Abstract
Bacterial biofilms, which consist of three-dimensional extracellular polymeric substance (EPS), not only function as signaling networks, provide nutritional support, and facilitate surface adhesion, but also serve as a protective shield for the residing bacterial inhabitants against external stress, such as antibiotics, antimicrobials, and host immune responses. Biofilm-associated infections account for 65-80% of all human microbial infections that lead to serious mortality and morbidity. Tremendous effort has been spent to address the problem by developing biofilm-dispersing agents to discharge colonized microbial cells to a more vulnerable planktonic state. Here, we discuss the recent progress of enzymatic eradicating strategies against medical biofilms, with a focus on dispersal mechanisms. Particularly, we review three enzyme classes that have been extensively investigated, namely glycoside hydrolases, proteases, and deoxyribonucleases.
Similar content being viewed by others
Introduction
Bacterial Biofilms
Bacterial biofilms consist of surface-attached, and sometimes non-surface attached, colonies embedded within a self-produced extracellular matrix known as the extracellular polymeric substance (EPS). The EPS is composed of extracellular proteins, lipids, nucleic acids (extracellular-DNA and extracellular-RNA), polysaccharides, and secondary metabolites1,2,3 Biofilms are not only capable of reversible surface attachment, but also serve to trap nutrients, as well as shield cells against host immune responses and antimicrobial treatments4 Besides these functional roles, the EPS also provides structural support and holds the bacterial cells in close proximity, thereby enabling the exchange of genetic material and facilitating quorum sensing5,6 Biofilm-associated infections are common and account for 65-80% of all human microbial infections7, such as vaginitis8, colitis9, conjunctivitis10, gingivitis11, urethritis12, and otitis13. Additionally, biofilms formed by adherent bacteria on medical implants and devices can result in serious mortality and morbidity14 Furthermore, sessile bacterial colonies covered by established biofilms are more difficult to eradicate than planktonic cells. Biofilms not only function as the physical shield against exogeneous stress, but also lower the metabolic rates of the inhabiting cells to survive harsh environments. As a result, biofilm-associated infections are difficult to eradicate and pose a danger to prevalence of chronic persistentillnesses15.
Biofilm formation
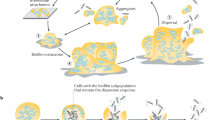
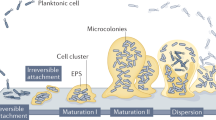
Biofilm formation proves to occur in diverse scenarios and the environment poses a significant influence on biofilm establishment through impacting gene expression and modulating bacteria behaviors. As shown in Fig. 1, an expanded biofilm model was proposed by Sauer et al. and is still growing to reflect all processes involved in the biofilm life cycle16. The most commonly accepted model of biofilm formation, typically based on the in vitro biofilm developed by Pseudomonas aeruginosa, can be subdivided into five major stages5. In the beginning, individual planktonic cells, or preformed aggregates in some cases, migrate and reversibly adhere to a surface. If the surface is suitable for growth, the newly adherent bacterial cells proliferate and initiate biofilm production on the surface. Then, the adherent cells irreversibly attach to the surface, facilitating cell aggregation and EPS production. Later, the biofilm reaches the first stage of maturation (maturation I) and starts to develop mushroom-like structures, which becomes more layered and develop three-dimensional microstructures, including nutrient and water channels. After that, the biofilm reaches a fully mature status (maturation II) with maximal cell density and is now regarded as a three-dimensional community17. In the final stage, the mature biofilm releases planktonic cells, with the help of hydrolase enzymes, to migrate and spread to new, unoccupied surfaces18,19. However, this model does not entirely represent the complex biofilms formed in the real world including those in industrial, clinical, and natural environments. Indeed, a more inclusive model involving three major events was recently proposed: aggregation, growth, and disaggregation16. Besides the in vitro model, biofilms also develop in vivo, in situ, and ex vivo, each of which follows different biofilm developmental pathways in response to diverse environmental factors16. In various settings such as on cell surfaces, in fluids, and on transplant devices, surface association is not required and diversely shaped microbial communities are observed. Additionally, in open systems like human gastrointestinal and circulatory systems, there tends to be a consistent influx of new microbial bodies or biofilm aggregates to microbial communities undergoing establishment16.
Bacteria can exist as both single cells and biofilm aggregates with regard to environment cues. In vivo, ex vivo, and in situ, bacteria can remain in a planktonic state or reside within non-surface-attached biofilms and these two existing forms are interchangeable depending on the environment. The commonly accepted biofilm formation model, typically the in vitro biofilm developed by P. aeruginosa, can be subdivided into five major stages consisting of reversible attachment, irreversible attachment, maturation I, maturation II, and dispersion16.
Biofilms serve as an effective protective shield for the encased bacterial cells, providing protection from antimicrobial treatments, host immune responses, bacteriophages, and other external stressors, which frequently results in persistent and chronic infections20. In fact, studies have shown that bacteria growing in biofilms are often thousands of times more tolerant to antibiotic treatment than their planktonic counterparts21. This is in part due to the limited diffusion of nutrients throughout the biofilm EPS resulting in cell heterogeneity (Fig. 2)22. Bacterial cells near the biofilm surface are highly metabolically active and more susceptible to antibiotic treatments, while cells in the core of the biofilm exist within a low-oxygen microenvironment, causing these cells to have a decreased metabolic rate, facilitating their resistance to antibiotics23. Furthermore, there is a small subpopulation of cells within the biofilm community, known as persister cells, that tend to adopt a dormant state with extreme antimicrobial tolerance24. Despite their small numbers, these persister cells contribute significantly to the pathogenesis of biofilm infections25,26. Studies indicate that small populations of persister cells are able to survive antimicrobial treatment regardless of the concentration of antibiotic utilized27. Once the antibiotic treatment ceases, these remaining persister cells repopulate the microbial community and ultimately lead to a relapsing biofilm infection28.
The biofilm EPS protects the residing bacteria against threats like antibiotics, bacteriophages, and host immune response. While metabolically active surface-residing cells in the nutrition-rich outer portion of the biofilm may be less resistant to environmental pressure, bottom-residing bacteria have greater resistance due to their low metabolic rate. Dormant persister cells can repopulate the bacterial community after antibiotic courses, leading to chronic infections.
Biofilm dispersal strategies
There are a number of promising biofilm eradication strategies that have been developed to hinder bacterial biofilm formation or disrupt maturation by dysregulating biofilm growth. These proactive approaches include the use of antimicrobial peptides and lipids29,30,31, medical device surface modifications32,33, quaternary ammonium compounds (QACs)34, nitric oxide-releasing compounds35,36, cell-signaling inhibitors37,38, antibiotic-conjugation39, and direct surgical removal of biofilm biomass40. Biofilm dispersal is an intense area of study that may lead to the development of novel agents that inhibit biofilm formation or promote biofilm cell detachment. Such agents may be useful for the prevention and treatment of biofilms in a variety of industrial and clinical settings41.
In clinical settings, enzymes, small molecules, surgical removal, and other strategies have been successfully applied to break down biofilms and release microbes to a more vulnerable planktonic state42. Thus, dispersal agents are utilized to improve therapeutic outcomes by increasing access of antimicrobials and host immune cells to the bacteria43. Compared with other biofilm dispersal strategies, enzymatic treatments have more advantages. Biofilm-dispersing enzymes are more effective on both growing and pre-existing biofilms, and relatively low concentrations are required to achieve high specificity and efficacy towards the targeted biofilms. Additionally, antibiotic resistance, the issue that many small molecule drugs face, is a less likely occurrence for biofilm-dispersing enzymes, which function extracellularly without the need to be transported across the outer membrane.
Biofilm dispersal enzymes
Extracellular enzymes can effectively disperse bacterial biofilms by degrading the EPS, specifically by targeting exopolysaccharides, extracellular DNA, and extracellular proteins within in the EPS. By hydrolyzing the microbe biofilm, these enzymes initiate the detachment of sessile bacterial cells and convert them to a planktonic state, which causes increased susceptibility to antibiotics and the host immune system. By laboratory approaches of isolation or over-expression in model organisms, biofilm-dispersing enzymes can be procured at high concentrations and added exogenously to microbial colonies to efficiently break down biofilms. Herein, we review the recent progress of biofilm disruption via three major enzyme classes: glycoside hydrolases44,45, deoxyribonucleases46,47, and proteases48,49.
Exopolysaccharides within the EPS
As the major component of the EPS, secreted extracellular polysaccharides are critical for biofilm integrity. Exopolysaccharides widely exist as structural components in microbial biofilms including poly-N-acetylglucosamine (dPNAG), alginate, Psl, Pel, amylose-like glucan, cellulose, galactosaminogalactan, β-(1,3)-glucan, levan, and inulin (Fig. 3)50,51,52,53,54.
dPNAG Exopolysaccharide
Many medically relevant microbial pathogens produce a common exopolysaccharide, partially de-N-acetylated poly β-(1,6)-N-acetyl-d-glucosamine (dPNAG), as a key component of their biofilm matrix (Fig. 3A)55. Both Gram-positive and Gram-negative bacteria have been confirmed to produce dPNAG (sometimes referred to as polysaccharide intercellular adhesin in Gram-positive strains) as a biofilm exopolysaccharide, including Staphylococcus aureus56, Escherichia coli57, Yersinia pestis58, Actinobacillus pleuropneumoniae59, Aggregatibacter actinomycetemcomitans60, Bordetella species61, Acinetobacter baumannii62, Burkoholderia species63, Klebsiella pneumoniae64, Vibrio parahaemolyticus65, and Bacillus subtilis66. Individual dPNAG polysaccharides are tens to hundreds of monosaccharide units in length67. In Gram-positive bacteria, the icaABCD locus is responsible for dPNAG production, whereas Gram-negative bacteria the homologous pgaABCD operon regulates its formation54,68,69. Chemical modifications of dPNAG, such as N- deacetylation and O-succinylation, play key roles in the adhesiveness and structural integrity of the biofilm matrices70. In Gram-positive bacteria, IcaB is responsible for the N-deacetylation of PNAG, and O-succinylation of PNAG is catalyzed by IcaC71. In Gram-negative bacteria, PgaB C-terminal domain functions as N-deacetylase towards PNAG polymers72.
Alginate exopolysaccharide
Alginate was the first and most thoroughly studied biofilm exopolysaccharide discovered and is produced by P. aeruginosa, a pathogenic bacterial species associated with lung infections in cystic fibrosis patients73. Alginate is composed of β-d-mannuronic acid and its C-5 epimer, α-l-guluronic acid, connected through (1,4)-glycosidic linkages (Fig. 3B). Most of the enzymes responsible for alginate biosynthesis are encoded by the alg operon (algACD844KEGXLIJF) in the P. aeruginosa genome74. The synthesis of the sugar-nucleotide precursors of alginate require the algACD operon; algA and algD are found on the alginate operon while algC is located in the genome at PA532275. Chemical modifications are commonly found at the C-2 and C-3 positions of mannuronate residues in alginate polymers. They are frequently acetylated, which is driven by the combined effect of the acetyltransferases AlgI, AlgJ, AlgF and AlgX with varied acetylation rates from 4 to 57%76,77. In addition to acetylation, AlgG also catalyzes the epimerization of β-d-mannuronic acid to α-l-guluronic acid78. Alginate can facilitate the formation of gel-like structures in the presence of cations, including sodium and calcium, with functional properties strongly correlated to the ManA/GulA ratio and sequence79.
Psl exopolysaccharide
Psl exopolysaccharide serves as structural scaffold, and plays a key role in surface attachment and eDNA interactions in the biofilm matrix of the opportunistic pathogen, P. aeruginosa80. The Psl exopolysaccharide contains a penta-saccharide repeating unit consisting of d-mannose, l-rhamnose, and deoxyglucose (Fig. 3C). The biosynthesis of the Psl exopolysaccharide occurs via a Wzx/Wzy-dependent mechanism and is accomplished by 12 proteins encoded by the pslABCDEFGHIJKL operon73.
Pel exopolysaccharide
Pel is one of the most phylogenetically widespread biofilm matrix determinants in both Gram-negative and Gram-positive bacteria (Fig. 3D)81. A recent study shows that Pel is a partially de-N-acetylated linear polymer of α-1,4-N-acetylgalactosamine, comprised predominantly of dimeric repeats of galactosamine and N-acetylgalactosamine82. Gram-negative bacteria, P. aeruginosa, forms Pel-dependent biofilms regulated by a seven gene operon (pelABCDEFG), whereas numerous Gram-positive bacterial species use a variant form of this gene cluster (pelDEADAFG) to produce Pel-like polysaccharide83,84,85. In P. aeruginosa biofilms, PelDEFG mediates sugar polymerization and transport across the cytoplasm, while PelBC is responsible for export83,86. PelA exhibits hydrolase and deacetylase activities and regulates the deacetylation of Pel polymers87.
Amylose-like glucan
Gram-negative bacterial species, such as Francisella tularensis and Pasteurella multocida, produce biofilm matrices containing amylose-like glucan, an exopolysaccharide made of α-d-glucose units connected through α-(1,4) glycosidic bonds (Fig. 3E)88,89. In the production of capsular polysaccharide (CPS) by P. multocida biofilms, which consist of amylose-like glucan, capsular polysaccharide production was found to be inversely related to biofilm formation89. Little is known about the genes of amylose exopolysaccharides; more work is needed to reveal its biosynthetic mechanism.
Cellulose exopolysaccharide
Cellulose, composed of β-(1,4)-d-glucose (Glc) monomer subunits (Fig. 3F), has been identified as a biofilm matrix component of several bacterial species including Agrobacterium tumefaciens90, Escherichia coli91, Pseudomonas flurescens92, and Gluconacetobacter xylinus51. The cellulose biosynthetic and secretive machineries of various bacteria are extremely diverse, and different bacteria utilize varying bacterial cellulose synthase (bcs) operons to produce this exopolysaccharide93. Multiple chains of cellulose can begin to form greater aggregates through hydrogen bonding interactions between cellulose polymer strands94. Besides this, phosphoethanolamine-modified cellulose generated by E. coli is required for extracellular matrix assembly and biofilm architecture95. The modification is catalyzed by phosphoethanolamine transferase, BcsG, in the presence of biofilm-promoting cyclic diguanylate monophosphate95.
Galactosaminogalactan
Galactosaminogalactan (GAG), commonly found in the biofilms of various fungal species, is a heteroglycan composed of galactose and N-acetylgalactosamine (GalNAc) linked by α-(1,4) glycosidic bonds (Fig. 3G)96. In biofilm-associated infections, GAG serves as an adhesion factor to the host, and mediates virulence by masking other pathogen-associated molecular patterns (PAMPs). The synthesis of GAG is regulated by a cluster of genes (gtb3, agd3, ega3, sph3, and uge3) encoding five eponymic, carbohydrate-active enzymes97. Agd3, categorized as a carbohydrate esterase family CE18 enzyme, deacetylates GAG in a metal-dependent manner98. Deacetylation of GAG serves as a key factor for adherence to hyphae and mediates biofilm formation97.
β-(1,3)-Glucans
β-(1,3)-glucans are glucose polymers mainly linked by β-(1,3)-glycosidic bonds with branched side chains attaching to the backbone through 1,6-linkages (Fig. 3H)99. Synthesis of the linear β-(1,3)-glucan polymer is catalyzed by UDP-glucose glucosyltransferase in many microbial species including Candida albicans, Aspergillus fumigatus and Cryptococcus neoformans100. β-(1,3)-glucans are the primary components of the C. albicans biofilm EPS and are important for C. albicans biofilm formation and stress resistance101.
Fructan exopolysaccharide
Levan and inulin (Fig. 3I-J) are two primary fructans discovered in many microbial biofilms including the genera Acetobacter, Bacillus, Erwinia, Gluconobacter, Halomonas, Microbacterium, Pseudomonas, Streptococcus, and Zymomonas102. Levan is composed of β-(2, 6) glycosidic fructosyl bonds with occasional β-(2, 1) branching103, while inulin is primarily comprised of β-(2, 1) fructosyl linkages and some β-(2, 6) linkages at the branching point104. Microbial levan is synthesized through transfructosylation by a secreted levansucase (EC: 2.4.1.10) from sucrose substrates in Bacillus species105.
These exopolysaccharides play important roles in biofilm establishment and persistence through enhancing structural stability, defense against environmental stress, adhesion and aggregation of cells, absorption of exogenous compounds, and providing a carbon source during starvation. Because of their indispensable function in biofilm integrity, glycosidase enzymes that target exopolysaccharides are emerging as an effective means to disperse biofilms106,107,108. The glycol-hydrolases discussed are in the same order as the introduced corresponding exopolysaccharides.
Glycoside hydrolase enzymes
dPNAG glycoside hydrolase—Dispersin B
Dispersin B (DspB) belongs to glycoside hydrolase family 20 (GH20) and was first isolated from Aggregatibacter actinomycetemcomitans109. DspB is known to hydrolyze the exopolysaccharide dPNAG in biofilm matrices through both endo- and exo-glycoside hydrolase activity110,111,112. DspB utilizes a substrate-assisted mechanism in dPNAG hydrolysis in which the substrate’s 2-acetamido group facilitates glycoside hydrolysis through formation of a characteristic oxazolinium ion intermediate113. Within the catalytic site, the amino acid residue, D183, serves as catalytic acid and D184 stabilizes the oxazolinium ion intermediate113. In vitro studies show that DspB can effectively disperse biofilms formed by bacteria like S. aureus, A. actinomycetemcomitans, S. epidermidis, A. baumannii, K. pneumoniae, E. coli, Burkholderia spp., A. pleuropneumoniae, Y. pestis, and P. fluorescens. In an in vivo study, DspB was prepared into DispersinB® wound gel by Kane Biotech Inc., which significantly accelerated the healing of both infected and non-infected dermal wounds compared to controls114. Compared with wild-type DspB, most DspB mutants present significantly reduced activity on synthetic PNAG probes106,107. However, DspBE248Q demonstrates remarkably increased dPNAG breakdown and effective dispersal of S. aureus preformed biofilms115.
dPNAG glycoside hydrolase—PgaB
PgaB is a glycoside hydrolase encoded by the PNAG biosynthetic operon, namely by the gene pgaB, and has the capability to degrade PNAG synthetic analogues, as well as disrupt PNAG-dependent biofilms formed by Bordetella pertussis, Staphylococcus carnosus, S. epidermidis, and E. coli116,117. PgaB is a two-domain periplasmic protein that contains an N-terminal deacetylase domain that regulates PNAG deacetylation and a C-terminal PNAG binding domain that modulates PNAG export118. Detailed analysis shows that PgaB contains a C-terminal CAZy GH153 family glycosyl hydrolase that catalyzes the endoglycosidic cleavage of dPNAG containing de-N-acetylated glucosamine (GlcN) in the −3 binding site116. The C-terminal domain of PgaB produced by Bordetella bronchiseptica has a central cavity within an elongated surface groove that preferably recognizes the GlcN-GlcNAc-GlcNAc motif (where GlcNAc is N-acetylglucosamine), and the catalytic site amino residue, D474, functions as a catalytic acid to digest the dPNAG substrate. After hydrolysis, mass spectrometry reveals the GlcN-GlcNAc-GlcNAc motif at the new reducing end116. This research shows that PgaB not only serves as a deacetylase within the PNAG biosynthetic machinery, but also possesses glycoside hydrolase activity and may be used as a therapeutic agent against PNAG-dependent biofilm infections116.
Alginate glycoside hydrolase
In addition to dPNAG hydrolases, alginate lyase enzymes have been shown to exhibit effective dispersal of mature biofilms119. Alginate lyases catalyze the degradation of alginate, and have been isolated from various organisms with different substrate specificities, including algae, marine mollusks, marine and terrestrial bacteria, and some viruses and fungi120. Many studies demonstrating the antibiofilm activity of alginases have used crude cell extracts from Flavobacterium multivorum, but the synergistic effect with antibiotics remains contradictory121. Two distinct alginate lyase enzymes in F. multivorum extract have been discovered and characterized: one of which exhibits degradation towards both poly-β-d-mannuronate (polyM) and poly-α-l-guluronate (polyG), while the other only has polyG degradation activity119. Only alginate lyase enzymes with polyM/G activity are effective in destroying preformed mature biofilms and have a synergistic effect with antibiotics119. A recent study shows that purified marine alginate lyase enzyme (AlyP1400) is able to degrade P. aeruginosa biofilms and enhances bactericidal activity of the antibiotic, tobramycin, while also modulating expression of efflux antibiotic resistance-related genes; bdlA, mexF, mexY, and ndvB; suggesting an increased susceptibility of P. aeruginosa biofilms to this combinatorial treatment122.
Psl glycoside hydrolase
PslG, a member of glycoside hydrolase family 39 (GH39), is periplasmic glycoside hydrolase encoded by the Psl exopolysaccharide biosynthetic operon123. After removal of the N-terminal transmembrane domain, PslGh (which has a soluble catalytically active glycoside hydrolase domain) can hydrolyze Psl in P. aeruginosa biofilms123. PslGh inhibits clinical and environmental isolates of P. aeruginosa biofilm formation over a 24-h period and is also capable of disrupting newly formed biofilms but is less potent to disperse mature biofilms. Further, PslGh can potentiate the antibacterial effect of colistin, an antibiotic used to treat Gram-negative multi-drug resistant infections124. PslGh is noncytotoxic and support immune defenses; the enzyme does not impact host cell morphology and enhances neutrophil killing activity124.
Pel glycoside hydrolase
PelA, a periplasmic glycoside hydrolase encoded in the Pel exopolysaccharide biosynthetic operons, contains at least two catalytic domains—a putative glycoside hydrolase domain and a CE4 deacetylase domain87. Based on a bioinformatic analysis. the N-terminal domain of PelA was removed, generating the PelA47–303 construct (referred as PelAh), was expressed and purified in a study of its glycoside hydrolase activity124. Prophylactic treatment with PelAh resulted in a 2.5-log reduction of P. aeruginosa bacterial colony-forming units, and application of PelAh to established biofilms resulted in significant biofilm dispersal within 24 h124. Furthermore biofilm disruption with PelAh is not sensitive to the maturation state of the biofilm124. PelAh also boosted the antibiotic efficacy of colistin and increased neutrophil killing by ~50%124.
Amylose glycoside hydrolase
Endo-acting α-amylase, of the glycoside hydrolase family 13 (GH13), cleaves α-(1,4)-d-glucosidic linkages at random sites of amylose exopolysaccharide in biofilm matrices leading to biofilm dispersing events125. Research shows that α-amylase from Aspergillus oryzae, Bacillus subtilis, human saliva, and sweet potato demonstrates a strong inhibiting effect on S. aureus biofilm buildup, as well as degrade existing pre-formed S. aureus biofilms126. However, a less severe inhibiting effect was observed for β-amylase from sweet potato (~50% inhibition versus 77-89% inhibtion from the others) because it is an exo-acting GH14 carbohydrolase which hydrolyzes the α-1,4-glucosidic linkages of amylose exopolysaccharide only from the nonreducing end126.
Cellulose glycoside hydrolase
Cellulase is a glycoside hydrolase produced chiefly by fungi, bacteria, and protozoans that acts specifically by breaking down the β-(1,4) linkages in polysaccharides, such as cellulose, an exopolysaccharide commonly found in the biofilm of several bacteria, including E. coli, Salmonella, Citrobacter, Enterobacter, and Pseudomonas as well as Agrobacterium tumefaciens127. Cellulase from various sources such as Penicillium funiculosum and Trichaderma reseei can inhibit biofilm formation of P. aeruginosa in a pH dependent manner, in which exogenously added cellulase is more effective at pH 5 than pH 7128. Treatment combining cellulase with ceftazidime, an antibiotic, can more effectively inhibit P. aeruginosa biofilm formation and attachment129. In vitro testing also shows that Levofloxacin, an antibiotic for severe infection, combined with cellulase can powerfully disperse mature biofilms formed by bacille CalmetteGuerin130.
Galactosaminogalactan glycoside hydrolase
Sph3 is encoded by the Sph3 gene, which belongs to the five gene cluster regulating GAG biosynthesis. The glycol-hydrolase domain (Sph3h) of Sph3 is classified as glycoside hydrolase family 135 (GH135)131. Sph3 has the (β/α)8 fold structure that many glycoside hydrolase enzymes possess, and contains putative catalytic amino acid residues (Asp-166, Glu-167, and Glu-222) in the active site131. The hydrolase domains of Sph3 and PelA (Sph3h and PelAh, respectively) share structural and functional similarities given their ability to degrade GAG and disrupt preformed Aspergillus fumigatus biofilms in vitro132. A mechanistic study revealed that both Sph3h and PelAh function as retaining endo-α-(1,4)-N-acetylgalactosaminidases producing a minimal substrate size of seven residues132. Ega3 is another gene in the GAG biosynthesis cluster encoding a putative α-(1,4)-galactosaminidase belonging to glycoside hydrolase family 114 (GH114) which also has the (β/α)8 fold structure; its activity depends on the conserved acidic residues, Asp-189 and Glu-247133. Recombinant Ega3 is an endo-acting α-(1,4)-galactosaminidase that disrupts GAG-dependent A. fumigatus and Pel polysaccharide-dependent P. aeruginosa preformed biofilms in vitro at nanomolar concentrations133.
β-(1,3) glucan glycoside hydrolase
β-(1,3) glucanases, which belong to the pathogenesis-related-2 family (PR-2), are abundant in nature and have been characterized from a wide range of species134. They successively cleave at the nonreducing end of β- (1,3) glucan producing oligosaccharides and glucose134. β-glucanase derived from Arthrobacter luteus is able to degrade poly-β-(1,3)-glucose in Candida albicans preformed biofilms in vitro but has no effect on planktonic growth or adhesion101.
Fructan glycoside hydrolase—levanase
Levanase, SacC, is an exo-fructosidase belonging to Glycoside Hydrolase Family 32 (GH32) and hydrolyzes the terminal β-(2,1)- d-fructofuranose residues of fructans from the non-reducing end135. Levanase SacC is able to hydrolyze both levan and inulin to produce fructose, and is also able to hydrolyze sucrose and raffinose135. Levanase cannot be detected in the wild-type Bacillus subtilis, but levanase SacC could be found in the culture medium of laboratory grown B. subtilis in the form of SacL mutated extracellular enzymes136. A recent study shows that extracellular levanase SacC from B. subtilis disrupts preformed P. aeruginosa biofilms in vitro, increasing the efficiency of conventional the antibiotics, ciprofloxacin and amikacin137.
Fructan glycoside hydrolase—inulinase
Inulinase is an enzyme that catalyzes the hydrolysis of β-(2,1)-d-fructosidic linkages in inulin and is part of a group of naturally occurring polysaccharides138. Inulinase can be subcategorized into exo-inulinase (EC 3.2.1.80) and endo-inulinase (EC3.2.1.7) based on hydrolysis patterns. Exo-inulinase hydrolyzes the terminal fructose residue of inulin from the non-reducing end, whereas endo-inulinase initiates hydrolysis at random positions within inulin to give fructooligosaccharides139. Inulinase is capable of degrading in vitro pre-formed biofilms on reverse osmosis RO membranes by composed by multiple bacterial species. The mechanism of its destructive process is degrading the β-(2, 6)-glucan fructosidic bonds of inulin140 (Table 1).
Proteases
Exoproteins, another major component of the EPS, is account for a considerable portion of the biomass of most biofilms. Exoproteins are crucial to bacterial cell aggregation, surface adhesion, and structural integrity of biofilm matrices141,142. Enzymatic degradation of EPS exoproteins is one of the most effective ways to eradicate biofilms. To date, a number of proteases capable of biofilm dispersal have been discovered and investigated.
Proteinase K
Proteinase K is a broad-spectrum serine protease with a wide pH tolerance (pH 4 - 12) and thermostability (37 - 60°C)143. It specifically cleaves peptide bonds in proximity to carboxylic groups of aliphatic and aromatic amino acids144. Proteinase K is capable of inhibiting S. aureus biofilm formation by hampering early adhesion, but also disperses 24-h- and 48-h-old biofilms144. Recent studies show that co-treatment of proteinase K with antibiotics has a synergistic effect that thoroughly degrades preformed biofilms produced by a range of bacteria, including S. aureus, E. coli, Staphylococcus lugdunensis, Staphylococcus heamolyticus, Listeria monocytogenes, Gardnerella vaginalis, and Bdellovibrio bacteriovorus144,145,146,147,148,149.
Trypsin
Trypsin, a pancreatic serine protease that specifically acts on the carboxyl side of lysine and arginine, has been applied to disperse biofilms formed on teeth and wounds150,151,152. Bovine trypsin can degrade mature biofilms of various Gram-positive and Gram-negative bacterial species153. Trypsin alone is able to reduce the biomass of the preformed 24-h-old biofilms of both P. aeruginosa and Enterococcus faecalis but cannot completely remove biofilms regardless of the treatment time and enzyme concentrations154. However, trypsin combined with pepsin and Carvacrol is able to fully disperse mature biofilms of P. aeruginosa and E. faecalis on various abiotic surfaces154.
Pepsin
Pepsin is a promiscuous endopeptidase with a catalytic aspartate in its active site to favorably cleave Phe and Leu residues; however, His, Lys, Arg, and Pro residues prohibit cleavage155. Pepsin reduces the biomass of the preformed 24-h-old biofilms of both P. aeruginosa and E. faecalis, but cannot completely remove biofilms from polystyrene surfaces regardless of the treatment time and enzyme concentrations used, like trypsin154. Co-administered with trypsin and carvacrol, it can effectively irradicate preformed P. aeruginosa and E. faecalis biofilms154,156.
Aureolysin
Aureolysin (Aur), an S. aureus expressed extracellular metalloprotease, down-regulates the formation of biofilms and allows for the mobility of bacteria by cleavage of surface binding proteins, such as clumping factor B which causes loss of fibrinogen binding in S. aureus157. Aur is a major contributor to bacterial pathogenicity via cleaving components of the innate host immune system and regulating bacterial toxins and cell wall proteins158,159. Aur is associated with the processing of other biofilm proteases, such as V8, SspB, and ScpA which together are known as the Staphylococcal proteolytic cascade. These proteases are secreted into the environment with the pro-peptide inhibiting their activation. Aur undergoes autocatalysis and becomes active by the degrading the pro-peptide, then mature aur cleaves the pro-peptide from V8 to generate active V8 protease. Finally, V8 will cleave the SspB pro-peptide to complete cascade48. Purified aur suppresses biofilm formation and disperses established biofilms of various S. aureus strains160.
V8 serine protease
The V8 serine protease, also known as SspA protease, is the major extracellular protease secreted by S. aureus. It is secreted as a proenzyme before being proteolytically cleaved by aur to become the mature V8 enzyme48. Research shows that V8 serine protease added at the beginning of cell culture prevents the S. epidermidis biofilm formation by degrading Bap protein, a surface-anchored protein161. Esp protease, produced by S. epidermidis, is structurally highly similar to that of V8. Purified Esp protease prevents biofilm formation, promotes disassembly of pre-established biofilms by cleaving autolysin (Atl)-derived murein hydrolases, and prevents staphylococcal release of extracellular DNA49.
Staphopain A
Staphopain A (ScpA), encoded by the scpAB operon, is also a participant of the staphylococcal proteolytic cascade. It is an extracellular cysteine protease generated by S. aureus and demonstrates a very broad range of substrate specificity162. Purified ScpA inhibits S. aureus formation and disperses established biofilms. The antibiofilm properties of ScpA are conserved across S. aureus strain lineages. Additionally, inhibition of ScpA restores the biofilm forming capacity of the biofilm-negative S. aureus mutant, the sigma factor B (ΔsigB) mutantt163. Purified ScpA enzyme inhibited S. aureus formation as well as to disperse the established biofilms, and the antibiofilm properties of ScpA were conserved across S. aureus strain lineages163.
Staphopain B
Staphopain B (SspB) is encoded by the sspABC operon and is a cysteine protease secreted by S. aureus. SspA cleaves proSspB to activate SspB in the last step of the staphylococcal proteolytic cascade48. Silencing expression of SspB can enable the biofilm-deficient S. aureus mutant (ΔsigB) to gain biofilm-forming abilities163. However, bacteria-derived proteinase could facilitate the bacterial colony survival by degrading antimicrobial peptides (AMPs) generated by the host immune system, which leads to chronic infections164. Fragments of the AMP, cathelicidin LL-37, have been discovered as part of the innate immune response in skin diseases such as atopic dermatitis and acne rosacea164. S. aureus derived proteinases aureolysin, V8, and SspB have been observed in staphylococcal isolates from atopic dermatitis patients and contributed to bacterial virulence through degradation of the AMP cathelicidin LL-37, resulting in loss of inhibitory activity on biofilm formation and ultimately leading to bacterial persistence in atopic dermatitis165.
SplABCDEF
Spl proteases consists of six serine proteases, SplABCDEF, encoded by the νSaβ gene in S. aureus166. Spl proteases modulate S. aureus physiology and virulence, and can induce disseminated lung damage during pneumonia likely by degrading surface-associated proteins in staphylococcal and human proteins166. In S. aureus, deletion of the genes encoding the extracellular proteases, aureolysin and Spl, encourages biofilm formation in planktonic cells167. These findings indicate that Spl proteases have the ability to disperse S. aureus biofilms, but more research is needed to elucidate the dispersal mechanism.
Surface protein-releasing enzyme (SPRE)
Endogenous surface protein-releasing enzyme (SPRE), produced by S. mutans NG8, can disperse the preformed monolayer biofilm of S. mutans and detach cells from colonized surfaces168. SPRE cleaves the bacterial surface anchoring protein, adhesin P1, by dissociating the bonds between the C-terminus of adhesin P1 and other cell surface components169. SPRE degrades preformed biofilms in a pH-dependent manner with the optimal pH range from 5 to 6, and can also detach biofilms of non-dividing cells, indicating that cells detached from biofilms were not daughter cells168.
Streptococcal cysteine protease (SpeB)
Streptococcal cysteine protease (SpeB) is secreted by Streptococcus pyogenes, an exclusively Gram-positive human pathogen that causes a wide spectrum of diseases such as pharyngitis, impetigo, toxic shock, and necrotizing fasciitis170. SpeB is a promiscuous enzyme displaying a broad range enzymatic activities including degradation of biofilm, cytokines, chemokines, complement components, immunoglobulins, and serum protease inhibitors. It is also capable of degrading and releasing other streptococcal proteins from the bacterial surface170. The constitutive production of SpeB by an S. pyogenes mutant strain is responsible for a significant reduction of biofilm formation. Beyond this, addition of purified SpeB to actively growing wild-type cultures significantly inhibits biofilm formation171. SpeB disperses biofilms and facilitates bacterial colonization and occupation of new areas, resulting in infections caused by S. pyogenes to vary from mild to severe172. However, a recent study shows that SpeB exhibits potent activity towards biofilm disruption at multiple stages of S. aureus biofilm formation by cleaving SdrC adhesin, which renders the bacteria more susceptible to antimicrobial agents and host immune components173.
Peptidase M16
Peptidase M16 is a Microbacterium sp. SKS10 secreted metalloprotease that exhibits optimal activity at 60°C, pH 12174. Peptidase M16 shows low cytotoxicity and excellent stability in the presence of various salts and organic solvents. Besides this, peptidase M16 can disperse mature S. aureus biofilms at concentrations lower than trypsin and α-amylase, and can be co-treated with kanamycin to enhance antimicrobial efficacy174 (Table 2).
Deoxyribonucleases
Extracellular DNA (eDNA) is a ubiquitous and vital structural component of the EPS with functions including microbial adhesion, cell signaling, gene transfer, and biofilm matrix stabilization175,176,177. Despite the importance of eDNA in bacterial biofilms, it had not attracted much attention until 2002 when Whitchurch et al. exogenously added DNase I to disperse biofilms and boost bactericidal efficiency when combined with antibiotics46. Since then, substantial work has been done to employ various DNases targeting eDNA to eradicate biofilm infections.
DNase I
DNase I is a widely used pancreatic endonuclease that specifically digests DNA. It is secreted in the extracellular environment to degrade both single-stranded and double-stranded DNA into oligonucleotides with 5′ monophosphate and 3′ hydroxyl DNA ends178,179. DNase I can disrupt the formation of both mono- and polymicrobial biofilms179. Biofilms formed in the presence of DNase I display reduced biofilm biomass, total bacterial biomass, decreased viability of bacteria, and decreased tolerance to antibiotics180. However, DNase I is more effective towards the destruction of rapidly growing biofilms. Newly established biofilms (up to 60-h old) were also dissolved by DNase I treatment, whereas more mature biofilms (over 84-h old) exhibited strong resistance to DNase I degradation. This is likely due to mature biofilms being strengthened by other substances such as exopolysaccharides and exoproteins, additionally, mature biofilms may have produced sufficient proteolytic exoenzymes to locally inactivate DNase I46. Besides this, recombinant human DNase I (rhDNase) has been clinically applied in cystic fibrosis patients to reduce the viscosity of purulent sputum181. rhDNase exhibits strong antibiofilm activity and reduces the antibiotic resistance of S. aureus and S. epidermidis182. Recent studies show that DNase I also presents a wide compatibility with various antimicrobial agents such as ceftazidime, proteinase K, and silver sulfadiazine183,184,185. DNase I-like protein 2 (DNase1L2), found in the human stratum corneum of the epidermis, is able to suppress P. aeruginosa and S. aureus biofilm formation, indicating that DNase1L2 is an innate antimicrobial defense of the epidermis186.
Nucleases Xds and Dns
Extracellular nucleases, Xds and Dns, are produced by Vibrio cholerae and act as virulence factors in an infant mouse cholera model187. Xds, a Mg2+ dependent nuclease, belongs to the protein family PF03372 and exerts both endo- and exonuclease activity188. Dns, also known as VcEndA, belongs to the endonuclease I superfamily and does not have a specific nucleic acid cleavage site189. These two extracellular nucleases can degrade both circular and linearized DNA within biofilms, and deletion of the genes encoding these nucleases results in increased biofilm formation47. It would be worth directly determining their biofilm dispersing activity since Xds and Dns are secreted enzymes with good stability.
Streptodornase
Streptodornase, also known as Varidase, is a commercial mixture of four DNase enzymes produced by P. aeruginosa, which reduces the viscosity of biofilm matrices by digesting the eDNA of biofilms190. An in vitro study found streptodornase is more active against the pre-formed biofilms of P. aeruginosa than DNase I, and has been successfully applied in P. aeruginosa focal infections, such as urinary tract infection191.
NucB
NucB, a biofilm-dispersing nuclease from the marine Bacillus licheniformis strain, also disperses newly formed biofilms by degradation of eDNA. It is a non-specific endonuclease belonging to the ββα metal-dependent nuclease subfamily192, and can degrade preformed biofilms of coagulase-negative staphylococci, S. aureus, and α-hemolytic streptococci isolated from chronic rhinosinusitis infections, offering a promising therapeutic approach for chronic rhinosinusitis patients193 (Table 3).
Current barriers and future directions of biofilm-dispersing enzymes
Utilization of enzymes to disperse biofilms has been a popular research topic for decades, and a number of dispersing enzymes have proven effective for inhibiting biofilm formation in diverse in vivo animal infection models including wound, nasal cavity, lung, and urinary tract108. Further, the rhDNase I, Dornase alfa, has been applied via inhalation in cystic fibrosis patients to reduce the viscosity of purulent sputum by preventing the establishment of chronic P. aeruginosa infection through inhibiting biofilm formation181. DispersinB® wound gel developed by Kane Biotech Inc. showed positive results in biocompatibility and in vivo preclinical studies and has been undergoing human clinical trials since 2022114. However, in practice most enzymatic biofilm eradication assays are carried out by in vitro testing against monomicrobial biofilms instead of multispecies-based biofilms that occur in nature. Thus, advancing the research of biofilm-dispersing enzymes is bottlenecked by the lack of the reliable biofilm models that mimic the true complexity of microbial colonization in humans and the world we live in.
Scientists must ascertain the compatibility of these enzymes and antibiotics with prudence. In order to enhance the potency of biofilm eradication, biofilm dispersing enzymes are always co-administered with antimicrobial agents, thereby providing an entryway to allow the antibiotics reach to the bacteria194. However, antibiotics can impact biofilm dispersing enzymes, for example, S. aureus micrococcal nuclease activity is modulated by sub-minimum inhibitory concentrations of antibiotics such as erythromycin and doxycycline195. Besides this, antibiotics bearing varied pKa values may also influence the activity of dispersing enzymes, thus robust enzymes with greater ranges of pH tolerance are more likely to be regarded as potential candidates for co-administration196. In the early stages of development, it is necessary to systematically characterize dispersing enzymes using artificial substrates that may need organic solvents in the testing buffer197. In preparations for manufacture and storage, enzymes with better thermostability will be favored for therapeutic development196. Therefore, it is necessary to consider enzymatic stability; including by not limited to thermostability, pH tolerance, and durability in organic solvents; when developing such dispersal agents for clinical application. Biofilm-dispersing enzymes, such as peptidase M16 and proteinase K, demonstrate remarkable stability in organic solvents, a wide pH tolerance, and great thermostability (37-60°C)143,174, but not all enzymes can maintain activity under harsh conditions. For example, DspB will denature when over 5% DMSO is present in solution106, and SPRE disperses biofilm in a pH-dependent manner, ranging between pH 5 – 6168.
Toxicity is another issue to be addressed in the development of enzymatic biofilm dispersal approaches, as these exogeneous agents could elicit strong immune responses or exhibit cellular toxicity. In a study on glycosidases to degrade biofilm, a total of 12 glycol-hydrolases including alginate lyase, amylase, amyloglucosidase, xylanase, cellulase, and pectinase were found to be cytotoxic towards human epithelial fibroblasts and human normal colonic cells108. Furthermore, xylanase displayed harmful effects in wound tissue at the wound site and even negatively impacted the spleen108.
Additionally, dispersing enzymes cannot achieve bactericidal activity and even facilitate bacterial colonization of new areas, leading to new or ongoing infections172. Research even shows that aureolysin and staphopain are able to degrade AMPs produced by the host immune system, resulting chronic atopic dermatitis172. Thus, biofilm dispersal enzymes are always combined with an antibiotic or other therapies to combat bacterial infections and may come into clinical applications in the near future, but careful consideration must be placed into the selection of agents for co-administration. Scientists have utilized enzyme engineering coupled with high-throughput screening to discover new enzymes as biofilm-dispersing agents106,107,140,198.
In summary, the capability of traditional antibiotics has been greatly compromised in recent years by increasing antibiotic tolerance of biofilm-embedded microbial pathogens. Clinically, biofilm-associated infections account for around 80% of human bacterial infections. Thus, effective biofilm dispersal strategies have been extensively sought after, and enzymatic dispersal stands out from other biofilm degrading methods due to its efficiency and specificity without causing selective pressure on bacteria. Biofilm-dispersing enzymes can effectively break down the EPS, leading to a collapse of biofilm matrices and making microbial cells accessible to antibiotic treatments or host immune responses. In this review, we have summarized the three major families of biofilm dispersal enzymes; glycosidases, proteases, and deoxyribonucleases; which target biofilm exopolysaccharides, extracellular proteins, and eDNA, respectively. Although numerous enzymes with biofilm-dispersing abilities have been discovered and demonstrate promising results in vitro, only a few in vivo studies have been performed, with clinical trials conducted for even less enzymes. Issues like toxicity, compatibility, and stability of biofilm-degrading enzymes have not yet been fully addressed. Further efforts are needed to develop robust, safe, and potent biofilm-dispersing enzymes for clinical applications.
References
Joo, H. S. & Otto, M. Molecular basis of in vivo biofilm formation by bacterial pathogens. Chem. Biol. 19, 1503–1513 (2012).
Taylor, P. K., Van Kessel, A. T. M., Colavita, A., Hancock, R. E. W. & Mah, T. F. A novel small RNA is important for biofilm formation and pathogenicity in Pseudomonas aeruginosa. PLoS ONE 12, e0182582 (2017).
Das, T., Kutty, S. K., Kumar, N. & Manefield, M. Pyocyanin facilitates extracellular DNA binding to Pseudomonas aeruginosa influencing cell surface properties and aggregation. PLoS ONE 8, e58299 (2013).
Flemming, H. C., Neu, T. R. & Wozniak, D. J. The EPS matrix: the “house of biofilm cells”. J. Bacteriol. 189, 7945–7947 (2007).
Schachter, B. Slimy business—the biotechnology of biofilms. Nat. Biotechnol. 21, 361–365 (2003).
Nadell, C. D., Xavier, J. B., Levin, S. A. & Foster, K. R. The evolution of quorum sensing in bacterial biofilms. PLoS Biol. 6, e14 (2008).
Davies, D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2, 114–122 (2003).
Machado, D., Castro, J., Palmeira-de-Oliveira, A., Martinez-de-Oliveira, J. & Cerca, N. Bacterial vaginosis biofilms: challenges to current therapies and emerging solutions. Front. Microbiol. 6, 1528 (2015).
von Rosenvinge, E. C., O’May, G. A., Macfarlane, S., Macfarlane, G. T. & Shirtliff, M. E. Microbial biofilms and gastrointestinal diseases. Pathog. Dis. 67, 25–38 (2013).
Behlau, I. & Gilmore, M. S. Microbial biofilms in ophthalmology and infectious disease. Arch. Ophthalmol. 126, 1572–1581 (2008).
Vieira Colombo, A. P. et al. Periodontal-disease-associated biofilm: a reservoir for pathogens of medical importance. Microb. Pathog. 94, 27–34 (2016).
Delcaru, C. et al. Microbial biofilms in urinary tract infections and prostatitis: etiology, pathogenicity, and combating strategies. Pathogens 5, https://doi.org/10.3390/pathogens5040065 (2016).
Post, J. C. Direct evidence of bacterial biofilms in otitis media. Laryngoscope 111, 2083–2094 (2001).
Francolini, I. & Donelli, G. Prevention and control of biofilm-based medical-device-related infections. FEMS Immunol. Med. Microbiol. 59, 227–238 (2010).
Fisher, R. A., Gollan, B. & Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 15, 453–464 (2017).
Sauer, K. et al. The biofilm life cycle: expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 20, 608–620 (2022).
Arjes, H. A. et al. Three-dimensional biofilm colony growth supports a mutualism involving matrix and nutrient sharing. Elife 10, https://doi.org/10.7554/eLife.64145 (2021).
Cherny, K. E. & Sauer, K. Untethering and degradation of the polysaccharide matrix are essential steps in the dispersion response of pseudomonas aeruginosa biofilms. J. Bacteriol. 202, https://doi.org/10.1128/JB.00575-19 (2020).
Rumbaugh, K. P. & Sauer, K. Biofilm dispersion. Nat. Rev. Microbiol. 18, 571–586 (2020).
Stewart, P. S. Antimicrobial tolerance in biofilms. Microbiol. Spectr. 3, https://doi.org/10.1128/microbiolspec.MB-0010-2014 (2015).
Stewart, P. S. & Costerton, J. W. Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138 (2001).
Brown, M. R., Allison, D. G. & Gilbert, P. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J. Antimicrob. Chemother. 22, 777–780 (1988).
Singh, S., Singh, S. K., Chowdhury, I. & Singh, R. Understanding the mechanism of bacterial biofilms resistance to antimicrobial agents. Open Microbiol. J. 11, 53–62 (2017).
Wood, T. K., Knabel, S. J. & Kwan, B. W. Bacterial persister cell formation and dormancy. Appl Environ. Microbiol. 79, 7116–7121 (2013).
Lewis, K. Persister cells and the riddle of biofilm survival. Biochemistry (Mosc.) 70, 267–274 (2005).
Lewis, K. Persister cells. Annu. Rev. Microbiol. 64, 357–372 (2010).
LaFleur, M. D., Kumamoto, C. A. & Lewis, K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob. Agents Chemother. 50, 3839–3846 (2006).
Lewis, K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45, 999–1007 (2001).
Baltzer, S. A. & Brown, M. H. Antimicrobial peptides: promising alternatives to conventional antibiotics. J. Mol. Microbiol. Biotechnol. 20, 228–235 (2011).
Mookherjee, N., Anderson, M. A., Haagsman, H. P. & Davidson, D. J. Antimicrobial host defence peptides: functions and clinical potential. Nat. Rev. Drug Discov. 19, 311–332 (2020).
Yoon, B. K., Jackman, J. A., Valle-Gonzalez, E. R. & Cho, N. J. Antibacterial free fatty acids and monoglycerides: biological activities, experimental testing, and therapeutic applications. Int. J. Mol. Sci. 19, 1114 (2018).
Arciola, C. R., Campoccia, D., Speziale, P., Montanaro, L. & Costerton, J. W. Biofilm formation in Staphylococcus implant infections. A review of molecular mechanisms and implications for biofilm-resistant materials. Biomaterials 33, 5967–5982 (2012).
Katsikogianni, M. & Missirlis, Y. F. Concise review of mechanisms of bacterial adhesion to biomaterials and of techniques used in estimating bacteria-material interactions. Eur. Cell Mater. 8, 37–57 (2004).
Russell, A. D. Biocide use and antibiotic resistance: the relevance of laboratory findings to clinical and environmental situations. Lancet Infect. Dis. 3, 794–803 (2003).
Barraud, N. et al. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 188, 7344–7353 (2006).
Barraud, N., Kelso, M. J., Rice, S. A. & Kjelleberg, S. Nitric oxide: a key mediator of biofilm dispersal with applications in infectious diseases. Curr. Pharm. Des. 21, 31–42 (2015).
Brackman, G. & Coenye, T. Quorum sensing inhibitors as anti-biofilm agents. Curr. Pharm. Des. 21, 5–11 (2015).
Poulin, M. B. & Kuperman, L. L. Regulation of biofilm exopolysaccharide production by cyclic di-guanosine monophosphate. Front. Microbiol. 12, 730980 (2021).
Zhou, C. et al. Pharmacokinetics and pharmacodynamics of DSTA4637A: a novel THIOMAB antibody antibiotic conjugate against Staphylococcus aureus in mice. MAbs 8, 1612–1619 (2016).
Buhmann, M. T. et al. Extraction of biofilms from ureteral stents for quantification and cultivation-dependent and -independent analyses. Front. Microbiol. 9, 1470 (2018).
Kaplan, J. B. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 89, 205–218 (2010).
Snarr, B. D., Howell, P. L. & Sheppard, D. C. Hoisted by their own petard: do microbial enzymes hold the solution to treating and preventing biofilm infections? Future Microbiol. 13, 395–398 (2018).
Fleming, D. & Rumbaugh, K. P. Approaches to Dispersing Medical Biofilms. Microorganisms 5, https://doi.org/10.3390/microorganisms5020015 (2017).
Izano, E. A., Wang, H., Ragunath, C., Ramasubbu, N. & Kaplan, J. B. Detachment and killing of Aggregatibacter actinomycetemcomitans biofilms by dispersin B and SDS. J. Dent. Res. 86, 618–622 (2007).
Fazekas, E., Kandra, L. & Gyemant, G. Model for beta-1,6-N-acetylglucosamine oligomer hydrolysis catalysed by DispersinB, a biofilm degrading enzyme. Carbohydr. Res. 363, 7–13 (2012).
Whitchurch, C. B., Tolker-Nielsen, T., Ragas, P. C. & Mattick, J. S. Extracellular DNA required for bacterial biofilm formation. Science 295, 1487 (2002).
Seper, A. et al. Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation. Mol. Microbiol. 82, 1015–1037 (2011).
Shaw, L., Golonka, E., Potempa, J. & Foster, S. J. The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology (Reading) 150, 217–228 (2004).
Marti, M. et al. Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect. 12, 55–64 (2010).
Ryder, C., Byrd, M. & Wozniak, D. J. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 10, 644–648 (2007).
Ross, P., Mayer, R. & Benziman, M. Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 55, 35–58 (1991).
Jackson, K. D., Starkey, M., Kremer, S., Parsek, M. R. & Wozniak, D. J. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186, 4466–4475 (2004).
Friedman, L. & Kolter, R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186, 4457–4465 (2004).
Heilmann, C. et al. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20, 1083–1091 (1996).
Rinehart, K. L. et al. Bioactive compounds from aquatic and terrestrial sources. J. Nat. Prod. 53, 771–792 (1990).
Vocadlo, D. J. & Withers, S. G. Detailed comparative analysis of the catalytic mechanisms of beta-N-acetylglucosaminidases from families 3 and 20 of glycoside hydrolases. Biochemistry 44, 12809–12818 (2005).
Wang, X., Preston, J. F. 3rd & Romeo, T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186, 2724–2734 (2004).
Jarrett, C. O. et al. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190, 783–792 (2004).
Izano, E. A. et al. Poly-N-acetylglucosamine mediates biofilm formation and antibiotic resistance in Actinobacillus pleuropneumoniae. Microb. Pathog. 43, 1–9 (2007).
Izano, E. A. et al. Poly-N-acetylglucosamine mediates biofilm formation and detergent resistance in Aggregatibacter actinomycetemcomitans. Microb. Pathog. 44, 52–60 (2008).
Parise, G., Mishra, M., Itoh, Y., Romeo, T. & Deora, R. Role of a putative polysaccharide locus in Bordetella biofilm development. J. Bacteriol. 189, 750–760 (2007).
Choi, A. H., Slamti, L., Avci, F. Y., Pier, G. B. & Maira-Litran, T. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 191, 5953–5963 (2009).
Yakandawala, N. et al. Characterization of the poly-beta-1,6-N-acetylglucosamine polysaccharide component of Burkholderia biofilms. Appl. Environ. Microbiol. 77, 8303–8309 (2011).
Chen, K. M. et al. The role of pgaC in Klebsiella pneumoniae virulence and biofilm formation. Microb. Pathog. 77, 89–99 (2014).
Ye, L., Zheng, X. & Zheng, H. Effect of sypQ gene on poly-N-acetylglucosamine biosynthesis in Vibrio parahaemolyticus and its role in infection process. Glycobiol 24, 351–358 (2014).
Roux, D. et al. Identification of poly-N-acetylglucosamine as a major polysaccharide component of the bacillus subtilis biofilm matrix. J. Biol. Chem. 290, 19261–19272 (2015).
Maira-Litran, T. et al. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 70, 4433–4440 (2002).
McKenney, D. et al. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect. Immun. 66, 4711–4720 (1998).
Cramton, S. E., Gerke, C., Schnell, N. F., Nichols, W. W. & Gotz, F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67, 5427–5433 (1999).
Little, D. J. et al. Structural basis for the De-N-acetylation of poly-beta-1,6-N-acetyl-D-glucosamine in Gram-positive bacteria. J. Biol. Chem. 289, 35907–35917 (2014).
Atkin, K. E., MacDonald, S. J., Brentnall, A. S., Potts, J. R. & Thomas, G. H. A different path: revealing the function of staphylococcal proteins in biofilm formation. FEBS Lett. 588, 1869–1872 (2014).
Nishiyama, T., Noguchi, H., Yoshida, H., Park, S. Y. & Tame, J. R. The structure of the deacetylase domain of Escherichia coli PgaB, an enzyme required for biofilm formation: a circularly permuted member of the carbohydrate esterase 4 family. Acta Crystallogr. D: Biol. Crystallogr. 69, 44–51 (2013).
Franklin, M. J., Nivens, D. E., Weadge, J. T. & Howell, P. L. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front. Microbiol. 2, 167 (2011).
Stover, C. K. et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964 (2000).
Zielinski, N. A., Chakrabarty, A. M. & Berry, A. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J. Biol. Chem. 266, 9754–9763 (1991).
Baker, P. et al. P. aeruginosa SGNH hydrolase-like proteins AlgJ and AlgX have similar topology but separate and distinct roles in alginate acetylation. PLoS Pathog. 10, e1004334 (2014).
Gheorghita, A. A. et al. Structure of the AlgKX modification and secretion complex required for alginate production and biofilm attachment in Pseudomonas aeruginosa. Nat. Commun. 13, 7631 (2022).
Wolfram, F. et al. Catalytic mechanism and mode of action of the periplasmic alginate epimerase AlgG. J. Biol. Chem. 289, 6006–6019 (2014).
Blasi, P. et al. Preparation and in vitro and in vivo characterization of composite microcapsules for cell encapsulation. Int J. Pharm. 324, 27–36 (2006).
Colvin, K. M. et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol 14, 1913–1928 (2012).
Whitfield, G. B. & Howell, P. L. The matrix revisited: opening night for the pel polysaccharide across eubacterial kingdoms. Microbiol Insights 14, 1178636120988588 (2021).
Le Mauff, F. et al. The Pel polysaccharide is predominantly composed of a dimeric repeat of alpha-1,4 linked galactosamine and N-acetylgalactosamine. Commun. Biol. 5, 502 (2022).
Whitfield, G. B. et al. Pel polysaccharide biosynthesis requires an inner membrane complex comprised of PelD, PelE, PelF, and PelG. J. Bacteriol. 202, https://doi.org/10.1128/JB.00684-19 (2020).
Whitfield, G. B. et al. Discovery and characterization of a Gram-positive Pel polysaccharide biosynthetic gene cluster. PLoS Pathog. 16, e1008281 (2020).
Bundalovic-Torma, C., Whitfield, G. B., Marmont, L. S., Howell, P. L. & Parkinson, J. A systematic pipeline for classifying bacterial operons reveals the evolutionary landscape of biofilm machineries. PLoS Comput. Biol. 16, e1007721 (2020).
Friedman, L. & Kolter, R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690 (2004).
Colvin, K. M. et al. PelA deacetylase activity is required for Pel polysaccharide synthesis in Pseudomonas aeruginosa. J. Bacteriol. 195, 2329–2339 (2013).
Champion, A. E., Catanzaro, K. C. F., Bandara, A. B. & Inzana, T. J. Formation of the Francisella tularensis biofilm is affected by cell surface glycosylation, growth medium, and a glucan exopolysaccharide. Sci. Rep. 9, 12252 (2019).
Petruzzi, B. et al. Capsular polysaccharide interferes with biofilm formation by Pasteurella multocida Serogroup A. mBio 8, https://doi.org/10.1128/mBio.01843-17 (2017).
Matthysse, A. G., Thomas, D. L. & White, A. R. Mechanism of cellulose synthesis in Agrobacterium tumefaciens. J. Bacteriol. 177, 1076–1081 (1995).
Zogaj, X., Nimtz, M., Rohde, M., Bokranz, W. & Romling, U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39, 1452–1463 (2001).
Gal, M., Preston, G. M., Massey, R. C., Spiers, A. J. & Rainey, P. B. Genes encoding a cellulosic polymer contribute toward the ecological success of Pseudomonas fluorescens SBW25 on plant surfaces. Mol. Ecol. 12, 3109–3121 (2003).
Romling, U. & Galperin, M. Y. Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol. 23, 545–557 (2015).
Shen, T. & Gnanakaran, S. The stability of cellulose: a statistical perspective from a coarse-grained model of hydrogen-bond networks. Biophys. J. 96, 3032–3040 (2009).
Thongsomboon, W. et al. Phosphoethanolamine cellulose: a naturally produced chemically modified cellulose. Science 359, 334–338 (2018).
Fontaine, T. et al. Galactosaminogalactan, a new immunosuppressive polysaccharide of Aspergillus fumigatus. PLoS Pathog. 7, e1002372 (2011).
Lee, M. J. et al. Deacetylation of fungal exopolysaccharide mediates adhesion and biofilm formation. mBio 7, e00252–00216 (2016).
Bamford, N. C. et al. Structural and biochemical characterization of the exopolysaccharide deacetylase Agd3 required for Aspergillus fumigatus biofilm formation. Nat. Commun. 11, 2450 (2020).
Matyjaszewski, K. & Möller, M. Polymer science: a comprehensive reference. Chapter 2, 111–184 (2012).
Douglas, C. Fungal β (1, 3)-D-glucan synthesis. Sabouraudia 39, 55–66 (2001).
Tan, Y., Ma, S., Leonhard, M., Moser, D. & Schneider-Stickler, B. beta-1,3-glucanase disrupts biofilm formation and increases antifungal susceptibility of Candida albicans DAY185. Int J. Biol. Macromol. 108, 942–946 (2018).
Oner, E. T., Hernandez, L. & Combie, J. Review of Levan polysaccharide: from a century of past experiences to future prospects. Biotechnol. Adv. 34, 827–844 (2016).
Han, Y. W. Microbial levan. Adv. Appl. Microbiol. 35, 171–194 (1990).
Zhang, W. et al. An overview of levan-degrading enzyme from microbes. Appl. Microbiol. Biotechnol. 103, 7891–7902 (2019).
Dedonder, R. In Methods in Enzymology Vol. 8, 500–505 (Elsevier, 1966). https://www.sciencedirect.com/science/article/abs/pii/0076687966080911.
Wang, S., Breslawec, A. P., Li, C. & Poulin, M. B. A colorimetric assay to enable high-throughput identification of biofilm exopolysaccharide-hydrolyzing enzymes. Chemistry 26, 10719–10723 (2020).
Wang, S., Breslawec, A. P. & Poulin, M. B. Multifunctional fluorescent probes for high-throughput characterization of hexosaminidase enzyme activity. Bioorg. Chem. 119, 105532 (2022).
Redman, W. K. et al. Efficacy and safety of biofilm dispersal by glycoside hydrolases in wounds. Biofilm 3, 100061 (2021).
Kaplan, J. B., Ragunath, C., Ramasubbu, N. & Fine, D. H. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J. Bacteriol. 185, 4693–4698 (2003).
Kerrigan, J. E. et al. Modeling and biochemical analysis of the activity of antibiofilm agent Dispersin B. Acta Biol. Hung. 59, 439–451 (2008).
Wang, S. et al. Differential recognition of deacetylated PNAG oligosaccharides by a biofilm degrading glycosidase. ACS Chem. Biol. 14, 1998–2005 (2019).
Breslawec, A. P., Wang, S., Monahan, K. N., Barry, L. L. & Poulin, M. B. The endoglycosidase activity of Dispersin B is mediated through electrostatic interactions with cationic poly-beta-(1–>6)-N-acetylglucosamine. FEBS J. 290, 1049–1059 (2023).
Manuel, S. G. et al. Role of active-site residues of dispersin B, a biofilm-releasing beta-hexosaminidase from a periodontal pathogen, in substrate hydrolysis. FEBS J. 274, 5987–5999 (2007).
Gawande, P. V. et al. Antibiofilm efficacy of DispersinB((R)) wound spray used in combination with a silver wound dressing. Microbiol Insights 7, 9–13 (2014).
Breslawec, A. P., Wang, S., Li, C. & Poulin, M. B. Anionic amino acids support hydrolysis of poly-beta-(1,6)-N-acetylglucosamine exopolysaccharides by the biofilm dispersing glycosidase Dispersin B. J. Biol. Chem. 296, 100203 (2021).
Little, D. J. et al. PgaB orthologues contain a glycoside hydrolase domain that cleaves deacetylated poly-beta(1,6)-N-acetylglucosamine and can disrupt bacterial biofilms. PLoS Pathog. 14, e1006998 (2018).
Forman, A., Pfoh, R., Eddenden, A., Howell, P. L. & Nitz, M. Synthesis of defined mono-de-N-acetylated beta-(1–>6)-N-acetyl-d-glucosamine oligosaccharides to characterize PgaB hydrolase activity. Org. Biomol. Chem. 17, 9456–9466 (2019).
Little, D. J. et al. Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine. Proc. Natl Acad. Sci. USA 111, 11013–11018 (2014).
Blanco-Cabra, N. et al. Characterization of different alginate lyases for dissolving Pseudomonas aeruginosa biofilms. Sci. Rep. 10, 9390 (2020).
Zhu, B. & Yin, H. Alginate lyase: Review of major sources and classification, properties, structure-function analysis and applications. Bioengineered 6, 125–131 (2015).
Jang, C. H. et al. Modeling and re-engineering of azotobacter vinelandii alginate lyase to enhance its catalytic efficiency for accelerating biofilm degradation. PLoS ONE 11, e0156197 (2016).
Daboor, S. M., Raudonis, R. & Cheng, Z. Characterizations of the viability and gene expression of dispersal cells from Pseudomonas aeruginosa biofilms released by alginate lyase and tobramycin. PLoS ONE 16, e0258950 (2021).
Baker, P. et al. Characterization of the Pseudomonas aeruginosa glycoside hydrolase PslG reveals that its levels are critical for Psl polysaccharide biosynthesis and biofilm formation. J. Biol. Chem. 290, 28374–28387 (2015).
Baker, P. et al. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci. Adv. 2, e1501632 (2016).
Paul, J. S., Gupta, N., Beliya, E., Tiwari, S. & Jadhav, S. K. Aspects and recent trends in microbial alpha-amylase: a review. Appl. Biochem. Biotechnol. 193, 2649–2698 (2021).
Craigen, B., Dashiff, A. & Kadouri, D. E. The use of commercially available alpha-amylase compounds to inhibit and remove Staphylococcus aureus biofilms. Open Microbiol J. 5, 21–31 (2011).
Karatan, E. & Watnick, P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73, 310–347 (2009).
Loiselle, M. & Anderson, K. W. The use of cellulase in inhibiting biofilm formation from organisms commonly found on medical implants. Biofouling 19, 77–85 (2003).
Kamali, E., Jamali, A., Izanloo, A. & Ardebili, A. In vitro activities of cellulase and ceftazidime, alone and in combination against Pseudomonas aeruginosa biofilms. BMC Microbiol. 21, 347 (2021).
Zhang, Z., Liao, H., Yang, M., Hu, C. & Du, Y. [Levofloxacin combined with cellulase can eradicate bacille Calmette-Guerin biofilm infection]. Nan Fang Yi Ke Da Xue Xue Bao 43, 257–264 (2023).
Bamford, N. C. et al. Sph3 is a glycoside hydrolase required for the biosynthesis of galactosaminogalactan in Aspergillus fumigatus. J. Biol. Chem. 290, 27438–27450 (2015).
Le Mauff, F. et al. Molecular mechanism of Aspergillus fumigatus biofilm disruption by fungal and bacterial glycoside hydrolases. J. Biol. Chem. 294, 10760–10772 (2019).
Bamford, N. C. et al. Ega3 from the fungal pathogen Aspergillus fumigatus is an endo-alpha-1,4-galactosaminidase that disrupts microbial biofilms. J. Biol. Chem. 294, 13833–13849 (2019).
Balasubramanian, V., Vashisht, D., Cletus, J. & Sakthivel, N. Plant beta-1,3-glucanases: their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol. Lett. 34, 1983–1990 (2012).
Wanker, E., Huber, A. & Schwab, H. Purification and characterization of the Bacillus subtilis levanase produced in Escherichia coli. Appl. Environ. Microbiol. 61, 1953–1958 (1995).
Shida, T., Mukaijo, K., Ishikawa, S., Yamamoto, H. & Sekiguchi, J. Production of long-chain levan by a sacC insertional mutant from Bacillus subtilis 327UH. Biosci. Biotechnol. Biochem. 66, 1555–1558 (2002).
Trizna, E., Bogachev, M. I. & Kayumov, A. Degrading of the Pseudomonas aeruginosa biofilm by extracellular levanase SacC from Bacillus subtilis. BioNanoScience 9, 48–52 (2019).
Liu, Y. et al. Synergistic effect between the recombinant exo-inulinase and endo-inulinase on inulin hydrolysis. J. Mol. Catal. B: Enzym. 128, 27–38 (2016).
Liu, G.-L., Chi, Z. & Chi, Z.-M. Molecular characterization and expression of microbial inulinase genes. Crit. Rev. Microbiol. 39, 152–165 (2013).
Møllebjerg, A. et al. Novel high-throughput screening platform identifies enzymes to tackle biofouling on reverse osmosis membranes. Desalination 554, 116485 (2023).
Alav, I., Sutton, J. M. & Rahman, K. M. Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 73, 2003–2020 (2018).
Erskine, E., MacPhee, C. E. & Stanley-Wall, N. R. Functional amyloid and other protein fibers in the biofilm matrix. J. Mol. Biol. 430, 3642–3656 (2018).
Ebeling, W. et al. Proteinase K from Tritirachium album Limber. Eur. J. Biochem. 47, 91–97 (1974).
Kumar Shukla, S. & Rao, T. S. Dispersal of Bap-mediated Staphylococcus aureus biofilm by proteinase K. J. Antibiot. (Tokyo) 66, 55–60 (2013).
Nguyen, U. T. & Burrows, L. L. DNase I and proteinase K impair Listeria monocytogenes biofilm formation and induce dispersal of pre-existing biofilms. Int J. Food Microbiol. 187, 26–32 (2014).
Cui, H., Ma, C. & Lin, L. Co-loaded proteinase K/thyme oil liposomes for inactivation of Escherichia coli O157:H7 biofilms on cucumber. Food Funct. 7, 4030–4040 (2016).
Richert, A. & Dabrowska, G. B. Enzymatic degradation and biofilm formation during biodegradation of polylactide and polycaprolactone polymers in various environments. Int J. Biol. Macromol. 176, 226–232 (2021).
Radwan, A. A., Darwesh, O. M., Emam, M. T., Mohamed, K. A. & Shady, H. M. A. A combined treatment of Proteinase K and biosynthesized ZnO-NPs for eradication of dairy biofilm of sporeformers. AIMS Microbiol. 8, 507–527 (2022).
Hu, W. S., Min Nam, D., Kim, J. S. & Koo, O. K. Synergistic anti-biofilm effects of Brassicaceae plant extracts in combination with proteinase K against Escherichia coli O157:H7. Sci. Rep. 10, 21090 (2020).
Thomas, R. E. & Thomas, B. C. Reducing biofilm infections in burn patients’ wounds and biofilms on surfaces in hospitals, medical facilities and medical equipment to improve burn care: a systematic review. Int. J. Environ. Res. Public Health 18, https://doi.org/10.3390/ijerph182413195 (2021).
Mugita, N., Nambu, T., Takahashi, K., Wang, P. L. & Komasa, Y. Proteases, actinidin, papain and trypsin reduce oral biofilm on the tongue in elderly subjects and in vitro. Arch. Oral. Biol. 82, 233–240 (2017).
Banar, M. et al. Evaluation of mannosidase and trypsin enzymes effects on biofilm production of Pseudomonas aeruginosa isolated from burn wound infections. PLoS ONE 11, e0164622 (2016).
Zhou, J. et al. An in vitro study on the degradation of multispecies biofilm of periodontitis-related microorganisms by bovine trypsin. Front. Microbiol. 13, 951291 (2022).
Mechmechani, S. et al. Pepsin and trypsin treatment combined with carvacrol: an efficient strategy to fight Pseudomonas aeruginosa and Enterococcus faecalis biofilms. Microorganisms 11, 143 (2023).
Hamuro, Y., Coales, S. J., Molnar, K. S., Tuske, S. J. & Morrow, J. A. Specificity of immobilized porcine pepsin in H/D exchange compatible conditions. Rapid Commun. Mass Spectrom. 22, 1041–1046 (2008).
Mechmechani, S. et al. Hurdle technology based on the use of microencapsulated pepsin, trypsin and carvacrol to eradicate Pseudomonas aeruginosa and Enterococcus faecalis biofilms. Biofouling 38, 903–915 (2022).
Potempa, J. & Shaw, L. N. In Handbook of Proteolytic Enzymes 3rd edn (eds Rawlings, N. D. & Guy, S.) 563-569 (Academic Press, 2013).
Laarman, A. J. et al. Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. J. Immunol. 186, 6445–6453 (2011).
Pietrocola, G., Nobile, G., Rindi, S. & Speziale, P. Staphylococcus aureus manipulates innate immunity through own and host-expressed proteases. Front. Cell Infect. Microbiol. 7, 166 (2017).
Loughran, A. J. et al. Impact of individual extracellular proteases on Staphylococcus aureus biofilm formation in diverse clinical isolates and their isogenic sarA mutants. Microbiologyopen 3, 897–909 (2014).
Chen, C. et al. Secreted proteases control autolysin-mediated biofilm growth of Staphylococcus aureus. J. Biol. Chem. 288, 29440–29452 (2013).
Barrett, A., Rawlings, N. & Woessner, J. Handbook of Proteolytic Enzymes (Academic Press, 1998).
Mootz, J. M., Malone, C. L., Shaw, L. N. & Horswill, A. R. Staphopains modulate Staphylococcus aureus biofilm integrity. Infect. Immun. 81, 3227–3238 (2013).
Yamasaki, K. et al. Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nat. Med. 13, 975–980 (2007).
Sonesson, A. et al. Identification of bacterial biofilm and the Staphylococcus aureus derived protease, staphopain, on the skin surface of patients with atopic dermatitis. Sci. Rep. 7, 1–12 (2017).
Paharik, A. E. et al. The Spl serine proteases modulate staphylococcus aureus protein production and virulence in a rabbit model of pneumonia. mSphere 1, https://doi.org/10.1128/mSphere.00208-16 (2016).
Boles, B. R. & Horswill, A. R. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4, e1000052 (2008).
Lee, S. F., Li, Y. H. & Bowden, G. H. Detachment of Streptococcus mutans biofilm cells by an endogenous enzymatic activity. Infect. Immun. 64, 1035–1038 (1996).
Lee, S. F. Identification and characterization of a surface protein-releasing activity in Streptococcus mutans and other pathogenic streptococci. Infect. Immun. 60, 4032–4039 (1992).
Nelson, D. C., Garbe, J. & Collin, M. Cysteine proteinase SpeB from Streptococcus pyogenes—a potent modifier of immunologically important host and bacterial proteins. Biol. Chem. 392, 1077–1088 (2011).
Roberts, A. L., Holder, R. C. & Reid, S. D. Allelic replacement of the streptococcal cysteine protease SpeB in a Δ srv mutant background restores biofilm formation. BMC Res. Notes 3, 1–7 (2010).
Connolly, K. L., Roberts, A. L., Holder, R. C. & Reid, S. D. Dispersal of Group A streptococcal biofilms by the cysteine protease SpeB leads to increased disease severity in a murine model. PLoS ONE 6, e18984 (2011).
Carothers, K. E. et al. The streptococcal protease SpeB antagonizes the biofilms of the human pathogen Staphylococcus aureus USA300 through cleavage of the staphylococcal SdrC protein. J. Bacteriol. 202, e00008–e00020 (2020).
Saggu, S. K., Jha, G. & Mishra, P. C. Enzymatic degradation of biofilm by metalloprotease from microbacterium sp. SKS10. Front. Bioeng. Biotechnol. 7, 192 (2019).
Panlilio, H. & Rice, C. V. The role of extracellular DNA in the formation, architecture, stability, and treatment of bacterial biofilms. Biotechnol. Bioeng. 118, 2129–2141 (2021).
Okshevsky, M. & Meyer, R. L. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 41, 341–352 (2015).
Alhede, M., Bjarnsholt, T., Givskov, M. & Alhede, M. Pseudomonas aeruginosa biofilms: mechanisms of immune evasion. Adv. Appl. Microbiol. 86, 1–40 (2014).
Baranovskii, A. G., Buneva, V. N. & Nevinsky, G. A. Human deoxyribonucleases. Biochemistry (Mosc.) 69, 587–601 (2004).
Sharma, K. & Pagedar Singh, A. Antibiofilm effect of DNase against single and mixed species biofilm. Foods 7, https://doi.org/10.3390/foods7030042 (2018).
Tetz, V. V. & Tetz, G. V. Effect of extracellular DNA destruction by DNase I on characteristics of forming biofilms. DNA Cell Biol. 29, 399–405 (2010).
Wagener, J. S. & Kupfer, O. Dornase alfa (Pulmozyme). Curr. Opin. Pulm. Med. 18, 609–614 (2012).
Kaplan, J. B. et al. Recombinant human DNase I decreases biofilm and increases antimicrobial susceptibility in staphylococci. J. Antibiot. (Tokyo) 65, 73–77 (2012).
Pakkulnan, R., Thonglao, N. & Chareonsudjai, S. DNase I and chitosan enhance efficacy of ceftazidime to eradicate Burkholderia pseudomallei biofilm cells. Sci. Rep. 13, 1059 (2023).
Fang, J. Y. et al. Facile biofilm penetration of cationic liposomes loaded with DNase I/Proteinase K to eradicate cutibacterium acnes for treating cutaneous and catheter infections. Int. J. Nanomed. 16, 8121–8138 (2021).
Patel, K. K. et al. Antibiofilm potential of silver sulfadiazine-loaded nanoparticle formulations: a study on the effect of DNase-I on microbial biofilm and wound healing activity. Mol. Pharm. 16, 3916–3925 (2019).
Eckhart, L., Fischer, H., Barken, K. B., Tolker-Nielsen, T. & Tschachler, E. DNase1L2 suppresses biofilm formation by Pseudomonas aeruginosa and Staphylococcus aureus. Br. J. Dermatol. 156, 1342–1345 (2007).
Focareta, T. & Manning, P. A. Distinguishing between the extracellular DNases of Vibrio cholerae and development of a transformation system. Mol. Microbiol. 5, 2547–2555 (1991).
Dlakic, M. Functionally unrelated signalling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem. Sci. 25, 272–273 (2000).
Niiranen, L. et al. Effects of salt on the kinetics and thermodynamic stability of endonuclease I from Vibrio salmonicida and Vibrio cholerae. FEBS J. 275, 1593–1605 (2008).
Tang, Y.-W., Liu, D., Sussman, M., Poxton, I. & Schwartzman, J. Molecular Medical Microbiology (Academic Press, 2014).
Nemoto, K. et al. Effect of Varidase (streptodornase) on biofilm formed by Pseudomonas aeruginosa. Chemotherapy 49, 121–125 (2003).
Basle, A. et al. Crystal structure of NucB, a biofilm-degrading endonuclease. Nucleic Acids Res. 46, 473–484 (2018).
Shields, R. C. et al. Efficacy of a marine bacterial nuclease against biofilm forming microorganisms isolated from chronic rhinosinusitis. PLoS ONE 8, e55339 (2013).
Fanaei Pirlar, R. et al. Combinatorial effects of antibiotics and enzymes against dual-species Staphylococcus aureus and Pseudomonas aeruginosa biofilms in the wound-like medium. PLoS ONE 15, e0235093 (2020).
Rosman, C. W. K., van der Mei, H. C. & Sjollema, J. Influence of sub-inhibitory concentrations of antimicrobials on micrococcal nuclease and biofilm formation in Staphylococcus aureus. Sci. Rep. 11, 13241 (2021).
Thapa, S. et al. Biochemical characteristics of microbial enzymes and their significance from industrial perspectives. Mol. Biotechnol. 61, 579–601 (2019).
Borooah, J., Leaback, D. H. & Walker, P. G. Studies on glucosaminidase. 2. Substrates for N-acetyl-β-glucosaminidase*. Biochem. J. 78, 106–110 (1961).
Ngara, T. R. & Zhang, H. Recent advances in function-based metagenomic screening. Genomics Proteom. Bioinform. 16, 405–415 (2018).
Acknowledgements
This work was supported by Henan Province in 2023 Key scientific research project plan of colleges and universities (Grant No. 23A310029) to Dr. Shaochi Wang.
Author information
Authors and Affiliations
Contributions
S.W., L.L.K., and Q.Y. took the lead in writing the manuscript. A.P.B., Y.Z., and Z.D. provided critical feedback and helped shape the manuscript. T.L. helped revise the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, S., Zhao, Y., Breslawec, A.P. et al. Strategy to combat biofilms: a focus on biofilm dispersal enzymes. npj Biofilms Microbiomes 9, 63 (2023). https://doi.org/10.1038/s41522-023-00427-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41522-023-00427-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.