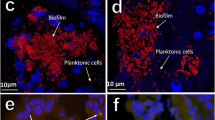
Abstract
The formation of microbial biofilms enables single planktonic cells to assume a multicellular mode of growth. During dispersion, the final step of the biofilm life cycle, single cells egress from the biofilm to resume a planktonic lifestyle. As the planktonic state is considered to be more vulnerable to antimicrobial agents and immune responses, dispersion is being considered a promising avenue for biofilm control. In this Review, we discuss conditions that lead to dispersion and the mechanisms by which native and environmental cues contribute to dispersion. We also explore recent findings on the role of matrix degradation in the dispersion process, and the distinct phenotype of dispersed cells. Last, we discuss the translational and therapeutic potential of dispersing bacteria during infection.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Costerton, J. W., Stewart, P. S. & Greenberg, E. P. Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322 (1999).
Flemming, H.-C. et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563 (2016).
Geesey, G. G., Richardson, W. T., Yeomans, H. G., Irvin, R. T. & Costerton, J. W. Microscopic examination of natural sessile bacterial populations from an alpine stream. Can. J. Microbiol. 23, 1733–1736 (1977).
Costerton, J. W. et al. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41, 435–464 (1987).
Stoodley, P., Sauer, K., Davies, D. G. & Costerton, J. W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209 (2002).
Petrova, O. E. & Sauer, K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 5, e1000668 (2009).
Petrova, O. E., Gupta, K., Liao, J., Goodwine, J. S. & Sauer, K. Divide and conquer: the Pseudomonas aeruginosa two-component hybrid SagS enables biofilm formation and recalcitrance of biofilm cells to antimicrobial agents via distinct regulatory circuits. Environ. Microbiol. 19, 2005–2024 (2017).
O’Toole, G. A. & Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28, 449–461 (1998).
Davey, M. E. & O’Toole, G. A. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64, 847–867 (2000).
Heacock-Kang, Y. et al. Spatial transcriptomes within the Pseudomonas aeruginosa biofilm architecture. Mol. Microbiol. 106, 976–985 (2017).
Williamson, K. S. et al. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J. Bacteriol. 194, 2062–2073 (2012).
Liao, J., Schurr, M. J. & Sauer, K. The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug-efflux pumps in Pseudomonas aeruginosa biofilms. J. Bacteriol. 195, 3352–3363 (2013).
Davies, D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2, 114 (2003).
de Carvalho, C. C. C. R. Marine biofilms: a successful microbial strategy with economic implications. Front. Mar. Sci. 5, 126 (2018).
Fitridge, I., Dempster, T., Guenther, J. & de Nys, R. The impact and control of biofouling in marine aquaculture: a review. Biofouling 28, 649–669 (2012).
Hauser, A. R., Jain, M., Bar-Meir, M. & McColley, S. A. Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin. Microbiol. Rev. 24, 29–70 (2011).
Kerr, K. G. & Snelling, A. M. Pseudomonas aeruginosa: a formidable and ever-present adversary. J. Hosp. Infect. 73, 338–344 (2009).
Lyczak, J. B., Cannon, C. L. & Pier, G. B. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15, 194–222 (2002).
Pirnay, J. P. et al. Pseudomonas aeruginosa population structure revisited. PLoS ONE 4, e7740 (2009).
Rosenberg, A. L. et al. The importance of bacterial sepsis in intensive care unit patients with acquired immunodeficiency syndrome: implications for future care in the age of increasing antiretroviral resistance. Crit. Care Med. 29, 548–556 (2001).
Rabello, L. S. et al. Clinical outcomes and microbiological characteristics of severe pneumonia in cancer patients: a prospective cohort study. PLoS ONE 10, e0120544 (2015).
Hassett, D. J., Borchers, M. T. & Panos, R. J. Chronic obstructive pulmonary disease (COPD): evaluation from clinical, immunological and bacterial pathogenesis perspectives. J. Microbiol. 52, 211–226 (2014).
Omar, A., Wright, J. B., Schultz, G., Burrell, R. & Nadworny, P. Microbial biofilms and chronic wounds. Microorganisms 5, 9 (2017).
van Loosdrecht, M. C. M., Picioreanu, C. & Heijnen, J. J. A more unifying hypothesis for the structure of microbial biofilms. FEMS Microb. Ecol. 24, 181–183 (1997).
Breyers, J. D. in Physiology Models in Microbiology Vol. 2 (eds Bazin, M. J. & Prosser, J. I.) 109–144 (CRC, 1988).
Sauer, K., Camper, A. K., Ehrlich, G. D., Costerton, J. W. & Davies, D. G. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184, 1140–1154 (2002).
Petrova, O. E. & Sauer, K. Escaping the biofilm in more than one way: desorption, detachment or dispersion. Curr. Opin. Microbiol. 30, 67–78 (2016).
Davies, D. G. in Biofilm Highlights (eds Flemming, H.-C., Wingender, J. & Szewzyk, U.) 1–28 (Springer, 2011).
Purevdorj-Gage, B., Costerton, W. J. & Stoodley, P. Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology 151, 1569–1576 (2005).
Otto, M. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 322, 207–228 (2008).
Kostakioti, M., Hadjifrangiskou, M. & Hultgren, S. J. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 3, a010306–a010306 (2013).
Webb, J. S. in The Biofilm Mode of Life: Mechanisms and Adaptations (eds Kjelleberg, S. & and Givskov, M.) 165–174 (Horizon Bioscience, 2007).
Bjarnsholt, T. et al. The in vivo biofilm. Trends Microbiol. 21, 466–474 (2013).
Serra, D. O. & Hengge, R. Stress responses go three dimensional–the spatial order of physiological differentiation in bacterial macrocolony biofilms. Environ. Microbiol. 16, 1455–1471 (2014).
Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R. & Lappin-Scott, H. M. Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745 (1995).
Sternberg, C. et al. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65, 4108–4117 (1999).
Anwar, H., Strap, J. L., Chen, K. & Costerton, J. W. Dynamic interactions of biofilms of mucoid Pseudomonas aeruginosa with tobramycin and piperacillin. Antimicrob. Agents Chemother. 36, 1208–1214 (1992).
Anderl, J. N., Zahller, J., Roe, F. & Stewart, P. S. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 47, 1251–1256 (2003).
Fux, C. A., Wilson, S. & Stoodley, P. Detachment characteristics and oxacillin resistance of Staphyloccocus aureus biofilm emboli in an in vitro catheter infection model. J. Bacteriol. 186, 4486–4491 (2004).
Nguyen, D. et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986 (2011).
Spoering, A. L. & Lewis, K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183, 6746–6751 (2001).
Keren, I., Kaldalu, N., Spoering, A., Wang, Y. & Lewis, K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230, 13–18 (2004).
Brooun, A., Liu, S. & Lewis, K. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 44, 640–646 (2000).
Keren, I., Shah, D., Spoering, A., Kaldalu, N. & Lewis, K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186, 8172–8180 (2004).
Shah, D. et al. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6, 53 (2006).
Campanac, C., Pineau, L., Payard, A., Baziard-Mouysset, G. & Roques, C. Interactions between biocide cationic agents and bacterial biofilms. Antimicrob. Agents Chemother. 46, 1469–1474 (2002).
Picioreanu, C., van Loosdrecht, M. C. M. & Heijnen, J. J. Two-dimensional model of biofilm detachment caused by internal stress from liquid flow. Biotechnol. Bioeng. 72, 205–218 (2001).
Thormann, K. M. et al. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 188, 2681–2691 (2006).
Anderl, J. N., Franklin, M. J. & Stewart, P. S. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 44, 1818–1824 (2000).
Lewis, K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45, 999–1007 (2001).
Stewart, P. S. & Costerton, J. W. Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138 (2001).
Stewart, P. S. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob. Agents Chemother. 40, 2517–2522 (1996).
Drenkard, E. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 5, 1213–1219 (2003).
Reichhardt, C. & Parsek, M. R. Confocal laser scanning microscopy for analysis of Pseudomonas aeruginosa biofilm architecture and matrix localization. Front. Microbiol. 10, 677 (2019).
Doroshenko, N. et al. Extracellular DNA impedes the transport of vancomycin in Staphylococcus epidermidis biofilms preexposed to subinhibitory concentrations of vancomycin. Antimicrob. Agents Chemother. 58, 7273–7282 (2014).
Tseng, B. S. et al. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ. Microbiol. 15, 2865–2878 (2013).
Chua, S. L. et al. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyle. Nat. Commun. 5, 4462 (2014). This study shows that P. aeruginosa dispersed cells displayed an altered transcriptome and physiology, and increased virulence, in comparison with planktonic and biofilm cells, but were highly sensitive to iron stress.
Chambers, J. R., Cherny, K. E. & Sauer, K. Susceptibility of Pseudomonas aeruginosa dispersed cells to antimicrobial agents is dependent on the dispersion cue and class of the antimicrobial agent used. Antimicrob. Agents Chemother. 61, e00846-17 (2017).
Davies, D. G. & Marques, C. N. H. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 191, 1393–1403 (2009).
Costerton, J. Introduction to biofilm. Int. J. Antimicrob. Agents 11, 217–221 (1999).
Stewart, P. S. & Franklin, M. J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6, 199 (2008).
Haussler, S. & Fuqua, C. Biofilms 2012: new discoveries and significant wrinkles in a dynamic field. J. Bacteriol. 195, 2947–2958 (2013).
Marques, C. N., Davies, D. G. & Sauer, K. Control of biofilms with the fatty acid signaling molecule cis-2-decenoic acid. Pharmaceuticals 8, 816–835 (2015). This study shows that the fatty acid molecule cis-DA functions as a native dispersion inducer that is capable of inducing dispersion of biofilms formed by Gram-negative, Gram-positive and fungal species.
Dow, J. M. et al. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl Acad. Sci. USA 100, 10995–11000 (2003).
Dean, S. N., Chung, M.-C. & van Hoek, M. L. Burkholderia diffusible signal factor signals to Francisella novicida to disperse biofilm and increase siderophore production. Appl. Environ. Microbiol. 81, 7057–7066 (2015).
Gjermansen, M., Ragas, P., Sternberg, C., Molin, S. & Tolker-Nielsen, T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 7, 894–904 (2005).
Delille, A., Quiles, F. & Humbert, F. In situ monitoring of the nascent Pseudomonas fluorescens biofilm response to variations in the dissolved organic carbon level in low-nutrient water by attenuated total reflectance-Fourier transform infrared spectroscopy. Appl. Environ. Microbiol. 73, 5782–5788 (2007).
Schleheck, D. et al. Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PloS ONE 4, e5513 (2009).
Hunt, S. M., Werner, E. M., Huang, B., Hamilton, M. A. & Stewart, P. S. Hypothesis for the role of nutrient starvation in biofilm detachment. Appl. Environ. Microbiol. 70, 7418–7425 (2004).
Stoodley, P., deBeer, D. & Lewandowski, Z. Liquid flow in biofilm systems. Appl. Environ. Microbiol. 60, 2711–2716 (1994).
Rasmussen, K. & Lewandowski, Z. Microelectrode measurements of local mass transport rates in heterogeneous biofilms. Biotechnol. Bioeng. 59, 302–309 (1998).
Petrova, O. E., Schurr, J. R., Schurr, M. J. & Sauer, K. Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol. Microbiol. 86, 819–835 (2012).
Goodwine, J. et al. Pyruvate-depleting conditions induce biofilm dispersion and enhance the efficacy of antibiotics in killing biofilms in vitro and in vivo. Sci. Rep. 9, 3763 (2019).
Eschbach, M. et al. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 186, 4596–4604 (2004).
Schreiber, K. et al. Anaerobic survival of Pseudomonas aeruginosa by pyruvate fermentation requires an Usp-type stress protein. J. Bacteriol. 188, 659–668 (2006).
Price-Whelan, A., Dietrich, L. E. P. & Newman, D. K. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J. Bacteriol. 189, 6372–6381 (2007).
Leibig, M. et al. Pyruvate formate lyase acts as a formate supplier for metabolic processes during anaerobiosis in Staphylococcus aureus. J. Bacteriol. 193, 952–962 (2011).
Yang, L. et al. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 153, 1318–1328 (2007).
Lanter, B. B., Sauer, K. & Davies, D. G. Bacteria present in carotid arterial plaques are found as biofilm deposits which may contribute to enhanced risk of plaque rupture. mBio 5, 01206–01214 (2014).
Thomas, V. C. & Hancock, L. E. Suicide and fratricide in bacterial biofilms. Int. J. Artif. Organs 32, 537–544 (2009).
Hentzer, M., Eberl, L. & Givskov, M. Transcriptome analysis of Pseudomonas aeruginosa biofilm development: anaerobic respiration and iron limitation. Biofilms 2, 37–61 (2005).
Sriramulu, D. D., Lünsdorf, H., Lam, J. S. & Römling, U. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 54, 667–676 (2005).
Webb, J. S. et al. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185, 4585–4592 (2003).
Rice, S. A. et al. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 3, 271–282 (2009).
Sutherland, I. W., Hughes, K. A., Skillman, L. C. & Tait, K. The interaction of phage and biofilms. FEMS Microbiol. Lett. 232, 1–6 (2004).
Secor, P. R. et al. Biofilm assembly becomes crystal clear–filamentous bacteriophage organize the Pseudomonas aeruginosa biofilm matrix into a liquid crystal. Microb. Cell 3, 49 (2016).
Sauer, K. et al. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 186, 7312–7326 (2004). This seminal study shows that dispersal of P. aeruginosa from biofilms can be induced by the addition of simple carbon sources, and that dispersed cells adopt a specific phenotype.
Barraud, N. et al. Nitric oxide-mediated dispersal in single- and multi-species biofilms of clinically and industrially relevant microorganisms. Microb. Biotechnol. 2, 370–378 (2009).
Purcell, E. B. & Tamayo, R. Cyclic diguanylate signaling in Gram-positive bacteria. FEMS Microbiol. Rev. 40, 753–773 (2016).
Hengge, R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273 (2009).
Rybtke, M. T. et al. Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 78, 5060–5069 (2012).
Nair, H. A., Periasamy, S., Yang, L., Kjelleberg, S. & Rice, S. A. Real time, spatial, and temporal mapping of the distribution of c-di-GMP during biofilm development. J. Biol. Chem. 292, 477–487 (2017).
Basu Roy, A., Petrova, O. E. & Sauer, K. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J. Bacteriol. 194, 2904–2915 (2012).
Gjermansen, M., Nilsson, M., Yang, L. & Tolker-Nielsen, T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol. Microbiol. 75, 815–826 (2010).
Morgan, R., Kohn, S., Hwang, S.-H., Hassett, D. J. & Sauer, K. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J. Bacteriol. 188, 7335–7343 (2006).
Li, Y. et al. BdlA, DipA and induced dispersion contribute to acute virulence and chronic persistence of Pseudomonas aeruginosa. PLoS Pathog. 10, e1004168 (2014).
Li, Y., Heine, S., Entian, M., Sauer, K. & Frankenberg-Dinkel, N. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by a MHYT-domain coupled phosphodiesterase. J. Bacteriol. 195, 3531–3542 (2013).
Barraud, N. et al. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J. Bacteriol. 191, 7333–7342 (2009).
Christensen, L. D. et al. Clearance of Pseudomonas aeruginosa foreign-body biofilm infections through reduction of the cyclic di-GMP level in the bacteria. Infect. Immun. 81, 2705–2713 (2013).
Baraquet, C., Harwood, C. S. & FleQ, D. N. A. Binding consensus sequence revealed by studies of FleQ-dependent regulation of biofilm gene expression in Pseudomonas aeruginosa. J. Bacteriol. 198, 178–186 (2016).
Häußler, S., Tümmler, B., Weißbrodt, H., Rohde, M. & Steinmetz, I. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin. Infect. Dis. 29, 621–625 (1999).
Häußler, S. et al. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J. Med. Microbiol. 52, 295–301 (2003).
Proctor, R. A. et al. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4, 295–305 (2006).
Proctor, R. A., van Langevelde, P., Kristjansson, M., Maslow, J. N. & Arbeit, R. D. Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis. 20, 95–102 (1995).
Drenkard, E. & Ausubel, F. M. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416, 740–743 (2002).
Chua, S. L. et al. Bis-(3′-5′)-cyclic dimeric GMP regulates antimicrobial peptide resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57, 2066–2075 (2013).
Basu Roy, A. & Sauer, K. Diguanylate cyclase NicD-based signalling mechanism of nutrient-induced dispersion by Pseudomonas aeruginosa. Mol. Microbiol. 94, 771–793 (2014).
Cutruzzolà, F. & Frankenberg-Dinkel, N. Origin and impact of nitric oxide in Pseudomonas aeruginosa Biofilms. J. Bacteriol. 198, 55–65 (2016).
Zhou, L., Zhang, L.-H., Cámara, M. & He, Y.-W. The DSF family of quorum sensing signals: diversity, biosynthesis, and turnover. Trends Microbiol. 25, 293–303 (2017).
Andrade, M. O. et al. The HD-GYP domain of RpfG mediates a direct linkage between the Rpf quorum-sensing pathway and a subset of diguanylate cyclase proteins in the phytopathogen Xanthomonas axonopodis pv citri. Mol. Microbiol. 62, 537–551 (2006).
Deng, Y. et al. Cis-2-dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proc. Natl Acad. Sci. USA 109, 15479–15484 (2012).
Abee, T., Kovács, Á. T., Kuipers, O. P. & Van der Veen, S. Biofilm formation and dispersal in Gram-positive bacteria. Curr. Opin. Biotechnol. 22, 172–179 (2011).
Chou, S.-H. & Galperin, M. Y. Cyclic di-GMP in Streptomycetes: a new conformation, new binding mode, new receptor, and a new mechanism to control cell development. Mol. Cell 77, 443–445 (2020).
Flemming, H. C. & Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 (2010).
Cherny, K. E. & Sauer, K. Pseudomonas aeruginosa requires the DNA-specific endonuclease EndA to degrade eDNA to disperse from the biofilm. J. Bacteriol. 201, e00059-19 (2019).
Cherny, K. E. & Sauer, K. Untethering and degradation of the polysaccharide matrix are essential steps in the dispersion response of Pseudomonas aeruginosa biofilms. J. Bacteriol. 202, e00575-19 (2020). This study suggests matrix degradation to induce dispersion is a multistep process.
Rybtke, M. et al. The LapG protein plays a role in Pseudomonas aeruginosa biofilm formation by controlling the presence of the CdrA adhesin on the cell surface. Microbiologyopen 4, 917–930 (2015).
Borlee, B. R. et al. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 75, 827–842 (2010).
Monds, R. D., Newell, P. D., Gross, R. H. & O’Toole, G. A. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 63, 656–679 (2007).
Reichhardt, C., Wong, C., Passos da Silva, D., Wozniak, D. J. & Parsek, M. R. CdrA interactions within the Pseudomonas aeruginosa biofilm matrix safeguard it from proteolysis and promote cellular packing. mBio 9, e01376-18 (2018).
Devaraj, A. et al. The extracellular DNA lattice of bacterial biofilms is structurally related to Holliday junction recombination intermediates. Proc. Natl Acad. Sci. USA 116, 25068–25077 (2019).
Steinberger, R. E. & Holden, P. A. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl. Environ. Microbiol. 71, 5404–5410 (2005).
Whitchurch, C. B., Tolker-Nielsen, T., Ragas, P. C. & Mattick, J. S. Extracellular DNA required for bacterial biofilm formation. Science 295, 1487 (2002).
Nijland, R., Hall, M. J. & Burgess, J. G. Dispersal of biofilms by secreted, matrix degrading, bacterial DNase. PLoS ONE 5, e15668 (2010).
Boles, B. R. & Horswill, A. R. Staphylococcal biofilm disassembly. Trends Microbiol. 19, 449–455 (2011).
Gödeke, J., Heun, M., Bubendorfer, S., Paul, K. & Thormann, K. M. Roles of two Shewanella oneidensis MR-1 extracellular endonucleases. Appl. Environ. Microbiol. 77, 5342–5351 (2011).
McDougald, D., Rice, S. A., Barraud, N., Steinberg, P. D. & Kjelleberg, S. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10, 39–50 (2012).
Pestrak, M. J. et al. Treatment with the Pseudomonas aeruginosa glycoside hydrolase PslG combats wound infection by improving antibiotic efficacy and host innate immune activity. Antimicrob. Agents Chemother. 63, e00234-19 (2019). This study demonstrates that biofilm disassembly, induced by the exogenous addition of a matrix-degrading protein, PslG, has a positive outcome in the treatment of biofilm-related infections.
Baker, P. et al. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci. Adv. 2, e1501632 (2016).
Yu, S. et al. PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res. 25, 1352–1367 (2015).
Kaplan, J. B., Ragunath, C., Ramasubbu, N. & Fine, D. H. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. J. Bacteriol. 185, 4693–4698 (2003).
Kaplan, J. B., Ragunath, C., Velliyagounder, K., Fine, D. H. & Ramasubbu, N. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 48, 2633–2636 (2004). This study features the characterization of one of the first biofilm-releasing enzymes produced by the resident biofilm bacteria to induce biofilm disassembly.
Chaignon, P. et al. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl. Microbiol. Biotechnol. 75, 125–132 (2007).
Boles, B. R. & Horswill, A. R. agr mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4, e1000052 (2008).
Chua, S. L. et al. In vitro and in vivo generation and characterization of Pseudomonas aeruginosa biofilm-dispersed cells via c-di-GMP manipulation. Nat. Protoc. 10, 1165–1180 (2015).
Jones, C. J. et al. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 10, e1003984 (2014).
Xu, B. et al. The Pseudomonas aeruginosa AmrZ C-terminal domain mediates tetramerization and is required for its activator and repressor functions. Environ. Microbiol. Rep. 8, 85–90 (2016).
Jones, C. J. AmrZ Is A Central Regulator of Biofilm Formation in Pseudomonas aeruginosa. Thesis, Ohio State Univ. (2013).
Woo, J. K., Webb, J. S., Kirov, S. M., Kjelleberg, S. & Rice, S. A. Biofilm dispersal cells of a cystic fibrosis Pseudomonas aeruginosa isolate exhibit variability in functional traits likely to contribute to persistent infection. FEMS Immunol. Med. Microbiol. 66, 251–264 (2012). This study provides evidence that the development of a chronic infection phenotype can be reversed to recover acute infection isolates and that growth within a biofilm facilitates diversification of P. aeruginosa, which is important for ecological adaptation.
Hall-Stoodley, L. & Stoodley, P. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 13, 7–10 (2005). This review summarizes our current knowledge of the role of biofilm formation in facilitating the transmission of pathogens by providing a stable protective environment and acting as a nidus for the dissemination of large numbers of microorganisms.
Rollet, C., Gal, L. & Guzzo, J. Biofilm-detached cells, a transition from a sessile to a planktonic phenotype: a comparative study of adhesion and physiological characteristics in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 290, 135–142 (2009). This study provides a summary of the physiological characteristics of cells detached from a P. aeruginosa biofilm relative to their sessile and planktonic counterparts.
Uppuluri, P. et al. Candida albicans dispersed cells are developmentally distinct from biofilm and planktonic cells. mBio 9, e01338-18. (2018). This seminal study shows that dispersal of P. aeruginosa from biofilms can be induced by the addition of simple carbon sources and that dispersed cells adopt a specific phenotype.
Liu, J., Ling, J. Q., Zhang, K. & Wu, C. D. Physiological properties of Streptococcus mutans UA159 biofilm-detached cells. FEMS Microbiol. Lett. 340, 11–18 (2013).
Guilhen, C. et al. Transcriptional profiling of Klebsiella pneumoniae defines signatures for planktonic, sessile and biofilm-dispersed cells. BMC Genomics 17, 237 (2016).
Vaysse, P. J., Sivadon, P., Goulas, P. & Grimaud, R. Cells dispersed from Marinobacter hydrocarbonoclasticus SP17 biofilm exhibit a specific protein profile associated with a higher ability to reinitiate biofilm development at the hexadecane-water interface. Env. Microbiol. 13, 737–746 (2011).
Fleming, D. & Rumbaugh, K. The consequences of biofilm dispersal on the host. Sci. Rep. 8, 10738 (2018). This study shows that P. aeruginosa biofilm cells dispersed from mouse chronic wound infections by glycoside hydrolases can cause fatal sepsis unless antibiotics are given in conjunction.
Fleming, D. & Rumbaugh, K. Approaches to dispersing medical biofilms. Microorganisms 5, 15 (2017).
Waryah, C. B. et al. In vitro antimicrobial efficacy of tobramycin against staphylococcus aureus biofilms in combination with or without DNase I and/or dispersin B: a preliminary investigation. Microb. Drug Resist. 23, 384–390 (2017).
Izano, E. A., Wang, H., Ragunath, C., Ramasubbu, N. & Kaplan, J. B. Detachment and killing of Aggregatibacter actinomycetemcomitans biofilms by dispersin B and SDS. J. Dent. Res. 86, 618–622 (2007).
Gawande, P. V., Leung, K. P. & Madhyastha, S. Antibiofilm and antimicrobial efficacy of DispersinB(R)-KSL-W peptide-based wound gel against chronic wound infection associated bacteria. Curr. Microbiol. 68, 635–641 (2014).
Itoh, Y., Wang, X., Hinnebusch, B. J., Preston, J. F. III & Romeo, T. Depolymerization of β-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187, 382–387 (2005). This study demonstrates for the first time that enzymatic hydrolysis of PGA disrupts biofilm formation by strains harbouring pgaABCD homologues, including Yersinia pestis and P. fluorescens.
Gawande, P. V. et al. Antibiofilm efficacy of DispersinB® wound spray used in combination with a silver wound dressing. Microbiol. Insights 7, 9–13 (2014).
Kaplan, J. B. et al. Extracellular polymeric substance (EPS)-degrading enzymes reduce staphylococcal surface attachment and biocide resistance on pig skin in vivo. PLoS ONE 13, e0205526 (2018).
Darouiche, R. O., Mansouri, M. D., Gawande, P. V. & Madhyastha, S. Antimicrobial and antibiofilm efficacy of triclosan and DispersinB combination. J. Antimicrob. Chemother. 64, 88–93 (2009).
Barraud, N., J Kelso, M., A Rice, S. & Kjelleberg, S. Nitric oxide: a key mediator of biofilm dispersal with applications in infectious diseases. Curr. Pharm. Des. 21, 31–42 (2015).
Sepehr, S., Rahmani-Badi, A., Babaie-Naiej, H. & Soudi, M. R. Unsaturated fatty acid, cis-2-decenoic acid, in combination with disinfectants or antibiotics removes pre-established biofilms formed by food-related bacteria. PLoS ONE 9, e101677 (2014).
Rahmani-Badi, A. et al. A combination of cis-2-decenoic acid and antibiotics eradicates pre-established catheter-associated biofilms. J. Med. Microbiol. 63, 1509–1516 (2014).
Rahmani-Badi, A., Sepehr, S. & Babaie-Naiej, H. A combination of cis-2-decenoic acid and chlorhexidine removes dental plaque. Arch. Oral. Biol. 60, 1655–1661 (2015).
Harris, M. A., Beenken, K. E., Smeltzer, M. S., Haggard, W. O. & Jennings, J. A. Phosphatidylcholine coatings deliver local antimicrobials and reduce infection in a murine model: a preliminary study. Clin. Orthop. Relat. Res. 475, 1847–1853 (2017).
Choi, M. et al. Chitosan-based nitric oxide-releasing dressing for anti-biofilm and in vivo healing activities in MRSA biofilm-infected wounds. Int. J. Biol. Macromol. 142, 680–692 (2019).
Jardeleza, C. et al. An in vivo safety and efficacy demonstration of a topical liposomal nitric oxide donor treatment for Staphylococcus aureus biofilm-associated rhinosinusitis. Transl. Res. 166, 683–692 (2015).
Wo, Y. et al. Reduction of thrombosis and bacterial infection via controlled nitric oxide (NO) release from S-nitroso-N-acetylpenicillamine (SNAP) impregnated CarboSil intravascular catheters. ACS Biomater. Sci. Eng. 3, 349–359 (2017).
Mihu, M. R. et al. Sustained nitric oxide-releasing nanoparticles interfere with methicillin-resistant Staphylococcus aureus adhesion and biofilm formation in a rat central venous catheter model. Antimicrob. Agents Chemother. 61, e02020-16 (2017).
Tao, F., Swarup, S. & Zhang, L. H. Quorum sensing modulation of a putative glycosyltransferase gene cluster essential for Xanthomonas campestris biofilm formation. Environ. Microbiol. 12, 3159–3170 (2010).
An, S., Wu, J. & Zhang, L. H. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl. Environ. Microbiol. 76, 8160–8173 (2010).
Petrova, O. E., Cherny, K. E. & Sauer, K. The diguanylate cyclase GcbA facilitates Pseudomonas aeruginosa biofilm dispersion by activating BdlA. J. Bacteriol. 197.1, 174–187 (2015).
Petrova, O. E. & Sauer, K. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc. Natl Acad. Sci. USA 109, 16690–16695 (2012). This study demonstrates the post-translational modification of BdlA in response to dispersion cue sensing.
Marks, L. R., Davidson, B. A., Knight, P. R. & Hakansson, A. P. Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. mBio 4, e00438-13 (2013).
Rice, S. et al. Biofilm formation and sloughing in Serratia marcescens are controlled by quorum sensing and nutrient cues. J. Bacteriol. 187, 3477–3485 (2005).
Barraud, N. et al. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 188, 7344–7353 (2006). This seminal study shows that dispersal of P. aeruginosa from biofilms can be induced by the addition of NO and that combined exposure to both NO and antimicrobial agents greatly enhanced the efficacy of antimicrobial compounds in the removal of established P. aeruginosa biofilms.
Schmidt, I., Steenbakkers, P. J., op den Camp, H. J., Schmidt, K. & Jetten, M. S. Physiologic and proteomic evidence for a role of nitric oxide in biofilm formation by Nitrosomonas europaea and other ammonia oxidizers. J. Bacteriol. 186, 2781–2788 (2004).
Musk, D. J., Banko, D. A. & Hergenrother, P. J. Iron salts perturb biofilm formation and disrupt existing biofilms of Pseudomonas aeruginosa. Chem. Biol. 12, 789–796 (2005).
Mikkelsen, H., Hui, K., Barraud, N. & Filloux, A. The pathogenicity island encoded PvrSR/RcsCB regulatory network controls biofilm formation and dispersal in Pseudomonas aeruginosa PA14. Mol. Microbiol. 89, 450–463 (2013).
Kragh, K. N. et al. Role of multicellular aggregates in biofilm formation. mBio 7, e00237-16 (2016).
Naumoff, D. Hierarchical classification of glycoside hydrolases. Biochemistry 76, 622–635 (2011).
Fleming, D., Chahin, L. & Rumbaugh, K. Glycoside hydrolases degrade polymicrobial bacterial biofilms in wounds. Antimicrob. Agents Chemother. 61, e01998-16 (2017).
Mulcahy, H., Charron-Mazenod, L. & Lewenza, S. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ. Microbiol. 12, 1621–1629 (2010).
Klausen, M., Aaes-Jørgensen, A., Molin, S. & Tolker-Nielsen, T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50, 61–68 (2003).
Acknowledgements
This work was supported by grants from the US National Institutes of Health (R21 AI137462-01A1 to K.P.R. and 2R01 AI080710 to K.S.) and the Ted Nash Long Life Foundation (to K.P.R.).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Microbiology thanks S. Kjelleberg, P. Stoodley and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Centers for Disease Control and Prevention programme announcement PA-03-047: https://grants.nih.gov/grants/guide/pa-files/pa-03-047.html
Centers for Disease Control and Prevention programme announcement PA-06-537: https://grants.nih.gov/grants/guide/pa-files/PA-06-537.html
Glossary
- Dispersion
-
Generally characterized as the terminal stage of biofilm development, dispersion is an active regulated event during which cells actively escape from the biofilm, leaving behind eroded biofilms and biofilms having central voids. Often referred to as ‘seeding dispersal’ or ‘dispersal’, as dispersion is assumed to lead to the translocation of bacteria to new sites for colonization.
- Detachment
-
Characterized as the passive release or loss of biofilm cells or biofilm particles due to mechanical, physical or frictional forces. Another term used to refer to detachment is ‘dissolution’.
- Shear
-
Strain in the structure of a substance produced by pressure.
- Abrasion
-
Loss of biofilm cells by collisions with particles from the environment; a form of detachment.
- Dissemination
-
The translocation of dispersed biofilm cells to new sites.
- RpoS
-
Also referred to as ‘σ38’, a primary regulator of stationary phase genes and a central regulator of the general stress response.
- Native dispersion
-
Dispersion in response to self-synthesized signalling molecules or cues that are likely to be the result of steep gradients within the biofilm.
- Diffusive and advective transport
-
Processes that move nutrients, waste, gases or other compounds through the biofilm and the surrounding environment. ‘Advection’ refers to transport of compounds with fluid flow, whereas diffusion eliminates sharp discontinuities of compounds through the action of random motions.
- Disassembly
-
The egress from biofilms and/or the disintegration of the biofilm structure in response to exogenously added matrix-degrading enzymes. The term is frequently used when it is unclear whether the released cell subpopulation retains the biofilm phenotype or adopts the dispersal phenotype.
- Quorum sensing
-
Also referred to as ‘cell-to-cell signalling’, it refers to the regulation of gene expression in response to the production and release of chemical signal molecules called ‘autoinducers’ that increase in concentration as a function of cell density.
Rights and permissions
About this article
Cite this article
Rumbaugh, K.P., Sauer, K. Biofilm dispersion. Nat Rev Microbiol 18, 571–586 (2020). https://doi.org/10.1038/s41579-020-0385-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-020-0385-0
This article is cited by
-
ZnO-incorporated chitin hydrogels for infected wound therapy
Cellulose (2024)
-
Biofilm-mediated infections by multidrug-resistant microbes: a comprehensive exploration and forward perspectives
Archives of Microbiology (2024)
-
Anti-Biofilm Effects of Melittin: Lessons Learned and the Path Ahead
International Journal of Peptide Research and Therapeutics (2024)
-
A Review of Challenges and Solutions of Biofilm Formation of Escherichia coli: Conventional and Novel Methods of Prevention and Control
Food and Bioprocess Technology (2024)
-
Characterization and environmental applications of soil biofilms: a review
Environmental Chemistry Letters (2024)