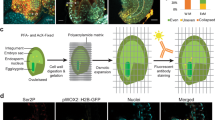
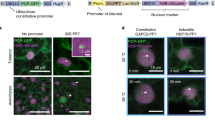
Abstract
Assessing cell proliferation dynamics is crucial to understand the spatiotemporal control of organogenesis. Here we have generated a versatile fluorescent sensor, PlaCCI (plant cell cycle indicator) on the basis of the expression of CDT1a-CFP, H3.1-mCherry and CYCB1;1-YFP, that identifies cell cycle phases in Arabidopsis thaliana. This tool works in a variety of organs, and all markers and the antibiotic resistance are expressed from a single cassette, facilitating the selection in mutant backgrounds. We also show the robustness of PlaCCI line in live-imaging experiments to follow and quantify cell cycle phase progression.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
Source data are provided with this paper.
References
Sakaue-Sawano, A. et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2007).
Sugiyama, M. et al. Illuminating cell-cycle progression in the developing zebrafish embryo. Proc. Natl Acad. Sci. USA 106, 20812–20817 (2009).
Zielke, N. et al. Fly-FUCCI: a versatile tool for studying cell proliferation in complex tissues. Cell Rep. 7, 588–598 (2014).
Bajar, B. T. et al. Fluorescent indicators for simultaneous reporting of all four cell cycle phases. Nat. Methods 13, 993–996 (2016).
Caro, E., Castellano, M. M. & Gutierrez, C. A chromatin link that couples cell division to root epidermis patterning in Arabidopsis. Nature 447, 213–217 (2007).
Caro, E. & Gutierrez, C. A green GEM: intriguing analogies with animal geminin. Trends Cell Biol. 17, 580–585 (2007).
Colon-Carmona, A., You, R., Haimovitch-Gal, T. & Doerner, P. Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin–GUS fusion protein. Plant J. 20, 503–508 (1999).
Adachi, S. et al. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl Acad. Sci. USA 108, 10004–10009 (2011).
Yin, K. et al. A dual-color marker system for in vivo visualization of cell cycle progression in Arabidopsis. Plant J. 80, 541–552 (2014).
Jones, A. R. et al. Cell-size dependent progression of the cell cycle creates homeostasis and flexibility of plant cell size. Nat. Commun. 8, 15060 (2017).
Desvoyes, B. et al. FBL17 targets CDT1a for degradation in early S-phase to prevent Arabidopsis genome instability. Preprint at https://www.biorxiv.org/content/10.1101/774109v1 (2019).
Castellano, M. M., Boniotti, M. B., Caro, E., Schnittger, A. & Gutierrez, C. DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner. Plant Cell 16, 2380–2393 (2004).
Otero, S., Desvoyes, B., Peiro, R. & Gutierrez, C. Histone H3 dynamics uncovers domains with distinct proliferation potential in the Arabidopsis root. Plant Cell 28, 1361–1371 (2016).
Ubeda-Tomas, S. et al. Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 19, 1194–1199 (2009).
Sarrion-Perdigones, A. et al. GoldenBraid: an iterative cloning system for standardized assembly of reusable genetic modules. PLoS ONE 6, e21622 (2011).
Montané, M.-H. & Menand, B. ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J. Exp. Bot. 64, 4361–4374 (2013).
Barbet, N. C. et al. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7, 25–42 (1996).
Park, J. A. et al. Retinoblastoma protein regulates cell proliferation, differentiation, and endoreduplication in plants. Plant J. 42, 153–163 (2005).
Desvoyes, B., Ramirez-Parra, E., Xie, Q., Chua, N. H. & Gutierrez, C. Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol. 140, 67–80 (2006).
Harashima, H. & Sugimoto, K. Integration of developmental and environmental signals into cell proliferation and differentiation through RETINOBLASTOMA-RELATED 1. Curr. Opin. Plant Biol. 29, 95–103 (2016).
Wildwater, M. et al. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123, 1337–1349 (2005).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Rueden, C. T. et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinforma. 18, 529 (2017).
Acknowledgements
We thank the Confocal Microscopy and the Cytometry services of CBMSO for support, E. Martinez-Salas for comments, S. Herrero-Desvoyes for help with cell size measurements, E. Caro for the terminator modules for GoldenBraid constructs, V. Mora-Gil for technical assistance and members of the laboratory for continuous feedback. Research in our laboratory is supported by grant nos. BIO2017-92329-EXP and RTI2018-094793-B-I00 from MICIU, and ERC-2018-AdG_833617 from European Union, and by institutional grants from Banco de Santander and Fundación Ramon Areces to the Centro de Biología Molecular Severo Ochoa.
Author information
Authors and Affiliations
Contributions
B.D. and C.G designed this work. A.A.-E. and M.D.B. participated in the initial cloning steps. B.D. performed all the experiments. B.D. and C.G. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Lieven De Veylder and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6 and Table 1.
Supplementary Video 1
Time lapse (7 h) of the root meristem of a PlaCCI line showing the switching on/off of various cell cycle markers.
Supplementary Data 1
Source data for Supplementary Fig. 4.
Supplementary Data 2
Source data for Supplementary Fig. 5a.
Supplementary Data 3
Source data for Supplementary Fig. 5b.
Source data
Source Data Fig. 1i
Raw data.
Source Data Fig. 1j
Raw data.
Source Data Fig. 1k
Raw data.
Source Data Fig. 1l
Raw data.
Source Data Fig. 2f
Raw data.
Source Data Fig. 2g
Raw data.
Rights and permissions
About this article
Cite this article
Desvoyes, B., Arana-Echarri, A., Barea, M.D. et al. A comprehensive fluorescent sensor for spatiotemporal cell cycle analysis in Arabidopsis. Nat. Plants 6, 1330–1334 (2020). https://doi.org/10.1038/s41477-020-00770-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-00770-4
This article is cited by
-
SHR and SCR coordinate root patterning and growth early in the cell cycle
Nature (2024)
-
Seeing is understanding
Nature Plants (2023)
-
Brassinazole represses tomato hypocotyl elongation via inhibition of cell division
Plant Growth Regulation (2022)