, we compared the patterns in control and treatment groups. We appreciate that the inclusion of two mice per group in each time point is not an ideal sample size. However, based on the criteria discussed in our previous study48, we believe that this gene expression analysis is reliable as we performed time-course measurements for both UUO and sham groups. Considering that the differences were consistently observed in all-time points for all the examined genes, the alterations in gene expression were interpreted as real signals. In addition, the magnitude of fold changes was fairly high and the differences reached a statistically significant threshold. Moreover, evaluations determined rhythmic patterns in ORs expression in sham groups somehow similar to the rhythmic patterns, which is not far from the oscillatory function of the kidney51,52. These findings are only an initial clue about the rhythmic expression patterns of OR genes and further investigations on the relation of ORs expression patterns with kidney function would be of interest.
Taken together, these in silico and in vivo investigations validate the expression changes of ORs during the progression of kidney fibrosis. The time-course evaluations determined rhythmic and robust patterns of ORs expression and highlighted the potential role of Olfr433 in the progression of kidney fibrosis. However, future studies are required for further validation of these findings. This study is a good example of the potential capacity of systems biology unsupervised top-down strategy to unravel the neglected aspects of disease pathogenesis.
Material and Methods
Microarray datasets analysis
The GSE36496 microarray dataset deposited by Wu et al.28 was downloaded from the gene expression omnibus (GEO) database53. The quality of the data was assessed by PCA and hierarchical clustering using ggplot254 and pheatmap55 packages of R software31, respectively. To determine significantly expressed genes in this time-course dataset, we applied LIMMA30, a package of R software. Using the multiple comparison method of LIMMA, the sham and UUO groups were compared with each other at different time points and DEGs were determined by False Discovery Rate <0.05 (Benjamini–Hochberg). The second dataset, GSE96571 deposited by Wu et al.29, was also analyzed similarly, and DEGs were determined according to the previously mentioned criteria.
Network construction and topological analysis
The DEGs were used for the construction of a gene-interaction network using the Cluepedia plugin56 (version 1.5.5) of Cytoscape software57 (version 3.7.2). The interaction data for activation, binding, inhibition, and post-translational modification with a confidence rate of 0.8, was retrieved from the search tool for the retrieval of interacting genes/proteins (STRING) database58 (STRING-ACTION-SCORE_v10.0_10090_09.06.2015). Based on the clustering coefficient parameter, the densely connected sites of the network with a cutoff point of 4 were determined by molecular complex detection (MCODE) plugin59 (version 1.5.1) as structural modules. The gene-interaction network for GSE96571 microarray dataset was also constructed, and the structural modules were investigated using the same method.
Animal model of unilateral ureteral obstruction
Male C57BL/6 mice aged 6–8 weeks were obtained from Pasteur Institute (Tehran, Iran). Animal use and care were according to the guide for the use and care of animals by National Institutes of Health. Also, this study was approved by the Iranian national committee for ethics in biomedical research (Approval ID: IR.MUI.MED.REC.1398.323). For anesthesia, Ketamine and Xylazine (Alfasan, Woerden, Netherland) were injected at the dose of 115 and 11.5 mg/kg intraperitoneally. During surgery, mice were kept on a 37.5 °C plate. After a mid-abdominal incision, the left ureter was isolated and then double ligated. Afterward, the incision was sutured. In the sham group, all the steps were performed except ligation of the left ureter. The kidneys were harvested 1, 3, 6, 9, 12, 15, 18, and 21 days after surgery. Two UUO and two sham-operated mice were allocated for each time point. Additionally, three untreated mice were used as normal controls. After sacrificing with cervical dislocation, the left kidney was harvested and coronal sections were prepared. The anterior part was kept in 3.7% formaldehyde in PBS for histopathological study and the posterior part was sustained in liquid nitrogen for RNA extraction. As the in vivo study was performed to confirm the in silico results, we tried to recapitulate our model with the findings of the two reanalyzed datasets. In this regard, the expression of two prominent genes from each study was evaluated by RT-qPCR (Supplementary Table 1).
Histopathological evaluation
The formalin-fixed kidney tissues were paraffin-embedded and 5 µm sections were prepared. Hematoxylin and eosin (H&E), as well as Masson trichrome (MT) staining, were carried out and histopathological evaluations were performed in a blinded manner. The existence of diffused glomerulosclerosis and mesangial cell proliferation were assessed between two experimental groups in random cortical fields using a X40 objective. Moreover, the percentage of cortex area affected by fibrosis including interstitial cortical fibrosis, tubular loss with minimal fibrosis, and periglomerular fibrosis were determined with the estimation of about five percent. Also, increased mesangial matrix was assessed by calculating the percentage of affected glomeruli in MT-stained sections. Measurements were repeated twice for each section.
Statistical analysis
To compare the significance of histopathological differences between sham and UUO groups, Friedman’s two-way ANOVA test was applied using the ‘friedman’ function of MATLAB software (MathWorks, 2020b).
Real-time quantitative polymerase chain reaction
Total RNA of the lower part of the left kidney was extracted using RNX-plus (CinnaGen, Tehran, Iran) according to the manufacturer’s instruction. Afterward, the concentration of the extracted RNA was measured by Epoch microplate spectrophotometer (BioTek, Winooski, Vermont). Since ORs sequence contains only the exon coding region, to eliminate DNA contamination, the samples were treated with DNase I (Thermo Fisher, Waltham, Massachusetts). To validate this procedure, mock controls were also employed (Supplementary Fig. 2). Subsequently, random hexamer primered cDNA synthesis was done using the first-strand cDNA synthesis kit (YektaTajhiz, Tehran, Iran). Specific primers for Olfr433, Olfr129, Olfr1393, Olfr161, Olfr622, Hnf4a, Cebpb, Serpina3n, Tgfb1, Hprt, and Tfrc were designed using AlleleID software60 (version 6.2) (Supplementary Table 2). RT-qPCR was performed using RealQ Plus 2x Master Mix Green with high ROXTM (Ampliqon, Odense, Denmark) by Rotor-gene 6000 cycler (Qiagen, Hilden, Germany). The expression of genes was normalized by considering Hprt and Tfrc as internal control genes. The results of RT-qPCR were analyzed using the Pfaffl method by relative expression software tool (REST) version 161.
Statistical analysis
To compare gene expression patterns in UUO and sham groups, Friedman’s two-way ANOVA test was applied using the ‘friedman’ function of MATLAB software (MathWorks, 2020b).
Sequencing of polymerase chain reaction products
PCR was performed by T100™ Thermal Cycler (Bio-Rad, Hercules, California) using Taq DNA Polymerase 2x Master Mix RED (Ampliqon, Odense, Denmark). The PCR products were cloned into the PTZ57R vector using TA Cloning™ kit (Thermo Fisher, Waltham, Massachusetts). Cloned products were transformed into competent TOP10 E. coli by incubating on ice with a subsequent heat-shock at 37 °C. Transformed colonies were cultured on LB-Agar (Sigma-Aldrich, St. Louis, Missouri) plate treated with ampicillin followed by overnight incubation at 37 °C. Afterward, plasmid extraction from bacteria was performed using Solg™ Plasmid Mini-prep Kit (SolGent, Daejeon, South Korea). Sanger sequencing of samples was done using both forward and reverse universal M13 (−40) primers by Bioneer biotech company (Daejeon, South Korea) (Supplementary Data 1).
Single-cell RNA-sequencing datasets analysis
The gene barcode matrix of each dataset was downloaded using accession numbers GSE11953138 and GSE14002339 from GEO database53 and used as raw data to work using Seurat (version 3) package62. All downstream analysis was performed in Seurat package and each dataset was analyzed separately. To preprocess the data, including removing bad quality cells, the instructions provided by reference papers were followed. Briefly, for Wu et al., 2019 study38, the cells that contained more than 200, less than 4000 identified genes, and also contained less than 5 percent of mitochondrial genome were kept. For Conway et al., 2020 study39, these parameters were set as minimum of 300 genes, maximum of 3000 genes, and mitochondrial genes as less than 50 percent (according to the main paper settings). Subsequently, the gene expression level of cells was log-normalized and scaled. Downstream analysis was limited to 2000 highly variable genes to uncover more detailed differences between captured cells. PCA was performed on the highly variable genes to reduce the dimensionality of the data and the first 20 principal components were selected for the clustering purpose. Louvain clustering algorithm was used several times to identify clusters at multiple different resolutions and the optimum resolution was obtained according to distribution of DEGs. Finally, the uniform manifold approximation and projection (UMAP) algorithm was used to visualize the clusters in two-dimensional space. The cell type annotation was performed based on the expression of lineage-specific markers retrieved from related articles39,63,64,65,66 (Supplementary Fig. 3).
Data availability
The UUO datasets analyzed during this study are available from GEO database using accession numbers GSE36496 and GSE96571. The results of PCR products sequencing are available at figshare repository (https://doi.org/10.6084/m9.figshare.12753749.v11). The expression value of ORs can be downloaded from figshare repository (https://doi.org/10.6084/m9.figshare.16755340.v2). ScRNA-seq datasets of normal and UUO samples are available in GEO database with the accession numbers GSE119531 and GSE140023.
Code availability
The source code for LIMMA and scRNA-seq datasets analysis are available on GitHub: https://github.com/alimotahharynia/ORs-article/tree/analysis-code.
References
Buck, L. & Axel, R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 65, 175–187 (1991).
Watts, G. Scientists receive Nobel prize for unravelling secrets of smell. BMJ 329, 815 (2004).
Massberg, D. & Hatt, H. Human Olfactory Receptors: Novel Cellular Functions Outside of the Nose. Physiol. Rev. 98, 1739–1763 (2018).
Parmentier, M. et al. Expression of members of the putative olfactory receptor gene family in mammalian germ cells. Nature 355, 453–455 (1992).
An, S. S. & Liggett, S. B. Taste and smell GPCRs in the lung: Evidence for a previously unrecognized widespread chemosensory system. Cell Signal 41, 82–88 (2018).
Kim, S. H. et al. Expression of human olfactory receptor 10J5 in heart aorta, coronary artery, and endothelial cells and its functional role in angiogenesis. Biochem Biophys. Res. Commun. 460, 404–408 (2015).
Jovancevic, N. et al. Medium-chain fatty acids modulate myocardial function via a cardiac odorant receptor. Basic Res. Cardiol. 112, 13 (2017).
Gelis, L. et al. Functional Characterization of the Odorant Receptor 51E2 in Human Melanocytes. J. Biol. Chem. 291, 17772–17786 (2016).
Tsai, T. et al. Two olfactory receptors-OR2A4/7 and OR51B5-differentially affect epidermal proliferation and differentiation. Exp. Dermatol 26, 58–65 (2017).
Wu, C. et al. Activation of OR1A1 suppresses PPAR-γ expression by inducing HES-1 in cultured hepatocytes. Int. J. Biochem. Cell Biol. 64, 75–80 (2015).
Braun, T., Voland, P., Kunz, L., Prinz, C. & Gratzl, M. Enterochromaffin cells of the human gut: sensors for spices and odorants. Gastroenterology 132, 1890–1901 (2007).
Kalbe, B. et al. Helional induces Ca2+ decrease and serotonin secretion of QGP-1 cells via a PKG-mediated pathway. J. Mol. Endocrinol. 57, 201–210 (2016).
Clark, A. A., Nurmukhambetova, S., Li, X., Munger, S. D. & Lees, J. R. Odorants specifically modulate chemotaxis and tissue retention of CD4+ T cells via cyclic adenosine monophosphate induction. J. Leukoc. Biol. 100, 699–709 (2016).
Li, J. J. et al. Activation of olfactory receptors on mouse pulmonary macrophages promotes monocyte chemotactic protein-1 production. PLoS One 8, e80148 (2013).
Pluznick, J. L. et al. Functional expression of the olfactory signaling system in the kidney. Proc. Natl Acad. Sci. USA 106, 2059–2064 (2009).
Shepard, B. D. & Pluznick, J. L. Saving the sweetness: renal glucose handling in health and disease. Am. J. Physiol. Ren. Physiol. 313, F55–F61 (2017).
Rajkumar, P., Aisenberg, W. H., Acres, O. W., Protzko, R. J. & Pluznick, J. L. Identification and characterization of novel renal sensory receptors. PLoS One 9, e111053 (2014).
Peti-Peterdi, J., Kishore, B. K. & Pluznick, J. L. Regulation of Vascular and Renal Function by Metabolite Receptors. Annu Rev. Physiol. 78, 391–414 (2016).
Halperin Kuhns, V. L. et al. Characterizing novel olfactory receptors expressed in the murine renal cortex. Am. J. Physiol. Ren. Physiol. 317, F172–F186 (2019).
Flegel, C., Manteniotis, S., Osthold, S., Hatt, H. & Gisselmann, G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS One 8, e55368 (2013).
Feldmesser, E. et al. Widespread ectopic expression of olfactory receptor genes. BMC Genomics 7, 121 (2006).
Pluznick, J. L. et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc. Natl Acad. Sci. USA 110, 4410–4415 (2013).
Shepard, B. D. et al. A Renal Olfactory Receptor Aids in Kidney Glucose Handling. Sci. Rep. 6, 35215 (2016).
Shepard, B. D., Koepsell, H. & Pluznick, J. L. Renal olfactory receptor 1393 contributes to the progression of type 2 diabetes in a diet-induced obesity model. Am. J. Physiol. Ren. Physiol. 316, F372–F381 (2019).
Shepard, B. D. & Pluznick, J. L. How does your kidney smell? Emerging roles for olfactory receptors in renal function. Pediatr. Nephrol. 31, 715–723 (2016).
Hommos, M. S. & Rule, A. D. Should We Always Defer Treatment of Kidney Disease When There Is Extensive Interstitial Fibrosis on Biopsy? Am. J. Nephrol. 44, 286–288 (2016).
Romagnani, P. et al. Chronic kidney disease. Nat. Rev. Dis. Prim. 3, 17088 (2017).
Wu, B. & Brooks, J. D. Gene expression changes induced by unilateral ureteral obstruction in mice. J. Urol. 188, 1033–1041 (2012).
Wu, B., Gong, X., Kennedy, W. A. & Brooks, J. D. Identification of transcripts associated with renal damage due to ureteral obstruction as candidate urinary biomarkers. Am. J. Physiol. Ren. Physiol. 315, F16–F26 (2018).
Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl Genet Mol. Biol. 3, Article3 (2004).
R Core Team. R: A language and environment for statistical computing. Available from: https://www.R-project.org/ (2019).
Moradzadeh, K., Moein, S., Nickaeen, N. & Gheisari, Y. Analysis of time-course microarray data: Comparison of common tools. Genomics 111, 636–641 (2019).
Choobdar, S. et al. Assessment of network module identification across complex diseases. Nat. Methods 16, 843–852 (2019).
Lecca, P. & Re, A. Detecting modules in biological networks by edge weight clustering and entropy significance. Front Genet. 6, 265 (2015).
Granata, S. et al. Mitochondrial dysregulation and oxidative stress in patients with chronic kidney disease. BMC Genomics 10, 388 (2009).
Jiang, Y., Li, Y. R., Tian, H., Ma, M. & Matsunami, H. Muscarinic acetylcholine receptor M3 modulates odorant receptor activity via inhibition of β-arrestin-2 recruitment. Nat. Commun. 6, 6448 (2015).
Ignatieva, E. V., Levitsky, V. G., Yudin, N. S., Moshkin, M. P. & Kolchanov, N. A. Genetic basis of olfactory cognition: extremely high level of DNA sequence polymorphism in promoter regions of the human olfactory receptor genes revealed using the 1000 Genomes Project dataset. Front. Psychol. 5, 247 (2014).
Wu, H., Kirita, Y., Donnelly, E. L. & Humphreys, B. D. Advantages of Single-Nucleus over Single-Cell RNA Sequencing of Adult Kidney: Rare Cell Types and Novel Cell States Revealed in Fibrosis. J. Am. Soc. Nephrol. 30, 23–32 (2019).
Conway, B. R. et al. Kidney Single-Cell Atlas Reveals Myeloid Heterogeneity in Progression and Regression of Kidney Disease. J. Am. Soc. Nephrol. 31, 2833–2854 (2020).
Moradzadeh, K. & Gheisari, Y. The analysis of a time-course transcriptome profile by systems biology approaches reveals key molecular processes in acute kidney injury. J. Res. Med. Sci. 24, 3 (2019).
Eardley, K. S. et al. The role of capillary density, macrophage infiltration and interstitial scarring in the pathogenesis of human chronic kidney disease. Kidney Int. 74, 495–504 (2008).
Nikolic-Paterson, D. J., Wang, S. & Lan, H. Y. Macrophages promote renal fibrosis through direct and indirect mechanisms. Kidney Int. Suppl. 4, 34–38 (2014).
Feng, Y. et al. The signaling protein Wnt5a promotes TGFβ1-mediated macrophage polarization and kidney fibrosis by inducing the transcriptional regulators Yap/Taz. J. Biol. Chem. 293, 19290–19302 (2018).
Feng, Y. et al. Wnt/ β -Catenin–Promoted Macrophage Alternative Activation Contributes to Kidney Fibrosis. J. Am. Soc. Nephrol. 29, 182–193 (2018).
Kim, M.-G. et al. The Role of M2 Macrophages in the Progression of Chronic Kidney Disease following Acute Kidney Injury. PLoS One. 10, e0143961 (2015).
Lin, S.-L., Kisseleva, T., Brenner, D. A. & Duffield, J. S. Pericytes and Perivascular Fibroblasts Are the Primary Source of Collagen-Producing Cells in Obstructive Fibrosis of the Kidney. Am. J. Pathol. 173, 1617–1627 (2008).
Kui Tan, T. et al. Matrix metalloproteinase-9 of tubular and macrophage origin contributes to the pathogenesis of renal fibrosis via macrophage recruitment through osteopontin cleavage. Lab. Investig. 93, 434–449 (2013).
Rabieian, R., Moein, S., Khanahmad, H., Mortazavi, M. & Gheisari, Y. Transcriptional noise in intact and TGF-beta treated human kidney cells; the importance of time-series designs. Cell Biol. Int. 42, 1265–1269 (2018).
Bar-Joseph, Z., Gitter, A. & Simon, I. Studying and modelling dynamic biological processes using time-series gene expression data. Nat. Rev. Genet. 13, 552–564 (2012).
Abedi, M., Fatehi, R., Moradzadeh, K. & Gheisari, Y. Big data to knowledge: common pitfalls in transcriptomics data analysis and representation. RNA Biol. 16, 1531–1533 (2019).
Hara, M. et al. Robust circadian clock oscillation and osmotic rhythms in inner medulla reflecting cortico-medullary osmotic gradient rhythm in rodent kidney. Sci. Rep. 7, 7306 (2017).
Firsov, D. & Bonny, O. Circadian rhythms and the kidney. Nat. Rev. Nephrol. 14, 626–635 (2018).
Barrett, T. et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. 41, D991–D995 (2013).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis. Available from: https://ggplot2.tidyverse.org (2016).
Kolde, R. Pheatmap: Pretty Heatmaps. Available from: https://CRAN.R-project.org/package (2019).
Bindea, G., Galon, J. & Mlecnik, B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics 29, 661–663 (2013).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Szklarczyk, D. et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 (2017).
Bader, G. D. & Hogue, C. W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinforma. 4, 2 (2003).
Apte, A. & Singh, S. AlleleID: a pathogen detection and identification system. Methods Mol. Biol. 402, 329–346 (2007).
Pfaffl, M. W., Horgan, G. W. & Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36 (2002).
Stuart, T. et al. Comprehensive Integration of Single-. Cell Data. Cell 177, 1888–1902.e21 (2019).
Nagamori, S. et al. Novel cystine transporter in renal proximal tubule identified as a missing partner of cystinuria-related plasma membrane protein rBAT/SLC3A1. Proc. Natl. Acad. Sci. 113, 775–780 (2016).
Vannella, K. M. et al. Incomplete Deletion of IL-4Rα by LysMCre Reveals Distinct Subsets of M2 Macrophages Controlling Inflammation and Fibrosis in Chronic Schistosomiasis. PLoS Pathog. 10, e1004372 (2014).
Takahashi, K. et al. Macrophage CD74 contributes to MIF-induced pulmonary inflammation. Respir. Res. 10, 33 (2009).
Langlais, D., Barreiro, L. B. & Gros, P. The macrophage IRF8/IRF1 regulome is required for protection against infections and is associated with chronic inflammation. J. Exp. Med. 213, 585–603 (2016).
Acknowledgements
This study was supported by Isfahan University of Medical Sciences (grant number: 398439).
Author information
Authors and Affiliations
Contributions
S.M., Y.G., and A.M. conceptualized the main idea. K.M. and S.M. analyzed microarray data. A.M. performed network construction and topological analysis. Y.G. and K.M. constructed the animal model. A.M. and F.K. performed the experiments. M.Y. analyzed scRNA-seq datasets. A.M. drafted the manuscript and prepared figures with the help of F.K. S.M. and Y.G. critically revised the manuscript. All authors made a substantial intellectual contribution to the work and approved the submitted version for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Motahharynia, A., Moein, S., Kiyanpour, F. et al. Olfactory receptors contribute to progression of kidney fibrosis. npj Syst Biol Appl 8, 8 (2022). https://doi.org/10.1038/s41540-022-00217-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41540-022-00217-w