Abstract
Plants initiate specific defense responses by recognizing conserved epitope peptides within the flagellin proteins derived from bacteria. Proteolytic cleavage of epitope peptides from flagellin by plant apoplastic proteases is thought to be crucial for the perception of the epitope by the plant receptor. However, the identity of the plant proteases involved in this process remains unknown. Here, we establish an efficient identification system for the target proteases in Arabidopsis apoplastic fluid; the method employs native two-dimensional electrophoresis followed by an in-gel proteolytic assay using a fluorescence-quenching peptide substrate. We designed a substrate to specifically detect proteolytic activity at the C-terminus of the flg22 epitope in flagellin and identified two plant subtilases, SBT5.2 and SBT1.7, as specific proteases responsible for the C-terminal cleavage of flg22. In the apoplastic fluid of Arabidopsis mutant plants deficient in these two proteases, we observe a decrease in the C-terminal cleavage of the flg22 domain from flagellin, leading to a decrease in the efficiency of flg22 epitope liberation. Consequently, defensive reactive oxygen species (ROS) production is delayed in sbt5.2 sbt1.7 double-mutant leaf disks compared to wild type following flagellin exposure.
Similar content being viewed by others
Introduction
Plants defend themselves against pathogenic infection by using innate immune systems that evolved to recognize the presence of pathogens. This immune response process begins with the perception of molecular signatures (termed pathogen- or microbe-associated molecular patterns; PAMPs or MAMPs) that are shared among whole classes of microbes; sensing of MAMPs by pattern-recognition receptors triggers a basal resistance called pattern-triggered immunity (PTI)1. PTI responses include the production of reactive oxygen species (ROS), activation of the mitogen-activated protein kinase cascade, defense gene expression, and callose deposition1.
One of the most extensively studied examples of MAMPs is flagellin, the major structural protein of the bacterial flagellum and a molecule that is distributed widely among distant bacterial species. Plants recognize flagellin monomers rather than polymerized flagellin. Notably, flg22, a 22-amino acid epitope peptide corresponding to the most conserved domain near the flagellin N-terminus, acts as a potent inducer of PTI in various plant species2. In Arabidopsis, the Leu-rich repeat transmembrane receptor kinase FLAGELLIN SENSING 2 (FLS2) directly recognizes flg22, leading to the initiation of immune signaling3,4,5,6.
The flg22-FLS2 pair has been used as a model system to study PTI triggered by flagellin perception. In nature, however, flagellin is polymerized to form the flagella, resulting in the flg22 epitope being buried in the flagellin polymer structure7. Plants are thought to recognize flagellin monomers that are released as a result of leaks or as spillover during the construction of the flagella8. Although intact flagellin monomer protein shows some interaction with FLS2, the binding affinity of FLS2 for the flagellin monomer is far weaker than that for flg22 peptide2,9, indicating that proteolytic release of the flg22 peptide from the flagellin protein is required for full induction of defense responses. However, in contrast to the well-characterized signaling cascade downstream of FLS2, little is known about the mechanisms of proteolytic release of the immunogenic peptide ligand flg22 from flagellin.
The importance of proteolytic digestion of flagellin in initiating the host immune response also is suggested by the finding that the secreted plant glycosidase β-galactosidase 1 (BGAL1) participates in flagellin deglycosylation and contributes to immunity in Nicotiana benthamiana10. BGAL1 removes O-glycans located on the outward-facing hydrophilic surface of the flagellin in flagellar filaments, facilitating exposure of the flg22 region to proteases that release the flg22 ligand. Consistent with this scenario, the addition of a protease inhibitor to apoplastic fluids containing BGAL1 blocks the release of immunogenic peptides from bacterial flagellin when the host is challenged with deglycanated flagellin protein, demonstrating an important role for proteases in immunogenic peptide release10.
Bacteria enter the plant apoplast through natural openings such as stomata or wounds. The apoplast is the site of the initial interaction between plants and bacteria; thus, the proteases that cleave flg22 from flagellin likely are present in the apoplast. Proteolytic release of the flg22 peptide from the flagellin protein requires cleavage at both the N- and C-termini of the 22-amino-acid epitope. The Arabidopsis genome contains hundreds of protease-encoding genes, among which nearly 300 encode secreted proteases carrying signal peptides11.
In the present study, we designed a fluorescence-quenching substrate to detect proteolytic activity at the C-terminus of the 22-amino-acid flg22 epitope in flagellin. Specifically, we established an efficient system for identification of the target proteases in Arabidopsis apoplastic fluid using native two-dimensional electrophoresis followed by an in-gel proteolytic assay and mass spectrometry-based proteomics. We identified two subtilases, SBT5.2 and SBT1.7, as specific proteases involved in C-terminal cleavage of flg22. The apoplastic fraction of Arabidopsis mutant plants deficient in these two proteases showed decreased C-terminal cleavage of flg22 from flagellin, resulting in a delay in apoplastic ROS production in leaf disks upon flagellin treatment.
Results
Proteolytic release of immunogenic peptides from bacterial flagellin by Arabidopsis apoplastic proteases
Proteolytic release of the immunogenic peptides from flagellin is thought to be required for full induction of host defense responses. To examine the temporal differences between the defense responses induced by the flagellin protein and the immunogenic flg22 peptide, we measured ROS production in wild-type (WT) Arabidopsis leaf disks treated with flagellin purified from Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) or synthetic flg22 peptide from Pst DC3000. When WT leaf disks were exposed to protein or peptide at 100 nM, ROS production was delayed for ≈5 min following flagellin protein exposure compared to that induced by flg22 peptide (Fig. 1a, b). An fls2 mutant (SAIL_691_C04) showed no ROS production upon exposure to either purified flagellin or synthetic flg22 (Supplementary Fig. 1a). We reasoned that this time lag represents the time taken for proteolytic release of the immunogenic peptide that acts as a ligand for FLS2.
a ROS burst in Arabidopsis leaves upon elicitor exposure. Leaf disks from WT plants were incubated with 100 nM flg22 peptide or flagellin protein. ROS production was measured every 30 s over 60 min. Values represent the mean ± SE (n = 8 biologically independent samples). b Comparison of time at which the maximum response (Tmax) was observed (n = 8 biologically independent samples). Boxes represent 25th to 75th percentile range, the line within the box marks the median and the whiskers represent the minimum and maximum values. P value was calculated by two-tailed non-paired Student’s t-test. c Representative fragmentation pattern of flagellin upon incubation in Arabidopsis submerged culture medium containing apoplastic proteases. Flagellin protein was incubated for 4 h in WT Arabidopsis submerged culture medium, and the resulting peptides were analyzed by nano-LC-MS/MS. Each PSM was mapped against the amino acid sequence of flagellin. The thick line at the top represents flagellin protein; the red domain indicates the flg22 epitope. PSMs that exactly match flg22 peptide are shown as thin dark-red lines. PSMs that match fragments cleaved at the C-terminus of the flg22 domain are shown as thin light-red lines. d Frequency at which each amino acid residue is the C-terminal end (upper panel) or N-terminal end (lower panel) of the peptides following digestion by WT apoplastic proteases. The gray box indicates the flg22 epitope. Values represent the mean ± SE (n = 3 biologically independent samples). Source data are provided as a Source Data file.
The apoplast, as the primary interface between plants and bacteria, serves as the crucial space where flagellin fragmentation is most likely to occur. To investigate fragmentation patterns of flagellin in plant apoplasts, purified flagellin protein was incubated for 4 h with spent medium from Arabidopsis whole-plant submerged culture (submerged culture medium), into which secreted proteins (including proteases) that are present in the apoplast diffuse12. Of the secreted proteins detected in the actual leaf apoplast, 75% also were detected in the submerged culture medium (Supplementary Data 1). Nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS/MS) of the digests yielded ≈ 1600 peptide spectrum matches (PSMs) for peptide fragments derived from flagellin (Fig. 1c). We mapped all of the identified peptides against the amino acid sequence of flagellin and observed that multiple peptide fragments derived primarily from the N-terminal region of flagellin, including the flg22 epitope peptide, are generated by the Arabidopsis apoplastic proteases. In comparison, few peptide fragments were detected in mock-treated flagellin (Supplementary Fig. 1b).
Detailed inspection of the digestion patterns indicated that one of the major cleavage sites within flagellin is the C-terminus of the flg22 epitope, representing 20.5% of the total PSMs that were detected (Fig. 1d). In contrast, complex cleavage pattern was observed around the N-terminus of the flg22 epitope. We also detected cleavage activity in the central region of the flg22 epitope, which potentially causes loss of the immunogenic activity of flg222; this observation suggested the existence of offensive and defensive interactions between plants and bacteria regarding cleavage of the flg22 epitope. Treatment of WT leaf disks with the synthetic flg22 peptide variant with a 6-residue C-terminal extension (flg22C6) resulted in delayed ROS production (Supplementary Fig. 1c, d); again, this observation suggested that the C-terminal cleavage of the flg22 epitope is important for activation of the defense response.
When flagellin was cleaved by the endoproteinase Asp-N, which cleaves peptide bonds N-terminal to Asp residues, peptide fragments were detected from across virtually the entire length of the protein, except for the Ser143-Ser201 domain to which carbohydrate chains are known to be attached (Supplementary Fig. 2a, b)13. These results indicated the presence of apoplastic proteases that predominantly cleave the flagellin N-terminal region, including those that release the flg22 epitope. Based on these results, we were interested in the molecular identity of the Arabidopsis endo-proteases that cleave flagellin to generate the C-terminal end of the flg22 epitope, and the extent to which these enzymes contribute to flagellin recognition by plants.
Specific detection of cleavage at the C-terminus of flg22 using a fluorescence-quenching substrate
To identify the proteases that cleave the C-terminus of the flg22 domain within flagellin, we designed a fluorescence-quenching substrate that specifically detects proteolytic activity at the C-terminus of the flg22 domain; specifically, we used a 10-amino-acid peptide spanning the cleavage site (flg[47–56]). Fluorescence-quenching technology employs a paired fluorophore and quencher, which are introduced on the opposite ends of the peptide14. Upon hydrolysis of an internal peptide bond, the distance between the donor and the acceptor increases, the fluorophore is no longer quenched, and an enhanced fluorescence signal is generated. We synthesized flg[47–56] by replacing Ser56 with a quencher, Nε−2,4-dinitrophenyl-lysine (Lys(Dnp)), followed by introduction of a fluorophore, N-methylanthranic acid (Nma)15, at the N-terminal amino group, to generate a fluorescence-quenching substrate, Nma-flg[47–56]-Dnp (Fig. 2a, b).
a The amino acid sequence near the N-terminus of Pst DC3000 flagellin. The underlining indicates the flg22 epitope domain; the dashed box indicates the flg[47–56] sequence used to construct the substrate. b Structure of the fluorescence-quenching substrate Nma-flg[47-56]-Dnp. c Fluorescence intensity of Nma-flg[47-56]-Dnp during incubation with Arabidopsis submerged culture medium (red circles) or heat-inactivated submerged culture medium (gray triangles). Fluorescent emission at 435 nm was measured every 1 h over 20 h. Values represent the mean ± SE (n = 3 biologically independent samples). d In vitro proteolytic assay of Nma-flg[47–56]-Dnp with Arabidopsis submerged culture medium. Representative selected ion chromatograms (SICs) are shown for Nma-GLQI, Nma-GLQIA, and Nma-GLQIAT. e MS/MS spectrum of the precursor ion at m/z 634.36 corresponding to Nma-GLQIA. Source data are provided as a Source Data file.
To test whether this substrate can detect the proteolytic activity at the C-terminus of flg22, we added 4 µM Nma-flg[47–56]-Dnp to the Arabidopsis submerged culture medium. The fluorescence intensity in the reaction increased over time during the 1–7 h after substrate addition, and the intensity maintained maximum levels for up to 20 h; in contrast, no increase was observed when we added the substrate to heat-inactivated submerged culture medium in which the proteases presumably had been denatured (Fig. 2c). Nano-LC-MS analysis of the digests confirmed that the major proteolytic product is a 5-amino-acid peptide cleaved at the C-terminus of the flg22 domain (Fig. 2d, e, Supplementary Fig. 3a, b). These results indicated that the fluorescence-quenching substrate Nma-flg[47-56]-Dnp can detect the Arabidopsis apoplastic endo-protease(s) that cleave the C-terminus of the flg22 domain within flagellin.
Identification of endo-proteases involved in C-terminal cleavage of flg22
The fluorescence-quenching substrate Nma-flg[47-56]-Dnp enabled high-throughput fluorogenic detection of proteolytic activity at the C-terminus of flg22. We employed this system for in-gel detection of endo-proteases on polyacrylamide gels. Arabidopsis submerged culture medium was concentrated and subjected to native two-dimensional electrophoresis. Proteins were separated in the first dimension by native isoelectric focusing (IEF) and in the second dimension by blue native polyacrylamide gel electrophoresis (BN-PAGE), followed by gel fractionation using a SAINOME-plate, which dices the gel into approximately 4.5-mm square pieces and drops these gel fragment into the individual wells of a 384-well plate16. We then performed an in-gel proteolytic assay in the 384-well plate using 40 µM Nma-flg[47-56]-Dnp as a substrate and detected any active spots with increased fluorescence (Fig. 3a).
a In-gel proteolytic assay of the diced two-dimensional gels using Nma-flg[47–56]-Dnp as a fluorescence-quenching substrate. The fluorescence intensity was measured 6 h after the addition of the substrate. The numbers and letters represent the columns and rows (respectively) of the microplate used in the assay. The fluorescence intensity is color-coded as indicated in the scale on the right. b Total count of PSMs of SBT5.2 and SBT1.7 identified in the fluorescence-positive gel fragments and in the fluorescence-negative gel fragment used as a control. The PSM count is color-coded as indicated in the scale on the right. c Agrobacterium-mediated transient expression of the His-tagged subtilases, SBT5.2-H6 and SBT1.7-H6, in N. benthamiana. Apoplastic fluids were extracted from agroinfiltrated leaves and subjected to SDS-PAGE followed by western blotting using HRP-conjugated anti-His tag antibody. The experiment was independently repeated three times with similar results. Source data are provided as a Source Data file.
To identify the proteins in this spot, we performed mass spectrometry-based proteomics of gel pieces at and around the spot at G9, which showed the highest activity; those at spot H14 were analyzed as a negative control. The resulting MS/MS data were used to search a database of Arabidopsis proteins, and proteases with an N-terminal signal peptide were extracted from among the identified proteins. This procedure identified two subtilases, SBT5.2 (At1g20160) and SBT1.7 (At5g67360; also known as ARA12), both of which were detected specifically in active gel fractions with high PSM scores (Fig. 3b, Supplementary Data 2).
SBT5.2 and SBT1.7 cleave the C-terminus of flg22 from flagellin
To test whether SBT5.2 and SBT1.7 cleave at the C-terminus of the flg22 domain, we transiently expressed C-terminally His-tagged SBT5.2 or SBT1.7 (SBT5.2-H6 or SBT1.7-H6, respectively) by agroinfiltration of N. benthamiana; we then isolated apoplastic fluid from the agroinfiltrated plants. The expression of SBT5.2-H6 and SBT1.7-H6 was confirmed by western blot analysis using anti-His-tag antibody (Fig. 3c). We captured SBT5.2-H6 or SBT1.7-H6 proteins on Ni-Sepharose beads for use in solid-phase protease assays. Incubation of flagellin protein with the beads for 1 h followed by nano-LC-MS/MS analysis confirmed that both SBT5.2-H6 and SBT1.7-H6 cleave precisely at the C-terminus of the flg22 domain (Ala51-Thr52) (Fig. 4a–e). Alanine-scanning experiments demonstrated that a residue at the P2 position (Ile50) is critical for efficient cleavage by SBT5.2-H6 at this site (Fig. 4f). When flagellin was digested by SAP2 (AT1G03220), a secreted protease detected nonspecifically in gel fractions (Supplementary Data 2), the Ala51-Thr52 site was not cleaved, indicating the substrate specificity of SBT5.2 and SBT1.7 (Supplementary Fig. 4a, b).
a Representative fragmentation pattern of flagellin digested with His-tagged SBT5.2 (SBT5.2-H6). Flagellin protein was incubated for 1 h with bead-captured SBT5.2-H6, and the resulting peptide fragments were analyzed by nano-LC-MS/MS. Each PSM was mapped against the amino acid sequence of flagellin. The thick line at the top represents flagellin protein; the red domain indicates the flg22 epitope. PSMs that matched fragments cleaved at the C-terminus of flg22 domain are shown as red lines. b Frequency at which each amino acid residue is the C-terminal end or N-terminal end of the peptides following digestion with SBT5.2-H6. The gray box indicates the flg22 epitope. Values represent the mean ± SE (n = 6 biologically independent samples). c Representative fragmentation pattern of flagellin by SBT1.7-H6. d Frequency at which each amino acid residue is the C-terminal end or N-terminal end of the peptides following digestion by SBT1.7-H6. Values represent the mean ± SE (n = 5 biologically independent samples). e Representative MS/MS spectrum of the flg22 epitope peptide cleaved from flagellin by SBT5.2-H6. f Effect of alanine-scanning substitution of the C-terminal cleavage site of flg22 on cleavage activity by SBT5.2. Each synthetic peptide was incubated for 1 h with bead-captured SBT5.2-H6, and the remaining undigested peptides were quantified by nano-LC-MS. Black arrowhead indicates cleavage site. Different letters indicate a significant difference with others (mean ± SE, P < 0.05, one-way ANOVA followed by Tukey’s test, n = 4 biologically independent samples). Source data are provided as a Source Data file.
In addition to the effects of the subtilases on Ala51-Thr52 cleavage, SBT5.2-H6 and SBT1.7-H6 provided proteolytic cleavage of flagellin at Asn39-Ser40, a site located in the central region of the flg22 domain (Fig. 4a–d). At this cleavage site, the residue at the P2 position is also Ile. Since the flg22 epitope loses its immunogenic activity when cleaved at Asn39-Ser40 2, the balance of the cleavage efficiency between Asn39-Ser40 and Ala51-Thr52 is critical for liberation of active immunogenic peptides.
We also detected minor but multiple cleavage sites within the N-terminal region of flagellin (Fig. 4a–d). We confirmed that flagellin does not fragment when incubated with Ni-Sepharose beads exposed to control apoplastic fluid collected from N. benthamiana plants expressing the empty vector (Supplementary Fig. 4c). We further confirmed that immobilized Arabidopsis SBT5.2-H6 and SBT1.7-H6 contain few or no detectable contamination by N. benthamiana proteases, thus excluding the possibility that multiple cleavage of the N-terminal region of flagellin reflects co-purified N. benthamiana proteases (Supplementary Data 3). The cleavage sequence preferences of SBT5.2 and SBT1.7 are similar, but not identical, to each other. Unlike SBT5.2-H6, SBT1.7-H6 was able to efficiently cleave the C-terminal region of flagellin at sites such as Asn207-Ser208, Gln217-Asn218 and Gln228-Asn229 (Fig. 4c, d). These results indicated that SBT5.2 and SBT1.7 have preferential cleavage target sequences, but selection of the cleavage site is not always stringent.
The flg22 domain is highly conserved in bacterial flagellin, but there is some sequence variation around the C-terminus of the flg22 domain depending on the bacterial species2. Therefore, we further investigated whether SBT5.2 cleaves the C-terminus of the flg22 domain of flagellins derived from bacterial species other than Pst DC3000. Flg22 peptides derived from the flagellin proteins of Pseudomonas aeruginosa and Xanthomonas axonopodis pv. citri both exhibit PTI-inducing activity2,17. Based on the flagellin sequences of these two bacterial species, two separate 10-amino-acid-long peptides, flg[47-56]Pa and flg[47-56]Xac (respectively) were synthesized; each peptide then was incubated for 1 h with SBT5.2-H6. Nano-LC-MS analysis confirmed that SBT5.2-H6 cleaves both flg[47-56]Pa and flg[47–56]Xac at the C-terminus of the flg22 domain, exhibiting cleavage efficiencies comparable to that observed with flg[47–56] (Supplementary Fig. 5a–f). These data suggested that SBT5.2 functions against substrates from across bacterial species.
Decrease in C-terminal cleavage of flg22 from flagellin in sbt5.2 sbt1.7 double mutant
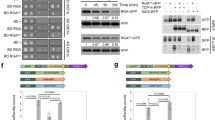
To determine the function of SBT5.2 and SBT1.7 in planta, an sbt5.2 sbt1.7 double mutant was generated by crossing the T-DNA insertion mutant lines for SBT5.2 (SALK_132812C, sbt5.2-2) and SBT1.7 (GABI_544E06, sbt1.7-2) (Fig. 5a). RT-PCR confirmed that full-length SBT5.2 and SBT1.7 transcripts were absent in the sbt5.2 sbt1.7 double mutant (Fig. 5b).
a- The T-DNA insertion sites of the sbt5.2-2 and sbt1.7-2 mutants. Exons are indicated as filled gray rectangles, introns are indicated as solid lines, and untranslated regions are indicated as white rectangles. Arrows indicate the sites of primers used for RT-PCR analysis. b Absence of SBT5.2 and SBT1.7 transcripts in the sbt5.2 sbt1.7 double mutant, as verified by RT-PCR. ACT3 was used as an internal control. Source data are provided as a Source Data file.
To investigate changes in flagellin fragmentation patterns in the apoplast of sbt5.2 sbt1.7 plants, purified flagellin protein was incubated for 4 h with submerged culture medium obtained from WT, sbt5.2 single, sbt1.7 single or sbt5.2 sbt1.7 double-mutant plants. Nano-LC–MS/MS of the digests exposed to the WT submerged culture medium revealed that as much as 15.8% PSMs were derived from peptide fragments cleaved at the C-terminus of the flg22 domain (Fig. 6a, b). In contrast, when incubated with submerged culture medium of the sbt5.2 sbt1.7 double mutant, only 0.6% of the total PSMs corresponded to fragments cleaved at the C-terminus of flg22 domain, confirming that proteolytic activity at the C-terminus of the flg22 domain is markedly decreased in the sbt5.2 sbt1.7 mutant (Fig. 6c, d).
a Representative fragmentation pattern of flagellin following incubation for 4 h in submerged culture medium from WT plants. b Frequency at which each amino acid residue is the C-terminal or N-terminal end of the peptides following digestion of flagellin by submerged culture medium from WT plants. Values represent the mean ± SE (n = 3 biologically independent samples). c Representative fragmentation pattern of flagellin following incubation for 4 h in submerged culture medium from sbt5.2 sbt1.7 plants. d Frequency at which each amino acid residue is the C-terminal or N-terminal end of the peptides following digestion of flagellin by submerged culture medium from sbt5.2 sbt1.7 plants. Values represent the mean ± SE (n = 3 biologically independent samples). Source data are provided as a Source Data file.
We also observed decreased proteolytic activity at the C-terminus of the flg22 domain in submerged culture medium from the sbt5.2 single mutant, compared to that of WT (Supplementary Fig. 6a–c). In contrast, the flagellin digestion pattern observed in submerged culture medium from the sbt1.7 single mutant was similar to that obtained from the WT (Supplementary Fig. 6a, b, d). Submerged culture medium from a complementation line, in which the SBT5.2 gene was introduced into the sbt5.2 sbt1.7 double mutant, showed proteolytic cleavage patterns comparable to those observed with the submerged culture medium from sbt1.7 single-mutant or WT plants (Supplementary Fig. 6a, b, e). These results indicated that SBT5.2 is the major apoplastic protease responsible for the cleavage of flagellin at the C-terminus of the flg22 domain in planta, and that SBT1.7 also functions redundantly in the generation of this immunogenic peptide.
Loss of SBT5.2 and SBT1.7 delays ROS production in leaf disks following flagellin treatment
To investigate the biological role of SBT5.2- and SBT1.7-mediated cleavage of flagellin during PTI, we measured ROS production in leaf disks of WT and sbt5.2 sbt1.7 double-mutant plants treated with 100 nM flagellin protein. We found that ROS production was delayed for ≈2 min in sbt5.2 sbt1.7 leaf disks, compared to WT leaf disks, following flagellin treatment, although the maximum signal intensity was comparable between the WT and sbt5.2 sbt1.7 double-mutant plants, presumably due to residual protease activity (Fig. 7a–d). This time lag between WT and sbt5.2 sbt1.7 in response to flagellin treatment is reminiscent of the time lag observed in leaf disks of WT plants in response to the synthetic flg22 peptide and flagellin (Fig. 1a, b). We confirmed that there was no time lag in ROS production between WT and sbt5.2 sbt1.7 when each was treated with 100 nM synthetic flg22 peptide, demonstrating that there is no difference in sensitivity to cleaved flg22 between WT and sbt5.2 sbt1.7 plants (Fig. 7b, c).
a ROS burst in WT and sbt5.2 sbt1.7 leaf disks following exposure to 100 nM flagellin. ROS production was measured every minute over 60 min. Values represent the mean ± SE (n = 12 biologically independent samples). b ROS burst in WT and sbt5.2 sbt1.7 leaf disks following exposure to 100 nM flg22. Values represent the mean ± SE (n = 12 biologically independent samples). c Comparison of time at which the maximum response (Tmax) observed in (a) and (b) (n = 12 biologically independent samples). d Delay in time to maximum RLU (ΔTmax RLU) of the sbt5.2 sbt1.7 double mutant compared to that of WT in seven independent experiments (n = 7 independent experiments). For all box plots, boxes represent 25th to 75th percentile range, the line within the box marks the median and the whiskers represent the minimum and maximum values. P values were calculated by two-tailed non-paired Student’s t test. Source data are provided as a Source Data file.
We interpreted this phenotype as being a consequence of the reduced proteolytic activity at the C-terminus of the flg22 domain in sbt5.2 sbt1.7. Loss of SBT5.2 and SBT1.7 would increase the time required for proteolytic release of the immunogenic peptides, resulting in delayed ROS production.
Discussion
In the present study, we analyzed the fragmentation patterns of flagellin in Arabidopsis thaliana apoplastic fluid using nano-LC–MS/MS to understand proteolytic release of the immunogenic peptide from flagellin protein. Detailed inspection of the flagellin digestion patterns revealed that one of the major endo-cleavage sites within flagellin protein is the C-terminus of the flg22 domain. Furthermore, by using a proteomic approach in combination with a fluorescence-quenching peptide substrate, we identified two subtilases, SBT5.2 and SBT1.7, as proteases involved in flagellin cleavage at the C-terminus of the flg22 domain. It has been reported in Nicotiana benthamiana that the secreted glycosidase BGAL1 is involved in the deglycosylation of flagellin, facilitating the exposure of the flg22 region to proteases that release the flg22 ligand10. Because At2g28470, the Arabidopsis homolog of tobacco BGAL1, is also detected in submerged culture medium (Supplementary Data 1), it may facilitate flg22 cleavage. However, since purified SBT5.2-H6 alone can cleave flagellin, it appears that flagellin is cleaved to some extent even in the absence of BGAL1.
The proteolytic activity of SBT5.2 and SBT1.7 in cleaving flagellin at the C-terminus of flg22 was confirmed not only for the amino acid sequence of flagellin from P. syringae pv. tomato DC3000, but also for those from P. aeruginosa and Xanthomonas axonopodis pv. citri, two additional species that generate flg22 peptides that induce PTI2,17. These data suggest that SBT5.2 and SBT1.7 possess activity against substrates from across bacterial species. The broad substrate specificity of the flg22 cleavage proteases against substrates from across bacterial species may ensure that FLS2 recognizes a wide range of pathogenic bacteria.
In sbt5.2 sbt1.7 double-mutant plants, the apoplastic C-terminal cleavage of flg22 from flagellin was decreased, resulting in delayed ROS production in leaf disks upon flagellin treatment. A recent study has highlighted the significance of apoplastic proteases in the release of immunogenic peptides10. The addition of protease inhibitors to the apoplastic fluid inhibits the release of immunogenic peptides from bacterial flagellin, leading to the suppression of ROS production in leaf disks10. Plants lacking SBT5.2 and SBT1.7, however, did not achieve complete suppression of ROS production activity. We attribute this incomplete attenuation to the presumed involvement of additional proteases, other than SBT5.2 and SBT1.7, in the release of immunogenic peptides from flagellin. Indeed, the flagellin cleavage activity at the C-terminus of the flg22 domain was decreased, rather than completely abrogated, in the sbt5.2 sbt1.7 double-mutant plants. These results suggest the existence of other proteases that cleave flagellin at the C-terminus of flg22. Furthermore, in the apoplast of Arabidopsis, particularly in that of sbt5.2 sbt1.7 double-mutant plants, peptide fragments with C-terminal extensions from the C-terminus of the flg22 domain also were detected. The elucidation of the potential ligand activity of these C-terminally extended peptides and the proteases involved in their cleavage remains a challenge. Additionally, the cleavage of the N-terminus of the flg22 domain is necessary for the release of the immunogenic peptides. The fragmentation pattern of flagellin in submerged culture medium obtained from sbt5.2 sbt1.7 plants suggests the presence of endo-proteases that cleave precisely at the N-terminus of the flg22 domain.
Previous work has shown that SBT5.2 and SBT1.7 contribute to various physiological processes by cleaving precursors of endogenous peptide hormones as substrates in the apoplast. SBT5.2 has been reported to be involved in the precursor processing and maturation of the IDA (INFLORESCENCE DEFICIENT IN ABSCISSION) peptide, which controls floral organ abscission18; the SBT5.2 enzyme also was identified as a protease (referred to as CRSP (CO2 RESPONSE SECRETED PROTEASE)) that cleaves and activates the pro-peptide EPF2 (EPIDERMAL PATTERNING FACTOR 2), which in turn represses stomatal density19. Separately, SBT1.7 has been reported to function in the precursor processing of the CLE40 (CLAVATA3/EMBRYO SURROUNDING REGION-RELATED 40) peptide, which controls stem cell maintenance in the root20. In addition to known functions of these enzymes in cleaving endogenous proteins, the present study demonstrates that these proteases cleave flagellin, an exogenous protein from pathogenic bacteria. This finding demonstrates the dual role of these subtilases in regulating both internal and external signals.
SBT5.2 has two splice variants, SBT5.2(a) and SBT5.2(b). SBT5.2(a) is secreted to the apoplast and exhibits protease activity; in contrast, SBT5.2(b) lacks a signal peptide, preventing its secretion, and this protein does not exhibit protease activity21. SBT5.2(b) localizes to the endosomes and interacts with the defense-related transcription factor MYB30 (MYELOBLASTOSIS-LIKE PROTEIN 30), resulting in the retention of MYB30 at the endosomes and exclusion from the nucleus. As a result, SBT5.2(b) negatively regulates the defense-related cell death induced by Pst AvrRpm121. Our study revealed that SBT5.2(a) functions upstream of flagellin perception and positively regulates PTI. Recent studies have demonstrated that PTI is required for effector-triggered hypersensitive cell death, and PTI enhances cell death induced by effector-triggered immunity (ETI)22,23. These findings suggest that SBT5.2(a) and SBT5.2(b) play opposing roles in defense responses associated with cell death. Presumably, defense responses need to be tightly controlled, given that such responses often are accompanied by growth inhibition. To maintain this balance, Arabidopsis may utilize two splice variants of the SBT5.2 gene with opposing functions.
Alternatively, another scenario can be used to explain the existence of these two opposing splice variants. It has been reported that during bacterial treatment, host expression of SBT5.2(a) is downregulated, while the expression of SBT5.2(b) is induced21. If this expression change is not a control mechanism employed by the plant to balance immunity but rather controlled by the bacteria, SBT5.2 may be acting as a suppressor of immunity through splicing regulation, which would benefit the bacteria. Therefore, the bacteria may not try to evade the cleavage of flagellin by SBT5.2(a) and the resulting recognition. This hypothesis may explain why splice variants of this protease with opposing functions have persisted evolutionarily.
It is also evolutionarily intriguing that SBT5.2 and SBT1.7 are not only involved in flg22 peptide liberation by cleavage of Ala51-Thr52, which is beneficial to the plant, but also in cleavage of Asn39-Ser40, which leads to inactivation of the flg22 peptide and benefits the bacterial side. An Ile residue required for SBT5.2 recognition is also found at the P2 position of the Asn39-Ser40 cleavage site, and this Ile-Asn-Ser sequence is conserved in the flg22 domain of many flagellin proteins2. Comprehensive amino acid substitution experiments in the flg22 domain of flagellin have revealed that 79.8% of the mutants lose motility, while 74.5% of the mutants still maintain binding activity to FLS224. This means that it would be difficult for bacteria to evolve the flg22 domain to escape recognition by FLS2 without losing motility. Therefore, rather than altering the overall conformation of the flg22 peptide to reduce its binding ability to FLS2, bacteria may have evolved to allow the plant enzyme to cleave one of the regions required for flg22 activity, thereby escaping FLS2 recognition and improving their own fitness. There seems to be an evolutionary “hide-and-seek” between plants and bacteria over which site of flg22, Asn39-Ser40 or Ala51-Thr52, is preferentially cleaved.
Even when a protein cleavage site has been identified or deduced, it is often the case that the corresponding protease has not yet been found. For example, although the proteolytic release of another flagellin epitope (flgII-28, which is recognized by FLS325,26,27), also is expected, the proteases responsible for this cleavage have not yet been identified. It is also known that a number of peptide hormones are produced from larger inactive precursor proteins by proteolytic processing. Recent reports have identified some of the proteases involved in these processes28,29, but there are still many examples where the corresponding enzymes have not yet been identified. The method employed in the present study, using fluorescence-quenching substrates in combination with native two-dimensional electrophoresis and proteomics, may be helpful for the identification and characterization of such proteases.
Methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia (Col) was used as the wild type (WT). The sbt5.2-2 (SALK_132812C), sbt1.7-2 (GABI_544E06), and fls2 (SAIL_691_C04) mutants, which were described previously4,21,30 and obtained from the Arabidopsis Biological Resource Center. The double mutant with homozygous T-DNA insertions in both SBT5.2 and SBT1.7 was generated by crossing the single mutants. The homozygosity of the mutation was confirmed by RT-PCR using total RNA isolated from Arabidopsis leaves using specific primers (Supplementary Data 4). Total RNA was extracted using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s protocol. Plants were grown from surface-sterilized Arabidopsis seeds that were vernalized for 2 d at 4 °C, sown on B5 medium containing 1.0% sucrose solidified with 0.7% agar, and incubated under continuous light at 22 °C. For the ROS production assay, plants were transplanted to soil and grown under a 8-h/16-h light/dark cycle at 22 °C. For the whole-plant submerged culture, forty 6-day-old seedlings were transplanted into 100 mL of B5 liquid medium containing 1.0% sucrose and incubated without shaking under continuous light at 22 °C for 7–11 days. For agroinfiltration, N. benthamiana plants were grown on soil under a 16-h/8-h light/dark cycle at 23 °C for 4–5 weeks.
Peptide synthesis
Epitope peptide flg22 from Pst DC3000 (TRLSSGLKINSAKDDAAGLQIA) and its variant flg22C6 (TRLSSGLKINSAKDDAAGLQIATKITSQ) were synthesized by fluorenylmethoxycarbonyl protecting group (Fmoc) solid-phase chemistry on Wang-PEG resin using an automated peptide synthesizer (Biotage Initiator+ Alstra). Using the same method, the peptide substrate flg[47–56] (GLQIATKITS), its Ala-substituted peptides, flg[47-56]Pa (GLQIANRLTS) and flg[47–56]Xac (GLAISERFTT) were synthesized. All peptides were purified by reverse-phase HPLC. Nma-flg[47–56]-Dnp was synthesized by the Peptide Institute, Inc. (Osaka, Japan).
Preparation of flagellin
Pst DC3000 was grown on King’s B agar plate containing 100 μg/mL rifampicin at 28 °C, then inoculated into King’s B liquid medium containing rifampicin and incubated overnight at 28 °C with shaking at 150 rpm. A portion of this culture was inoculated into ≈40 volumes of fresh King’s B liquid medium containing rifampicin and incubated overnight at 28 °C with shaking at 150 rpm. The bacterial pellet was collected by centrifugation at 3000 × g for 10 min and washed three times with 20 mM Tris-HCl (pH 8.0). To shear the flagella, the cell suspension was intensively blended for 2 min at 8000 rpm using a Polytron PT 10/35 homogenizer, and then centrifuged at 5000 × g for 10 min to pellet the cells31. The resulting supernatant was centrifuged at 100,000 × g for 1 h to pellet the flagella, and the resulting pellet then was suspended in 0.1 M glycine-HCl buffer (pH 2.0) for dissociation32. The flagellin protein was separated by a second round of centrifugation at 100,000 × g for 1 h, and the pH of the resulting supernatant (flagellin solution) was adjusted to 7.0 with NaOH.
Measurement of ROS production
ROS production was measured as described previously33. Briefly, using a 4-mm-diameter cork borer, leaf disks were excised from soil-grown Arabidopsis leaves and individual punches were floated, adaxial side up, in 200 µL water per well in a 96-well white plate; the plate then was incubated overnight at room temperature. On the following day, the water was replaced with 100 µL of an aqueous suspension containing 100 nM flg22, flg22C6 or flagellin, 34 μg/mL luminol (Sigma-Aldrich), and 20 μg/mL horseradish peroxidase (HRP; Sigma-Aldrich). The relative luminescence unit (RLU) of each well was measured using a VICTOR Nivo Multimode Plate Reader (PerkinElmer) for 60 min. For the analysis of ROS production delay, Tmax RLU was defined as the time to reach the maximum RLU, and ΔTmax RLU was obtained by subtracting Tmax RLU of WT from that of sbt5.2 sbt1.7. Statistical analyses were performed using Prism software (version 9; GraphPad).
Flagellin fragmentation in Arabidopsis submerged culture medium
Submerged culture medium was collected by decantation. Flagellin (final concentration, 100 nM) was added to 1 mL of submerged culture medium, and the mixture was incubated without shaking under continuous light at 22 °C for 4 h. Digested peptides were extracted from the incubated mixture using o-chlorophenol as described previously12, with some modifications. Briefly, N-ethylmorpholine (NEM) (final concentration, 1.0%) and 500 µL of o-chlorophenol saturated with 1% NEM were added to each sample. After shaking for 1 min at room temperature, each sample was centrifuged at 9390 × g for 20 min. The aqueous phase was removed and the phenolic phase was recentrifuged at 9390 × g for 10 min. The phenolic phase then was collected, and peptides were precipitated with 10 volumes of acetone and 0.1% Ficoll at −30 °C overnight. After centrifugation at 9390 × g for 10 min, the resulting pellet was rinsed twice with acetone and then dried in vacuo. The precipitate then was dissolved in 10 µL of water. This solution was diluted 20-fold with 2% acetonitrile (containing 0.1% TFA) and then desalted using a GL-Tip SDB (GL Science) according to the manufacturer’s instructions. After vacuum concentration, the samples were dissolved in 20 µL of 2% acetonitrile (containing 0.1% TFA) and entire 20 µL was analyzed by nano-LC-MS/MS.
Flagellin fragmentation by Asp-N
For Asp-N digestion, 3 µg of flagellin was reconstituted in 17 µL of 2 M urea containing 50 mM ammonium bicarbonate, and digested by 0.08 µg of Asp-N (Promega) at 37 °C for overnight. The digestion was stopped by adding 1/20 volume of 20% TFA, and digested peptides were desalted using a GL-Tip SDB (GL Science) according to the manufacturer’s instructions. After vacuum concentration, the samples were dissolved in 20 µL of 2% acetonitrile (containing 0.1% TFA) and 1.5-µL aliquots were analyzed by nano-LC-MS/MS.
Proteomic comparison between Arabidopsis submerged culture medium and leaf apoplastic fluid
The Arabidopsis leaf apoplastic fluid was collected as described previously34 with some modifications. Briefly, 12 completely expanded leaves were vacuum-infiltrated with water. The infiltrated leaf samples were packed in a syringe and apoplastic fluid was collected by centrifugation at 410 × g for 8 min. Aliquots (50 µL) of leaf apoplastic fluid were mixed with equal volume of 8 M urea containing 250 mM Tris-HCl (pH 8.5). For proteomic analysis of Arabidopsis submerged culture medium, proteins were extracted from 1 mL of submerged culture medium by o-chlorophenol extraction followed by acetone precipitation as described above, and the pellets were resuspended in 10 µL of water. 4-µL aliquots were diluted 5-fold with 8 M urea containing 250 mM Tris-HCl (pH 8.5). Protein sample solutions were reduced with 25 mM tris(2-carboxyethyl)phosphine (TCEP) at 37 °C for 15 min, alkylated using 25 mM iodoacetamide at 37 °C for 30 min in the dark with gentle shaking at 1000 rpm following the standard protocol. Proteins were then digested with 2 µg of Lys-C (FUJIFILM Wako, Japan) at 37 °C for 3 h. This Lys-C digest was diluted to a urea concentration of 2 M with 100 mM Tris-HCl (pH 8.5), followed by digestion with 2 µg of trypsin (Promega) at 37 °C overnight. The digestion was stopped by adding 1/20 volume of 20% TFA, and digested peptides were desalted using a MonoSpin C18 spin column (GL Science) according to the manufacturer’s instructions. After vacuum concentration, the samples were dissolved in 20 µL of 2% acetonitrile (containing 0.1% TFA) and 7.25-µL aliquots were analyzed by nano-LC–MS/MS.
Detection of proteolytic activity at the C-terminus of the flg22 domain using Nma-flg[47-56]-Dnp
A submerged culture medium was prepared as described above. Where applicable, the submerged culture medium was subjected to heat inactivation by incubation at 100 °C for 30 min. Nma-flg[47-56]-Dnp was added to the submerged culture medium or heat-inactivated submerged culture medium to a final concentration of 4 µM in a total volume of 100 µL per well in a 96-well white plate. The fluorescence of each well then was measured using a VICTOR Nivo Multimode Plate Reader equipped with a 340/20 nm excitation filter and a 435/20 nm emission filter; measurements were conducted every hour for 20 h.
Native two-dimensional electrophoresis
A volume (100 mL) of submerged culture medium from WT Arabidopsis was concentrated 10-fold by rotary evaporation. The concentrated sample was centrifuged at 11,900 × g for 5 min and the resulting supernatant was dialyzed (overnight at 4 °C) against 3 L of RO water using a Spectra/Por 7 membrane (molecular weight cut-off (MWCO), 10 kDa; Repligen). The water then was replaced with fresh water, and the sample was dialyzed for another night. The dialyzed sample was centrifuged at 11,900 × g for 5 min, and the resulting supernatant was lyophilized. The lyophilized powder was used for the native two-dimensional electrophoresis. The first dimension and gel equilibration were performed as described previously35, with minor modifications. Briefly, separation in the first dimension, using native isoelectric focusing (IEF), was performed using an Ettan IPGphore II (Cytiva). IPG strip (Immobiline DryStrip gel pH 3-10 NL, 7 cm; Cytiva) that was rehydrated in 125 µL rehydration solution for 14 h. The lyophilized powder derived from 100 mL of spent medium from submerged culture of WT Arabidopsis was added to the rehydration solution before use in rehydration of the strip. IEF was run using the following program: 500 V for 30 min, 1000 V for 30 min, and 5000 V for 100 min. After the first-dimensional separation, the IPG strip was equilibrated for 15 minutes at 4 °C. The equilibrated IPG strip then was rinsed with Native PAGE Running Buffer (prepared from Native PAGE Running Buffer (20×) stock (Thermo Fisher Scientific)) and placed onto the second-dimensional gel. The second-dimensional gel was prepared by removing the wells from a commercially obtained gel (Native PAGE 4 to 16%, Bis-Tris, 1.0 mm, Mini Protein Gel, 15 well; Thermo Fisher Scientific). The anode buffer was Native PAGE Running Buffer; the cathode buffer was Native PAGE Running Buffer supplemented with 20 µg/mL Coomassie Brilliant Blue (CBB) G-250. Separation in the second dimension, by blue native PAGE, was conducted at 150 V for 90 min.
In-gel proteolysis assay using fluorescence-quenching peptide substrate
The two-dimensional electrophoresis gel was rinsed in 25 mM 2-(N-morpholino)ethanesulfonic acid (MES) -KOH (pH 5.7) and diced using a SAINOME plate as described previously16. The gel was separated into 224 fragments. A solution of 40 µM Nma-flg[47-56]-Dnp in 25 mM MES-KOH (pH 5.7) then was dispensed at 60 µL/well into the SAINOME-plate, and the fluorescence of each well was measured as described above.
In-gel protein digestion
The fluorescence-positive gel fragments, as well as control fluorescence-negative gel fragments, were collected from the respective wells of the SAINOME plate and fixed in 20% methanol and 7.5% acetic acid for 1 h. The fixed gel fragments were used for in-gel protein digestion, performed as described previously36, with some modifications. Briefly, the fixed gel fragments were washed with MilliQ water, then with 50 mM ammonium bicarbonate in 50% acetonitrile and dried with 100% acetonitrile. The fragments were incubated in 50 mM tris(2-carboxyethyl)phosphine (TCEP) at 60 °C for 10 min. Iodoacetamide was added to a final concentration of 100 mM, and the gel fragments were incubated at room temperature in the dark for 1 h. After two washes, the gel fragments were dried with acetonitrile, and a solution of Lys-C/Trypsin (0.01 mg/mL; Promega) was added to swell the gel fragments. Then, 50 mM ammonium bicarbonate was added, and the fragments were incubated at 37 °C for 16 h. The resulting supernatants were collected, acidified to pH 2 with 20% TFA, then desalted using a Mono Spin C18 column (GL Sciences). After vacuum concentration, the samples were dissolved in 20 µL of 2% acetonitrile (containing 0.1% TFA) and 7.25-µL aliquots were analyzed by nano-LC–MS/MS.
Mass spectrometry
Nano-LC–MS/MS analysis was performed using a system combining a Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific) with a Dionex U3000 gradient pump (Thermo Fisher Scientific). For proteomic analysis, samples were loaded onto a trap column (L-column ODS [300 µm I.D. × 5 mm], CERI, Japan) and washed with 2% acetonitrile (0.1% TFA). Peptides were subsequently eluted from the trap column and separated on a nano-LC capillary column (NTCC-360/100 [100 µm I.D. × 125 mm], Nikkyo Technos, Japan) with a gradient of 5–40% acetonitrile (containing 0.5% acetic acid) over 100 min at a flow rate of 500 nL/min. For identification of in-gel digested proteins, nano-LC was operated with a gradient of 5–40% acetonitrile (containing 0.5% acetic acid) over 40 min at a flow rate of 500 nL/min. The Q Exactive mass spectrometer was operated in data-dependent acquisition mode with dynamic exclusion enabled (5 s for flagellin fragmentation analysis and identification of in-gel digested proteins; 10 s for identification of on-beads digested proteins; 20 s for proteomic analysis of Arabidopsis submerged culture medium and leaf apoplastic fluid) with a capillary temperature of 250 °C. Survey full-scan MS spectra (mass range 350–1800) were acquired at a resolution of 70,000 with the maximum injection time set at 60 ms. The 10 most-intense peptide ions in each survey scan with a charge state between 2+ and 4+ were selected for MS/MS fragmentation. MS/MS scans were performed by higher-energy collisional dissociation (HCD) with the normalized collision energy set to 27 (for conventional proteomics) or 35 (for flagellin fragmentation analysis). The MS/MS raw files were processed and analyzed with Proteome Discoverer 2.3 (Thermo Fisher Scientific) using the SEQUEST HT algorithm, searching against the TAIR10 Arabidopsis protein database, Nbe.v1 N. benthamiana protein database37, or flagellin. Precursor mass tolerance and fragment mass tolerance were set at 10 ppm and 0.02 Da, respectively, and for up to 2 missed cleavages were allowed. Oxidation (M) was set as the variable modification, and carbamidomethylation (C) as the fixed modification. For the identification of apoplastic proteases, proteases with signal peptides were extracted from the resulting high-confidence master protein list using TAIR Gene Ontology (GO) annotations and SignalP 6.038.
For peptide cleavage assay, nano-LC–MS analysis was performed using a system combining the LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) with a DiNa-M splitless nano-HPLC system (KYA Technologies). A trap column (HiQ sil C18W-3, 500 µm I.D. × 1 mm; KYA Technologies) and an analytical column (MonoCap C18 Fast-Flow, 100 µm I.D. × 150 mm; GL Sciences) were used. Nano-LC was operated with a gradient of 2–50% acetonitrile (containing 0.1% formic acid) over 30 min at a flow rate of 500 nL/min. For proteolytic assay of Nma-flg[47-56]-Dnp, nano-LC was operated with a gradient of 30–80% acetonitrile (containing 0.1% formic acid) over 30 min. MS analysis was performed in positive ion mode with a capillary temperature of 160 °C. Mass spectra were obtained by scanning from m/z 350 to m/z 1800. For structural confirmation, MS/MS analysis was performed in positive ion mode with HCD at 35 V.
Transient expression of proteases in N. benthamiana
For the transient expression of the His-tagged proteases SBT5.2-H6, SBT1.7-H6, and SAP2-H6, individual cDNAs were obtained by RT-PCR using total RNA isolated from Arabidopsis leaves and gene-specific primers (Supplementary Data 4). The resulting fragments were cloned separately into the binary vector pBI121 using the In-Fusion HD cloning kit (Clontech). The resulting constructs were transformed into Agrobacterium tumefaciens strain C58C1 (pMP90). Agrobacterium strains containing plasmids encoding SBT5.2-H6 or SBT1.7-H6, or a strain containing a p19 silencing suppressor expression construct, was grown at 28 °C on LB agar containing 10 µg/mL rifampicin, 50 µg/mL gentamycin, and 50 µg/mL kanamycin. For each strain, following growth, an isolated colony was inoculated into 5 mL LB liquid containing 100 µM rifampicin, 100 µM gentamycin, and 100 µM kanamycin and incubated overnight at 28 °C with shaking. Bacteria were collected by centrifugation at 2000 × g for 5 min at room temperature, and cells from the resulting pellet were employed for agroinfiltration in N. benthamiana leaves, performed as described previously39. Five days after agroinfiltration, apoplastic fluids were collected by vacuum infiltration with 50 mM sodium phosphate buffer (pH 7.4) containing 300 mM NaCl; the resulting fluids were cleared by centrifugation at 1500 × g for 10 min. An aliquot (5 µL) of each resulting supernatant (the apoplastic fluid) was used for western blotting. The proteins were separated by SDS-PAGE on a 10% acrylamide gel (REAL GEL PLATE, 10%, 16-well; Bio Craft) and transferred to a polyvinylidene difluoride (PVDF) membrane (Immobilon-P, Millipore). The membrane was blocked with Blocking One (Nacalai Tesque) at room temperature for 1 h, then incubated at room temperature for 30 min with HRP-conjugated anti-His-tag antibody (1:5000; Anti-His-Tag mAb-HRP-DirecT; MBL). The HRP signal was detected using ECL Prime Western Blotting Detection Reagent (Cytiva) and an ImageQuant LAS 4000 mini camera system (Cytiva).
Flagellin fragmentation by SBT5.2-H6 or SBT1.7-H6
Aliquots (200 µL) of apoplastic fluids from agroinfiltrated leaves were diluted in buffer A (20 mM sodium phosphate buffer (pH 7.4), 500 mM NaCl, and 10 mM imidazole in MilliQ water) to a volume of 500 µL. Each sample then was loaded onto a Ni-Sepharose-packed-tip column equilibrated with 1 mL of Buffer A. The tip columns were prepared as follows: A chelating Sepharose-packed column (HiTrap chelating HP 1 mL; Cytiva) was washed with 5 mL of water, and 0.5 mL of water containing 0.1 M NiSO4 was loaded onto the column. After washing with 5 mL of water, the column was equilibrated with 10 mL of buffer A. Ni-Sepharose beads were recovered from the dismantled column and suspended in 1.5 mL of Buffer A. To prepare protease-immobilized beads, a small amount of cotton was packed into a 200-µL micropipette tip, onto which 50 µL of suspended Ni-Sepharose beads were loaded. These micropipette-tip columns were loaded with the apoplast samples, then washed with 1 mL of Buffer A, and the Ni-Sepharose beads carrying immobilized proteases were collected from the micropipette-tip columns. Beads harboring immobilized SBT5.2-H6, SBT1.7-H6 or SAP2-H6 were used for the proteolysis assay. The assays were performed in a 200-µL reaction mixture containing 25 mM MES-KOH (pH 5.7), 2 µM flagellin or 500 nM synthetic peptides, and 10 µL of protease immobilized beads. The reaction mixtures were rotated for 1 h at room temperature. Reactions were terminated by the addition of 1/10 volume of 10% TFA. For flagellin proteolysis assay, aliquots (50 μL) of the quenched reaction mixtures were desalted using a GL-Tip SDB (GL Science) according to the manufacturer’s instructions. After vacuum concentration, the samples were dissolved in 20 µL of 2% acetonitrile (containing 0.1% TFA) and 3.33-µL aliquots were analyzed by nano-LC-MS/MS. For peptide proteolysis assay, aliquots (5 μL) of the quenched reaction mixtures then were analyzed by nano-LC-MS/MS.
Protein purity of SBT5.2-H6 and SBT1.7-H6
The protease-immobilized Ni-Sepharose beads were suspended in 20 μl of 8 M urea containing 250 mM Tris-HCl (pH8.5), reduced with 25 mM tris(2-carboxyethyl)phosphine (TCEP) at 37 °C for 15 min and alkylated using 25 mM iodoacetamide at 37 °C for 30 min in the dark, both with shaking at 1200 rpm following the standard protocol. After dilution to a urea concentration of 2 M with 50 mM Tris-HCl (pH 8.5) followed by addition of 1 mM CaCl2, proteins on beads were directly digested with 0.1 μg trypsin (Promega) with shaking at 1200 rpm at 37 °C overnight. The digestion was stopped by adding 1/20 volume of 20% TFA, and then digested peptides were desalted using a GL-Tip SDB (GL Science) according to the manufacturer’s instructions. After vacuum concentration, the samples were dissolved in 20 µL of 2% acetonitrile (containing 0.1% TFA) and 7.25-µL aliquots were analyzed by nano-LC–MS/MS.
Statistics and reproducibility
All statistical analyses were performed using Prism software (version 9; GraphPad). No statistical method was used to predetermine sample size. No data were excluded from the analysis. Samples were grown under the same conditions and randomly allocated in the growth chamber. Experimental plant material was collected randomly without any bias. Investigators were not blinded to allocation during the experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the results of this study are available in the paper and in the Supplementary Information. All other data that supporting the results of this study are available from the corresponding author upon request. The raw MS data generated in this study have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository under accession code PXD044549. Source data are provided with this paper.
References
Boller, T. & Felix, G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406 (2009).
Felix, G., Duran, J. D., Volko, S. & Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276 (1999).
Gómez-Gómez, L. & Boller, T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011 (2000).
Zipfel, C. et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767 (2004).
Chinchilla, D., Bauer, Z., Regenass, M., Boller, T. & Felix, G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18, 465–476 (2006).
Sun, Y. et al. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342, 624–628 (2013).
Fliegmann, J. & Felix, G. Immunity: flagellin seen from all sides. Nat. Plants 2, 1–2 (2016).
Komoriya, K. et al. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol. Microbiol. 34, 767–779 (1999).
Meindl, T., Boller, T. & Felix, G. The bacterial elicitor flagellin activates its receptor in tomato cells according to the address-message concept. Plant Cell 12, 1783–1794 (2000).
Buscaill, P. et al. Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides. Science 364, eaav0748 (2019).
Rawlings, N. D. et al. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 46, D624–D632 (2018).
Ohyama, K., Ogawa, M. & Matsubayashi, Y. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 55, 152–160 (2008).
Yamamoto, M. et al. Identification of genes involved in the glycosylation of modified viosamine of flagellins in Pseudomonas syringae by mass spectrometry. Genes 2, 788–803 (2011).
Matayoshi, E. D., Wang, G. T., Krafft, G. A. & Erickson, J. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science 247, 954–958 (1990).
Bickett, D. M. et al. A high throughput fluorogenic substrate for interstitial collagenase (MMP-1) and gelatinase (MMP-9). Anal. Biochem. 212, 58–64 (1993).
Ino, Y. et al. Protein fractionation for proteomics using the SAINOME-plate. J. Electrophor. 62, 11–15 (2018).
Chinchilla, D. et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500 (2007).
Schardon, K. et al. Precursor processing for plant peptide hormone maturation by subtilisin-like serine proteinases. Science 354, 1594–1597 (2016).
Engineer, C. B. et al. Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature 513, 246–250 (2014).
Stührwohldt, N., Ehinger, A., Thellmann, K. & Schaller, A. Processing and formation of bioactive CLE40 peptide are controlled by posttranslational proline hydroxylation. Plant Physiol. 184, 1573–1584 (2020).
Serrano, I. et al. A non canonical subtilase attenuates the transcriptional activation of defence responses in Arabidopsis thaliana. eLife 5, e19755 (2016).
Ngou, B. P. M., Ahn, H. K., Ding, P. & Jones, J. D. G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115 (2021).
Yuan, M. et al. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109 (2021).
Parys, K. et al. Signatures of antagonistic pleiotropy in a bacterial flagellin epitope. Cell Host Microbe 29, 620–634.e9 (2021).
Cai, R. et al. The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathog. 7, e1002130 (2011).
Clarke, C. R. et al. Allelic variation in two distinct Pseudomonas syringae flagellin epitopes modulates the strength of plant immune responses but not bacterial motility. N. Phytol. 200, 847–860 (2013).
Hind, S. R. et al. Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat. Plants 2, 16128 (2016).
Stührwohldt, N. & Schaller, A. Regulation of plant peptide hormones and growth factors by post-translational modification. Plant Biol. 21, 49–63 (2019).
Stintzi, A. & Schaller, A. Biogenesis of post-translationally modified peptide signals for plant reproductive development. Curr. Opin. Plant Biol. 69, 102274 (2022).
Rautengarten, C. et al. A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J. 54, 466–480 (2008).
Nishiyama, M. & Kojima, S. Bacterial motility measured by a miniature chamber for high-pressure microscopy. Int. J. Mol. Sci. 13, 9225–9239 (2012).
Taguchi, F. et al. Differential effects of flagellins from Pseudomonas syringae pv. tabaci, tomato and glycinea on plant defense response. Plant Physiol. Biochem. 41, 165–174 (2003).
Chakravarthy, S., Velasquez, A. C., Ekengren, S. K., Collmer, A. & Martin, G. B. Identification of Nicotiana benthamiana genes involved in pathogen-associated molecular pattern-triggered immunity. Mol. Plant Microbe Interact. 23, 715–726 (2010).
Pajerowska-Mukhtar, K. M. et al. The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr. Biol. 22, 103–112 (2012).
Sevcenco, A. M. et al. Development of a generic approach to native metalloproteomics: application to the quantitative identification of soluble copper proteins in Escherichia coli. J. Biol. Inorg. Chem. 14, 631–640 (2009).
Rosenfeld, J., Capdevielle, J., Guillemot, J. C. & Ferrara, P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203, 173–179 (1992).
Kurotani, K. et al. Genome sequence and analysis of Nicotiana benthamiana, the model plant for interactions between organisms. Plant Cell Physiol. 64, 248–257 (2023).
Teufel, F. et al. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 40, 1023–1025 (2022).
Doll, N. M. et al. A two-way molecular dialogue between embryo and endosperm is required for seed development. Science 367, 431–435 (2020).
Acknowledgements
This research was supported by a Grant-in-Aid for Scientific Research (S) (No. 23H05477 to Y.M.), Grant-in-Aid for Transformative Research Areas (A) (No. 20H05907 to Y.M.) and the Nagoya University Interdisciplinary Frontier Fellowship (to S.M.) supported by Nagoya University and Japan Science and Technology Agency (JST) (No. JPMJFS2120).
Author information
Authors and Affiliations
Contributions
Y.M. and H.S. conceived this project and S.M., H.S., and Y.M. designed the experiments. S.M. performed all the biological experiments and S.M., H.S., and Y.M. interpreted the results. S.N. and K.K. performed the nano-LC-MS/MS analysis. M.N. and Y.T. provided support for handling Pst DC3000 and collecting apoplastic fluid from Arabidopsis leaves. S.M. and Y.M. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Yuhta Nomura, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Matsui, S., Noda, S., Kuwata, K. et al. Arabidopsis SBT5.2 and SBT1.7 subtilases mediate C-terminal cleavage of flg22 epitope from bacterial flagellin. Nat Commun 15, 3762 (2024). https://doi.org/10.1038/s41467-024-48108-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-48108-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.