Abstract
Increasing nitrogen use efficiency (NUE) is critical to improve crop yield, reduce N fertilizer demand and alleviate environmental pollution. N remobilization is a key component of NUE. The nitrate transporter NRT1.7 is responsible for loading excess nitrate stored in source leaves into phloem and facilitates nitrate allocation to sink leaves. Under N starvation, the nrt1.7 mutant exhibits growth retardation, indicating that NRT1.7-mediated source-to-sink remobilization of stored nitrate is important for sustaining growth in plants. To energize NRT1.7-mediated nitrate recycling, we introduced a hyperactive chimeric nitrate transporter NC4N driven by the NRT1.7 promoter into the nrt1.7 mutant. NRT1.7p::NC4N::3′ transgenic plants accumulated more nitrate in younger leaves, and 15NO3− tracing analysis revealed that more 15N was remobilized into sink tissues. Consistently, transgenic Arabidopsis, tobacco and rice plants showed improved growth or yield. Our study suggests that enhancing source-to-sink nitrate remobilization represents a new strategy for enhancing NUE and crop production.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the article and Extended Data Figs. 1–9, or from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Schroeder, J. I. et al. Using membrane transporters to improve crops for sustainable food production. Nature 497, 60–66 (2013).
Hristov, A. N. et al. Invited review: nitrogen in ruminant nutrition: a review of measurement techniques. J. Dairy Sci. 102, 5811–5852 (2019).
Mulvaney, R. L., Khan, S. A. & Ellsworth, T. R. Synthetic nitrogen fertilizers deplete soil nitrogen: a global dilemma for sustainable cereal production. J. Environ. Qual. 38, 2295–2314 (2009).
McAllister, C. H., Beatty, P. H. & Good, A. G. Engineering nitrogen use efficient crop plants: the current status. Plant Biotechnol. J. 10, 1011–1025 (2012).
Godfray, H. C. et al. Food security: the challenge of feeding 9 billion people. Science 327, 812–818 (2010).
Masclaux-Daubresse, C. et al. Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. 105, 1141–1157 (2010).
Good, A. G., Shrawat, A. K. & Muench, D. G. Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci. 9, 597–605 (2004).
Xu, G., Fan, X. & Miller, A. J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63, 153–182 (2012).
Have, M., Marmagne, A., Chardon, F. & Masclaux-Daubresse, C. Nitrogen remobilization during leaf senescence: lessons from Arabidopsis to crops. J. Exp. Bot. 68, 2513–2529 (2017).
Hirel, B., Le Gouis, J., Ney, B. & Gallais, A. The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. J. Exp. Bot. 58, 2369–2387 (2007).
Tegeder, M. & Masclaux-Daubresse, C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 217, 35–53 (2018).
Tegeder, M. Transporters involved in source to sink partitioning of amino acids and ureides: opportunities for crop improvement. J. Exp. Bot. 65, 1865–1878 (2014).
Avin-Wittenberg, T. et al. Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J. Exp. Bot. 69, 1335–1353 (2018).
Wang, Y. Y., Hsu, P. K. & Tsay, Y. F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 17, 458–467 (2012).
Hsu, P. K. & Tsay, Y. F. Two phloem nitrate transporters, NRT1.11 and NRT1.12, are important for redistributing xylem-borne nitrate to enhance plant growth. Plant Physiol. 163, 844–856 (2013).
Geelen, D. et al. Disruption of putative anion channel gene AtCLC-a in Arabidopsis suggests a role in the regulation of nitrate content. Plant J. 21, 259–267 (2000).
De Angeli, A. et al. The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 442, 939–942 (2006).
Von der Fecht-Bartenbach, J., Bogner, M., Dynowski, M. & Ludewig, U. CLC-b-mediated NO-3/H+ exchange across the tonoplast of Arabidopsis vacuoles. Plant Cell Physiol. 51, 960–968 (2010).
Fan, S. C., Lin, C. S., Hsu, P. K., Lin, S. H. & Tsay, Y. F. The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21, 2750–2761 (2009).
Wang, Y. Y., Cheng, Y. H., Chen, K. E. & Tsay, Y. F. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 69, 85–122 (2018).
Liu, K. H., Huang, C. Y. & Tsay, Y. F. CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11, 865–874 (1999).
Liu, K. H. & Tsay, Y. F. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22, 1005–1013 (2003).
Ho, C. H., Lin, S. H., Hu, H. C. & Tsay, Y. F. CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194 (2009).
Huang, N. C., Liu, K. H., Lo, H. J. & Tsay, Y. F. Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11, 1381–1392 (1999).
Kanno, Y. et al. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl Acad. Sci. USA 109, 9653–9658 (2012).
Liang, K. et al. Nitrogen losses and greenhouse gas emissions under different N and water management in a subtropical double-season rice cropping system. Sci. Total Environ. 609, 46–57 (2017).
Wada, S. et al. Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol. 168, 60–73 (2015).
Li, H., Hu, B. & Chu, C. Nitrogen use efficiency in crops: lessons from Arabidopsis and rice. J. Exp. Bot. 68, 2477–2488 (2017).
Richard-Molard, C. et al. Plant response to nitrate starvation is determined by N storage capacity matched by nitrate uptake capacity in two Arabidopsis genotypes. J. Exp. Bot. 59, 779–791 (2008).
Crawford, N. M. & Glass, A. D. M. Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3, 389–395 (1998).
Kirk, G. J. & Kronzucker, H. J. The potential for nitrification and nitrate uptake in the rhizosphere of wetland plants: a modelling study. Ann. Bot. 96, 639–646 (2005).
Wong, M. T. F. & Nortcliff, S. Seasonal fluctuations of native available N and soil management implications. Fertil. Res. 42, 13–26 (1995).
Wu, W. & Ma, B. Integrated nutrient management (INM) for sustaining crop productivity and reducing environmental impact: a review. Sci. Total Environ. 512–513, 415–427 (2015).
Weigel, D. et al. Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1013 (2000).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Gallois, P. & Marinho, P. Leaf disk transformation using Agrobacterium tumefaciens—expression of heterologous genes in tobacco. Methods Mol. Biol. 49, 39–48 (1995).
Sallaud, C. et al. Highly efficient production and characterization of T-DNA plants for rice (Oryza sativa L.) functional genomics. Theor. Appl. Genet. 106, 1396–1408 (2003).
Lin, S. H. et al. Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20, 2514–2528 (2008).
Lee, Y. J., Kim, D. H., Kim, Y. W. & Hwang, I. Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13, 2175–2190 (2001).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2, 1565–1572 (2007).
Thayer, J. R. & Huffaker, R. C. Determination of nitrate and nitrite by high-pressure liquid chromatography: comparison with other methods for nitrate determination. Anal. Biochem. 102, 110–119 (1980).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Sun, J. et al. Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507, 73–77 (2014).
Acknowledgements
We thank S.-H. Lin for assistance with the 15N analysis; S.-P. Li and S.-M. Huang at the Confocal Microscope Facility, Institute of Molecular Biology, Academia Sinica, for assistance with the acquisition of confocal microscopy images; L.-Y. Kuang at the Transgenic Plant Core Facility, Academia Sinica, for generating transgenic tobacco and rice; C.-S. Suen at the Institute of Biomedical Sciences, Academia Sinica, for assistance with the homology modelling; and C.-D. Hsiao and T.-W. Lin at the Institute of Molecular Biology, Academia Sinica, for providing suggestions relating to the three-dimensional model analysis. This work was supported by the Center for Sustainability Science, Academia Sinica, Taiwan (AS-103-SS-A03 and AS-SS-106-A03).
Author information
Authors and Affiliations
Contributions
K.-E.C. and Y.-F.T. conceived of and designed the experiments. K.-E.C. performed most of the molecular, genetic and physiological experiments. H.-Y.C. constructed the NC4N chimera and characterized the function of NC4N in Xenopus oocytes. C.-S.T. provided expert suggestions and assisted with field trials on rice. K.-E.C. and Y.-F.T. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
For the new strategy described in this manuscript, a United States patent (patent application number: US 9,228,196 B2) entitled “Method for changing nitrogen utilization efficiency in plants” was issued to the inventors Y.-F.T., S.-C. Fan, H.-Y.C. and K.-E.C. on 5 January 2016. In addition, a Taiwan (ROC) patent with the same title was issued to the inventors Y.-F.T., S.-C. Fan and H.-Y.C. on 11 December 2013.
Additional information
Peer review information Nature Plants thanks Chengcai Chu, Yong Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Intracellular view of the putative nitrate-binding pockets of CHL1, NRT1.2 and NC4N.
Homology modeling of NRT1.2 and NC4N was performed using the CHL1 crystal structure (PDB ID 4OH3) as template. Distances (Å) between the nitrogen atom of the modelled nitrate and selected amino acid atoms are shown along respective dashed lines. The red dashed lines represent potential hydrogen bonds and green dashed lines represent potential Van der Waals forces. Transmembrane domains are numbered in the black circles.
Extended Data Fig. 2 NC4N and CHL1 are co-localized in the plasma membrane.
NC4N-GFP or CHL1-RFP driven by the 35S promoter was transiently expressed in Arabidopsis mesophyll protoplasts. Green, red and blue signals represent GFP, RFP and chlorophyll fluorescence, respectively. Scale bars, 10 μm. Similar results were obtained in five independent cells.
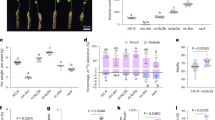
Extended Data Fig. 3 Leaf area and cell number of leaves are increased in NRT1.7p::NC4N::3’ transgenic Arabidopsis plants.
a, Plants grown hydroponically under the condition of N fluctuation described in the legend of Fig. 4a. The biggest leaf (the 5th leaf) of Col-0, nrt1.7 mutant and transgenic plants was collected for analysis. Scale bar, 2 cm. b–d, Leaf area (b), epidermal cell size (c), and cell number per leaf (d) of Col-0, nrt1.7 mutant and transgenic plants. Values are means ± s.d. of three leaves and different letters represent significant differences (p<0.05 by one-way ANOVA with Tukey’s post-hoc test). e, Confocal images of the epidermal cells stained with propidium iodide (PI). Three biological repeats showed similar results. Scale bar, 50 μm. The details of statistical analysis are provided in Source Data.
Extended Data Fig. 4 Expression levels of nitrate assimilation and senescence-marker genes in Col-0, nrt1.7 mutant and transgenic plants.
a–h, Quantitative PCR analysis of the three nitrate assimilation genes, NIA1 (a, e), NIA2 (b, f), NiR (c, g), and the senescence-maker gene SEN1 (d, h). Plants grown hydroponically with 2 mM KNO3 for 26 d (a–d), or 2 mM KNO3 for 25 d and then transferred to N-free medium for a further 1 d (e–h). Values are means ± s.d. of three biological repeats and each repeat comprised pooled leaves of two plants. Different letters represent significant differences (p<0.05 by one-way ANOVA with Tukey’s post-hoc test). The details of statistical analysis are provided in Source Data.
Extended Data Fig. 5 Nitrate uptake activity of Col-0, nrt1.7 mutant and transgenic plants.
a, b, Nitrate uptake activity of plants grown hydroponically with 2 mM KNO3 for 16 d and kept in the same medium of 2 mM KNO3 (a) or transferred to N-free medium for a further 3 d (b). Values are means ± s.d. of six plants and letter represent statistical analysis by one-way ANOVA with Tukey’s post-hoc test (p<0.05). The details of statistical analysis are provided in Source Data.
Extended Data Fig. 6 Flowering time of Col-0, nrt1.7 mutants and transgenic plants.
Arabidopsis plants grown hydroponically with 2 mM KNO3 (n = 6 independent plants for Col-1, n = 7 independent plants for nrt1.7 and #2, and n = 8 independent plants for #1 and #3) (a), N fluctuation (n = 12 independent plants) (b), or 2 mM KNO3 for 18 d and transferred to N-free medium (n = 5 independent plants for nrt1.7 and 7 independent plants for Col-0 and #1 - #3) (c), as described in the legend of Fig. 4. Values are means ± s.d. and different letters represent significant differences (p<0.05 by one-way ANOVA with Tukey’s post-hoc test). The details of statistical analysis are provided in Source Data.
Extended Data Fig. 7 Leaf size and cell number of the leaf are increased in NRT1.7p::NC4N::3’ transgenic tobacco plants.
a, Tobacco plants grown hydroponically under the condition of N fluctuation with alternation of 5 mM KNO3 for 3.5 d and N-free for 3.5 d. The biggest leaf (the 6th leaf) of W38 and transgenic tobacco was collected for analysis. Scale bar: 10 cm. b–d, Leaf area (b), epidermal cell size (c), and cell number per leaf (d) of W38 and transgenic tobacco. Values are means ± s.d. of three leaves and the asterisk above bar indicates a significant difference (p<0.05 by two-tailed Student’s t-test) compared to the W38 control. e, Confocal images of the epidermal cells stained with PI. Three biological repeats showed similar results. Scale bar: 50 μm. The details of statistical analysis are provided in Source Data.
Extended Data Fig. 8 Agronomic traits in field trials of NRT1.7p::NC4N::3’ transgenic rice and TNG67 control.
a-c, Grain number per panicle (a), 1000-kernel weight (b) and grain weight per panicle (c) of TNG67 and transgenic line Q-5 in the first crop season of 2019 (n = 5 biological repeats). d–i, Grain weight per plot (d, g), panicle number per clump (e, h), and grain number per clump (f, i) of TNG67 and transgenic line Q-5. d–f, Values are means ± s.d. of the first and second seasons in 2018 (n = 3 biological repeats) and asterisks above bars indicate significant differences (p<0.05 by two-tailed Student’s t-test) compared with the corresponding crop season of TNG67 control. g–i, Values are means ± s.d. of three seasons (2018 I, 2018 II and 2019 I), presented as a percentage relative to that of the TNG67 line (100%) in the corresponding season and asterisks above bars indicate significant differences (p<0.05 by two-tailed Student’s t-test) compared with the TNG67 control. The details of statistical analysis are provided in Source Data.
Extended Data Fig. 9 Transgenic lines grow better than Col-0 and nrt1.7 mutant plants even after long-term N starvation.
Relative biomass of Col-0, nrt1.7 and three transgenic lines during 10 d of N starvation (a), and the first 4 d (b) and last 4 d of N starvation (c). Plants were grown hydroponically with 2 mM KNO3 for 18 d and then transferred to N starvation for a further 10 d. The biomass at each time-point is normalized with the biomass of the same genotype at the beginning of N starvation (D0). Values of each time-point are means ± s.d. of six plants. The numbers in parentheses represent the slope of the growth curve (growth rate).
Supplementary information
Supplementary Information
Supplementary Table 1.
Source data
Source Data Fig. 1
Statistical source data for Fig. 1.
Source Data Fig. 1
Unprocessed western blot for Fig. 1b.
Source Data Fig. 2
Statistical source data for Fig. 2.
Source Data Fig. 3
Statistical source data for Fig. 3.
Source Data Fig. 4
Statistical source data for Fig. 4.
Source Data Fig. 5
Statistical source data for Fig. 5.
Source Data Fig. 6
Statistical source data for Fig. 6.
Source Data Fig. 7
Statistical source data for Fig. 7.
Source Data Extended Data Fig. 3
Statistical source data for Extended Data Fig. 3.
Source Data Extended Data Fig. 4
Statistical source data for Extended Data Fig. 4.
Source Data Extended Data Fig. 5
Statistical source data for Extended Data Fig. 5.
Source Data Extended Data Fig. 6
Statistical source data for Extended Data Fig. 6.
Source Data Extended Data Fig. 7
Statistical source data for Extended Data Fig. 7.
Source Data Extended Data Fig. 8
Statistical source data for Extended Data Fig. 8.
Source Data Extended Data Fig. 9
Statistical source data for Extended Data Fig. 9.
Rights and permissions
About this article
Cite this article
Chen, KE., Chen, HY., Tseng, CS. et al. Improving nitrogen use efficiency by manipulating nitrate remobilization in plants. Nat. Plants 6, 1126–1135 (2020). https://doi.org/10.1038/s41477-020-00758-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-00758-0
This article is cited by
-
Overexpression of rice OsNRT1.1A/OsNPF6.3 enhanced the nitrogen use efficiency of wheat under low nitrogen conditions
Planta (2024)
-
Vegetable Plant Canopy Inhibits Soil Urease Activity and Reduces NH3 Losses Under p-Phenylenediamine (PPD)-Coated Urea Application
Journal of Soil Science and Plant Nutrition (2024)
-
Study of vacuole glycerate transporter NPF8.4 reveals a new role of photorespiration in C/N balance
Nature Plants (2023)
-
Timing and Splitting of Nitrogen Compensated for the Loss in Grain Yield of Dual-Purpose Wheat Under Varied Cutting Heights
Gesunde Pflanzen (2023)
-
Trait Improvement of Solanaceae Fruit Crops for Vertical Farming by Genome Editing
Journal of Plant Biology (2023)