Abstract
MIKCC-type MADS-box (MIKCC) genes encode transcription factors that have crucial roles in controlling floral organogenesis and flowering time in plants. Although this gene family has been well characterized in many plant species, its evolutionary and comprehensive functional analysis in rose is lacking. In this study, 58 non-redundant MIKCC uni-transcripts were extensively identified from rose transcriptomes. Phylogenetic analysis placed these genes into 12 clades with their Arabidopsis and strawberry counterparts, and revealed that ABCDE model (including AP1/FUL, AP3/PI, AG, and SEP clades), and SOC1 and AGL6 clade genes have remarkably expanded in Rosa chinensis, whereas genes from the FLC and AGL17 clades were undetectable. Sequence alignments suggest that the AP3/PI clade may contribute to more specific functions in rose due to a high variation of amino acid residues within its MADS-box domains. A comparative analysis of gene expression in specific floral organ differentiation stages and floral organs between R. chinensis cv. Old Blush and the closely related mutant genotype R. chinensis cv. Viridiflora (floral organs mutated into leaf-like structures) further revealed the roles of ABCDE model genes during floral organogenesis in rose. Analysis of co-expression networks provided an overview of the regulatory mechanisms of rose MIKCC genes and shed light on both the prominent roles of AP3/PI clade genes in floral organogenesis and the roles of RcAGL19, RcAGL24, and RcSOC1 in regulating floral transition in rose. Our analyses provide an overall insight of MIKCC genes in rose and their potential roles in floral organogenesis.
Similar content being viewed by others
Introduction
MADS-box genes, which are defined by the presence of a highly conserved DNA-binding MADS-box domain in the N-terminal region of a protein sequence, encode a family of transcription factors (TFs) that are involved in various aspects of plant growth and developmental processes, especially floral organogenesis and flowering time1,2,3. Based on phylogenetic analysis of MADS-box genes in eukaryotes, two major lineages, termed type I and type II, were distinguished1. MIKCC-type MADS-box (MIKCC) genes are plant specific and some, which were initially and are best known as floral organ identity genes, comprise the majority of genes from the type II lineage4,5. In the model plant Arabidopsis thaliana, MIKCC genes can be phylogenetically clustered into 12 clades, i.e., AP1/FUL, AP3/PI, AG, SEP, SOC1, SVP, AGL6, AGL12, AGL15, AGL17, FLC, and BS clades, along with their counterparts from other plants6,7. According to the well-known ABCDE model or ‘floral quartets’ model of floral development8,9, the floral organ identity genes, which are mainly derived from the AP1/FUL, AP3/PI, AG, and SEP clades of the MIKCC gene family, can be further divided into five different classes, termed class A (corresponding to the AP1 genes from the AP1/FUL clade), B (AP3/PI), C (AG), D (STK genes from the AG clade), and E (SEP) genes, based on their different homeotic functions in floral organogenesis. In this model, A refers to sepals, A + B + E refers to petals, B + C + E refers to stamens, C + E refers to carpels, and D (sister clade of the C class) refers to ovules5,10. ABCDE model genes have been functionally characterized, mainly through homeotic flower mutants11, including the celebrated homeotic mutants from A. thaliana and Antirrhinum majus5,12. Loss of function or ectopic expression of A, B, C, D, and E genes usually leads to homeotic conversion of floral organs and floral aberrations2,3,12,13,14,15. For example, the loss of SEP clade genes in sep quadruple mutants (sep1/2/3/4) can turn the flower into leaf-like structures16,17, whereas ectopic expression of AG genes can lead to partial homeotic transformations of the sepals and petals toward carpels and stamens, respectively13. In addition, among the floral organ identity genes, several crucial MIKCC genes have been found to modulate flowering time in Arabidopsis2,3,18,19,20,21,22,23. Specifically, FLC, SOC1, AGL19, and AGL24 genes have been found to be vital components of the vernalization pathway23. Nevertheless, the regulatory mechanisms of MIKCC genes in regulating flowering transition and floral organogenesis have not been fully elucidated.
MADS-box genes are believed to have originated from gene duplications that initially occurred in the lineage of the most recent common ancestor of extant eukaryotes24. Thus far, MADS-box genes have been identified in nearly all groups of eukaryotes, including plants, animals, and fungi1. However, the number and functional diversity of MADS-box genes appear to have increased remarkably during the evolution of land plants, especially flowering plants (angiosperms), with more than 100 genes in Arabidopsis (A. thaliana), rice (Oryza sativa), and poplar (Populus trichocarpa)7,25,26,27. The expansion and functional diversification of plant MADS-box genes are tightly linked to the evolution of plant body plans and life history strategies1,3,28,29,30. Novel MADS-box genes and/or new functions have typically arisen in specific lineages of plants or even specific species3 and are closely linked to the evolution of novel structures (such as seeds, flowers, and fruits) and habitat-specific adaptations31,32. For instance, most subclades of MIKCC genes appear to have originated from ancestral seed plants1. In addition, within this group, the AP1/SQUA and SEP subclades seem to be the first to have duplicated or expanded in flowering plants, suggesting their key roles in the origination and subsequent morphological evolution of flowers1,3,33. In addition, six DORMANCY-ASSOCIATED MADS-BOX (DAM) genes that originated from a series of tandem duplications in peach (Rosaceae) have been associated with floral bud dormancy and thereby seasonal flowering34. Contrarily, multiple losses of MIKCC genes have also been identified, especially in TM8-like and FLC-like gene clades that might have been adaptive during plant evolution35. As a consequence of the frequent expansion, and gene loss and diversification of the MADS-box gene family, a full understanding of the biological functions of MADS-box genes in flower development will not only require understanding of the regulatory mechanisms of the MADS-box genes but will also require their identification and functional characterizations in a variety of species.
Roses (genus Rosa), which belong to the large Rosaceae family36, are one of the most commonly cultivated ornamental plants worldwide and are highly popular as cut flowers with a wide range of flower types, fragrances, and colors. The continuous flowering (CF) trait of Rosa chinensis implies differences in its flowering mechanisms and may subsequently lead to differences in floral organ development, as these are closely related proceses36,37. Although a few orthologs of the MIKCC genes, such as RhAPETALA3 (RhAP3), RhPISTILLATA (RhPI), RhAGAMOUS (RhAG), and RhSHATTERPROOF, which are mainly involved in floral organogenesis, have been isolated from Rosa hybrida cultivars and functionally characterized to some extent38,39,40,41,42,43, a comprehensive understanding of the role of MIKCC genes in controlling rose flowering transition and floral organogenesis at the genome level is still lacking.
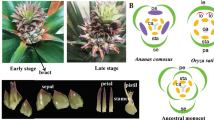
Commonly known as the “green rose,” R. chinensis cv. Viridiflora is a homeotic flower mutant of Chinese old rose44, with the floral organs mutated into leaf-like structures (called ‘phyllody’) instead of true petals, stamens, and pistils45 (Supplementary File S1). Similar phenotypes have also been observed in Arabidopsis16,17 and Catharanthus roseus46,47—these phenotypes can be caused by quadruple mutants (sep1/2/3/4) of SEP clade genes or by phytoplasma infection. However, both of these potential causes of phyllody were contradicted by the research of Yan et al.48, in which reciprocal grafting experiments between Viridiflora and R. chinensis cv. Old Blush did not cause the phyllody phenotype and expressions of RcSEP1 and RcSEP3 were detected. Thus, the mechanism of floral organogenesis of Viridiflora remains a mystery. In addition to having an interesting floral phenotype, R. chinensis cv. Viridiflora is closely related to R. chinensis cv. Old Blush48 (Supplementary File S1), which is a very important common ancestor of commercial modern roses and is the original source of the CF and tea scent traits that exist in modern roses. R. chinensis cv. Old Blush is also widely used in many breeding programs and has been suggested as a model cultivar for rose research36,43. These two closely related genotypes provide an excellent system for studying the homeotic functions of MIKCC genes in the floral organogenesis of rose.
In our study, a comprehensive identification of MIKCC genes was performed based on high-quality transcriptome datasets for rose (two datasets were previously produced by our group)43,48,49,50. Phylogenetic analysis revealed the remarkable expansion of rose ABCDE model genes, whereas FLC and AGL17 clade genes were not detected. Comparative gene expression analyses between Old Blush and its homeotic flower mutant Viridiflora across the major floral organ differentiation stages and different floral organs confirmed the roles of MIKCC genes in rose floral organogenesis. Co-expression networks of MIKCC genes were generated from transcriptome data, revealing the prominent roles of AP3/PI clade genes in floral organogenesis and of RcAGL19, RcAGL24, and RcSOC1 in regulating floral transition in rose. Our analyses provide a fundamental framework for the biological functions of MIKCC genes in rose floral organogenesis and flowering transition, and will facilitate the functional characterization of individual MIKCC genes in rose.
Results
Transcriptomic identification of MIKCC genes in rose
After extensively searching the integrated transcriptome database with two different HMM profiles of the MADS-box domain (see Materials and Methods section), a total of 58 non-redundant uni-transcripts were assigned as rose MIKCC genes (Supplementary File S2). This number is 1.49 (58/39) times and 1.38 (58/42) times higher than that in Arabidopsis and strawberry, respectively, indicating that MIKCC genes have expanded in rose. The rose MIKCC genes were assigned names consistent with Arabidopsis MIKCC gene names, based mainly on their phylogenetic relationships (Supplementary File S2). As the draft genome sequence of a wild rose, Rosa multiflora (once-flowering), was very recently released51, we searched the best hit orthologs for the MIKCC genes we identified from R. chinensis (CF). Based on the E-value and careful checking of the sequence similarity, we believe that some obvious differences exist in the MIKCC gene family between those two species. For example, RcAP1.1, RcAP1.2, RcAGL24.2, RcAGL6.1, RcAGL6.2, and RcAGL104.2 have low E-values with respect to their best hit in R. multiflora, suggesting they might be genes specific to R. chinensis (Supplementary File S2). In addition, reciprocal best hit orthologs of rose MIKCC genes were identified in Arabidopsis and rice (O. sativa). Eighteen and 14 rose MIKCC genes were detected to have their reciprocal best hit orthologs in Arabidopsis and rice, respectively, suggesting that these genes might be functionally conserved between plant species. Furthermore, the highest count number and fragments per kilobase of transcript per million mapped reads (FPKM) values from the currently available RNA sequencing (RNA-seq) data were presented to provide an overview of the expression levels of the rose MIKCC genes identified in our study (Supplementary File S2).
Phylogenetic analysis of rose MIKCC proteins
To examine the phylogenetic relationships among MIKCC proteins and to group them within established clades, a phylogenetic tree was constructed for MIKCC proteins from Arabidopsis, strawberry, and rose using MAFFT v7.037b with the specific sequence alignment parameter (E-INS-I) (see Materials and Methods section) (Fig. 1a). All the proteins were phylogenetically classified into 12 clades along with the established clades in Arabidopsis (the MIKC* were placed as the root branch), which included the AP1/FUL, AP3/PI, AG, SEP, SOC1, SVP, AGL6, AGL12, AGL15, AGL17, FLC, and BS clades. Interestingly, there is no rose MIKCC protein classified into the AGL17 clade present in Arabidopsis and strawberry, and neither rose nor strawberry has genes in the FLC clade (Fig. 1b, I cluster), indicating that these clades might have suffered extensive gene loss during their evolution. In contrast, the numbers of MIKCC proteins in eight clades, i.e., the AP1/FUL (7/4), AP3/PI (9/2), AG (7/4), SEP (9/4), AGL6 (5/2), and SOC1 (12/6) clades in rose (Fig. 1b, II cluster), and AGL15 (9/2) and SVP (8/2) in strawberry (Fig. 1b, III cluster), were much higher than their counterparts in Arabidopsis, indicating that these clades expanded remarkably after the split of Arabidopsis and rosaceae. By contrast, the number of AGL12 (1) and BS (2) clades (Fig. 1b, IV cluster) was the same in all three species we examined, suggesting that these clades could be functionally conserved in plants.
a Maximum likelihood (ML) phylogenetic trees of rose, Arabidopsis and strawberry MIKCC proteins (see Materials and Methods section for the parameters of tree construction). b Heatmap of the number of different clade genes from rose, Arabidopsis and strawberry. I–IV denote different clusters and the numbers in the colored box denote the number of MIKCC genes. At, A. thaliana, Fv, Fragaria vesca, Rc, R. chinensis. c, d Gene duplication events of the MIKCC gene family detected based on the phylogenetic relationships among rose, Arabidopsis and strawberry. The orange explosive shape denotes the gene duplication event that might have occurred before the split of Arabidopsis and rosaceae, whereas the green one denotes the gene duplication event that might have occurred within rosaceae
Examination of the phylogenetic relationships among MIKCC proteins from Arabidopsis, strawberry and rose revealed 16 closely related paralogous gene pairs in rose, suggesting recent duplication events. Two obvious imprints of gene duplication events were detected in the phylogeny of MIKCC proteins. One such event was detected in the SEP clade (Fig. 1c), from which two clades emerged containing MIKCC proteins from Arabidopsis, strawberry, and rose, suggesting a gene duplication event that contributed to the occurrence of the two groups before the split of Arabidopsis and rosaceae. A second instance of gene duplication was found in the AGL6 clade (Fig. 1d), from which two clades emerged, but they contained only the MIKCC proteins from strawberry and rose, suggesting that a gene duplication event may occurred within rosaceae and contributed to the two groups.
The structural diversity of MADS-box domains of AP1/FUL, AP3/PI, AG, and SEP clade genes in rose
To gain more insights into the functional diversity of AP1/FUL, AP3/PI, AG, and SEP clade genes in rose, which correspond to the homology of A-, B-, C/D-, and E-class genes, respectively, in the ABCDE model, multiple sequence alignments of MADS-box domain proteins were generated for these clades (Fig. 2a) and a homo-dimer tertiary structure of MADS-box domain proteins (RcPI4 as an example) was constructed to provide an overview of their structural features (Fig. 2b). We found that the divergence rates of amino acid residues and the Ka/Ks value within MADS-box domains from AP3/PI clade genes (corresponding to the homology of B- class genes) were much higher than those from AP1/FUL, AG, and SEP clades across different species (Fig. 2c), suggesting that important functional divergence of AP3/PI clade genes may contribute to rose floral organogenesis and development. Furthermore, three amino acid residues in the MADS-box domains of AP3/PI clade genes, i.e., lysine (K) at site 25, asparagine (N) at site 33, and phenylalanine (F) at site 58, were found to be rose specific (Fig. 2a, b), suggesting that these sites could contribute to DNA-binding and dimerization properties of MADS-box TFs in rose. Furthermore, the homo-/heterodimeric states of all the MADS-box domain proteins from Arabidopsis, strawberry, and rose were predicted to provide more insight into their dimerization properties (Supplementary File S3).
a Sequence alignments of MADS-box domain proteins of the AP1/FUL, AP3/PI, AG, and SEP clades. The amino acid residues in the red boxes are rose specific, and those in the green boxes appear to be specific to rosaceae. b The homo-dimer tertiary structure of MADS-box domain proteins; RcPI4 is shown as an example. c Ka/Ks rates within the MADS domains of the AP1/FUL, AP3/PI, AG, and SEP clades
Expression patterns of rose MIKCC genes in developmental processes
MIKCC genes have been suggested to be involved mainly in the regulation of reproductive organ identity and flowering time in plants. To further associate their biological function with specific developmental processes in rose, we re-analyzed their expression patterns across all the typical organs (six vegetative and seven reproductive organs) of Old Blush using the public RNA-seq datasets contributed by Dubois et al.43. As shown in Fig. 3a, five major clusters of gene expression patterns were distinguished: cluster I, containing RcAGL24.1 and RcAGL24.2, was expressed in all tissues except the floral meristem and early floral organs (IMO); cluster II, containing RcAGL42.2, RcSVP2, RcAGL19.3, RcSOC1.1, RcSOC1.4, and RcSOC1.5, was highly expressed in vegetative tissues, the floral meristem (IFL) and IMO but was reduced or nearly undetectable (ND) in the completely differentiated flower (BFL, DET, OFT, SEN) and rose hip from pollination to early pigmentation (CYN); cluster III, containing only one gene (RcAGL104.2), was detected only in open flowers (OFT); cluster IV, containing RcFUL3, RcFUL4, RcSEP4.1, RcSEP4.3, RcSEP4.4, and RcAGL6.5, was mainly characterized by high expression in the floral meristem, and IMO and CYN; and cluster V, containing RcAP3.2, RcAP3.3, RcPI3, RcPI4, RcPI5, RcAG2, RcAG3, RcSHP1.1, RcSHP1.2, RcSEP2.1, and RcSEP3.1, was characterized by high expression in the floral meristem, IMO, and the completely differentiated flower but was relatively reduced or ND in CYN. These gene expression patterns suggest the functional importance and diversification of rose MIKCC genes in flowering transition and floral organogenesis.
a Heatmap of the expression patterns of rose MIKCC genes. The heatmap was generated using MeV software v. 4.9.0 and is accompanied by a color scale corresponding to the gene expression level (FPKM values) from undetected (0.0) to ≥ 6. The sample names correspond to the previous descriptions43—BFL, closed flower; CYN, rose hip from pollination up to early pigmentation; DBO, active axillary buds (vegetative meristem); DET, stamens at microsporogenesis and microgametogenesis stages; FTB, leaves infected with Botrytis cinerea LR18; FTN, young leaves and stems; FTS, leaves from water-stressed plants; IFL, floral bud at floral meristem transition; IMO, floral meristem and early floral organ (sepals, petals, stamens and carpels) development; NDB, dormant axillary buds (vegetative meristem); OFT, open flower; RAC represents white young roots; and SEN, senescent flower. The black box marks the two most important stages (IFL and IMO) of floral transition. b The percentage of all MIKCC genes detected in the transcriptome dataset of Dubois et al.43 The percentages were cumulated according to the total number (58) of the MIKCC genes we identified in our study. The top three clades with a higher percentage of detected genes are highlighted in red
To better understand the biological functions of MIKCC genes in different clades, the number of detected MIKCC genes in the transcriptome dataset of Dubois et al.43 was counted for all 12 clades (58 genes) that we identified from all the transcriptome datasets. Only 7 (58.3%) out of 12 clades were detected to contain MIKCC genes from this dataset (Fig. 3b), and among them the proportion of detected MIKCC genes in the AG, AP3/PI, and SEP clades ranked in the top three. These results may indirectly reflect the remarkable expansion of ABCDE model genes (Fig. 1b) and may provide an indication of the role of MIKCC genes in floral organogenesis and development.
The expression profiles of rose AP1/FUL, AP3/PI, AG, and SEP clade genes in floral organogenesis
To compare the developmental difference between Old Blush and Viridiflora (in which petals, stamens, and pistils are converted into leaf-like organs and the flowers are fully sterile), the dissected structures of floral buds from different floral developmental stages were observed by stereo microscope (see Materials and Methods section). Six developmental stages for rose floral organogenesis were then defined in both the Old Blush (Fig. 4a) and Viridiflora (Fig. 4b) cultivar, including the active axillary bud stage (DBO, named according to the study by Dubois et al.43) and five morphologically distinct flower developmental stages (stage 1–5) named according to another study conducted by Dubois et al.52 Among these stages, DBO represents the vegetative meristem stage, whereas stages 1–4 represent the initiation of sepals (stage 1), petals/petal-like (stage 2), stamens/stamen-like (stage 3), and carpels/carpel-like (stage 4), respectively; in stage 5, all floral organs are apparent, and the hypanthium starts to sink below the perianth and stamens52. A comparison of the corresponding developmental stages between Old Blush and Viridiflora showed that while the order and timing of floral organogenesis between Old Blush and Viridiflora are very similar, the morphological structures of their floral organs are different, especially the morphological structures during the stamen initiation stage (stage 4).
Old Blush and R. chinensis cv. Viridiflora. a, b Rose flower development stages of cultivars Old Blush and Viridiflora, respectively. L, leaves; DBO, vegetative meristem stage; Stage 1–4, initiation stages of sepals (stage 1), petals/petal-like (stage 2), stamens/stamen-like (stage 3), and carpels/carpel-like (stage 4) in Old Blush and Viridiflora; Stage 5, hypanthium starts to sink below the perianth and stamens; Se, sepals; Pe, petals; St, stamens; Pi, pistils. c, d Heatmaps of the relative gene expression levels of MIKCC genes in R. chinensis cv. Old Blush and R. chinensis cv. Viridiflora, respectively, as determined by qRT-PCR. The relative gene expression levels were normalized with the RcTUBULIN, RcGAPDH, and RcTCTP reference genes that were used in previous studies43,48. e Fold change of gene expression for the sepal, petal, stamen, and pistil differentiation phases, which were calculated via comparison with the previous development phase
To further specify the function of the identified ABCDE model genes in floral organogenesis, the expression profiles of 21 selected rose MIKCC genes (the primers of the remaining genes were not specific to our effort) from the AP1/FUL, AP3/PI, AG, and SEP clades were determined using quantitative real-time PCR (qRT-PCR) across different floral organ initiation stages of buds from Old Blush and Viridiflora, with the leaf as a control (Fig. 4c, d). The results show that almost all of the selected genes were observably up-regulated during floral organ initiation in both Old Blush and Viridiflora. However, some genes displayed different expression patterns, e.g., the expression levels of RcFUL2, RcFUL3, and RcFUL5 were much higher in Viridiflora than in Old Blush during the floral organ initiation stages, especially the carpel initiation stage (Fig. 4c, d, stage 4). In addition, the expression of RcAP3.3 was lost in the floral DBO and sepal initiation stages (Fig. 4d, DBO and stage 1) in Viridiflora. Lastly, RcAP1.2, RcAG2 and RcSEP4.3 were expressed only in the leaves of Viridiflora, and no expression of these genes was detected in Old Blush. The different expression profiles of these genes suggest their important roles in floral organ development in rose. Furthermore, cluster analysis of gene expression showed that both clusters II and III contained AP3/PI clade genes (Fig. 5a,b), and AG and AP1 genes were always separated into cluster II and cluster III, respectively, in both Old Blush and Viridiflora. Interestingly, four genes, i.e., RcAP3.2, RcAP3.4, RcPI3 and RcSEP2.1, changed their positions from cluster III to cluster II when the floral organs mutated into leaf-like structures in Viridiflora, reflecting their specific roles in rose floral organ development.
a Floral whorls of R. chinensis cv. Old Blush flowers, including sepals, petals, stamens and carpels. b Floral whorls of R. chinensis cv. Viridiflora. c, d Heatmaps of MIKCC gene expression profiles of Old Blush and Viridiflora, respectively. The white boxes denote cases where expression was not detected
To clarify the temporal accumulation requirement of rose AP1/FUL, AP3/PI, AG, and SEP clade genes for floral organogenesis, the fold changes of gene expression in the sepal (stage 1), petal (2), stamen (3), and carpel (4) initiation stages were calculated via comparison with the previous development stage (Fig. 4e). In Old Blush, RcSEP4.3 (14.6-fold), RcSEP4.4 (9.7), and RcSHP1.1 (3.5) were strongly upregulated during sepal initiation, whereas RcAG2 (− 5.9-fold), RcAG3 (− 4.6), and RcAG4 (− 7.5) were significantly downregulated. Five genes, i.e., RcPI3 (3.8-fold), RcAG4 (3.8), RcSEP3.1 (3.4), RcSEP4.4 (3.2), and RcAP3.3 (2.9), were upregulated during petal initiation; all four genes from the AP3/PI and AG clades, and RcSEP3.1, RcSEP2.1, RcSEP4.3 (4.5-fold), and RcSEP4.4 (4.0) from the SEP clade strongly accumulated during stamen initiation; eight genes, i.e., RcAP1.1 (10.2-fold), RcAP1.2 (4.2), RcAP3.2 (8.2), RcAP3.4 (10.7), RcPI3 (8.8), RcPI5 (6.5), RcAG3 (4.0), and RcAG4 (3.8), were significantly increased during carpel initiation. However, when comparing the MIKCC gene expression patterns of Old Blush with those of Viridiflora (Fig. 4e), we found that the accumulation states of most of the genes we surveyed in Viridiflora were dramatically changed. For example, the prominent expressions of RcAP1.1 and RcAP1.2 changed from the carpel-like initiation stage to the sepal- and stamen-like initiation stages, the prominent expressions of RcAP3.2, RcAP3.4, RcPI3, and RcPI5 were lost during the carpel-like initiation stage and the prominent expressions of RcSEP4.1, RcSEP4.3, and RcSEP4.4 expanded during the petal- and stamen-like initiation stages. These shifts in expression patterns not only indicate the important roles of MIKCC genes in regulating the mutant floral phenotypes in Viridiflora but also highlight their potential homeotic functions in floral organogenesis and development.
The expression profiles of AP1/FUL, AP3/PI, AG, and SEP clade genes in different whorls of rose flowers
To gain more insight into the homeotic functions of the rose homologs of A-, B-, C-, D-, and E-class genes involved in floral organogenesis, the relative expression levels of AP1/FUL, AP3/PI, AG, and SEP clade genes in different floral whorls of both Old Blush and Viridiflora were determined using qRT-PCR (Fig. 5a, b). In the normal flower genotype of Old Blush, three gene clusters (I–III) were formed based on their expression patterns (Fig. 5c). Specifically, cluster I, containing RcAG2, RcAG3, RcAG4, and RcSHP1.1, strongly accumulated in whorl 3 (stamens) and was expressed at very low or undetectable levels in the other whorls of the rose flower (the expressions of RcAG2 and RcAG4 were lost in whorl 1). Cluster II, which contained all of the analyzed genes from clade AP3/PI, most of the SEP clade genes (except RcSEP4.1), and RcFUL2, RcFUL3, and RcFUL5 from the AP1/FUL clade, exhibited relatively higher expression levels in the second (petals) and third whorls but lower expression in the two other whorls (sepals and carpels). The expression level of B-class genes (AP3/PI clade) was remarkably higher in the second and third whorls, indicating that these rose B-class genes may be required for petal and stamen morphogenesis and development. In contrast, cluster III, which contained RcAP1.1, RcAP1.2, RcFUL1, and RcAP4.1, showed a very low expression in whorl 1 (sepals) and whorl 4 (carpels), and no expression in whorls 2 and 3, indicating the specific function of these genes in sepal and carpel morphogenesis and development.
Although the petals, stamens and pistils of Viridiflora have been converted into leaf-like organs, the different whorls of the floral organs can still be distinguished (Fig. 6b). A comparative analysis of gene expression in the different whorls of the floral organs of both Old Blush and Viridiflora (Fig. 6c, d) showed that the regional expression restrictions of the rose A-, B-, C-, D-, and E-class genes in the different whorls of floral organs shifted widely in Viridiflora vs. Old Blush (Fig. 5d). Specifically, the elevated expression levels of four genes, i.e., RcAG2, RcAG3, RcAG4, and RcSHP1.1, which are homologs of C/D-class genes, were extended from whorl 3 to whorl 4 in Viridiflora. Simultaneously, the expressions of RcAG3 and RcAG4 were lost in whorl 2 of Viridiflora, indicating their involvement in petal morphogenesis and development. Similarly, the expression superiority of RcAP3.3, RcPI3, and RcPI5 seemed to be extended to whorl 4 from petals (whorl 2 and whorl 3). Furthermore, the expression peaks of ten genes, i.e., RcAP3.2, RcAP3.4, RcSEP3.1, RcFUL2, RcFUL3, RcFUL5, RcSEP2.1, RcSEP2.2, RcSEP4.3, and RcSEP4.3, changed dramatically from whorl 2 and whorl 3 to whorl 4 in Viridiflora compared with Old Blush, whereas RcAP1.1, RcAP1.2, RcFUL1, RcSEP4.1, and RcSOC1.1 showed increased expression in whorl 2 and whorl 3 in Viridiflora vs. Old Blush. Interestingly, the cluster positions of seven genes (red gene names), i.e., RcAP3.2, RcAP3.3, RcAP3.4, RcPI3, RcPI5, RcSEP2.1, and RcSEP3.1, were changed in Viridiflora, further highlighting their role in floral organogenesis. Taken together, there data collectively demonstrate the expression patterns and important roles of A-, B-, C-, D-, and E-class genes in rose floral organogenesis.
a The distribution of Pearson’s correlation coefficient (PCC) values—PCC 0.77 was selected as the effective threshold in our study (see Materials and Methods section for details). b Co-expression networks of MIKCC genes constructed at a transcriptome-wide level. MIKCC genes are colored in green, and their adjacent genes are colored in orange. c The interactive graph of GO term enrichments of the adjacent genes from co-expression networks
Co-expression networks of rose MIKCC genes at the transcriptome level
To gain more insight into the regulatory relationships of rose MIKCC genes, we constructed co-expression networks of all rose genes at the transcriptome level. An effective PCC threshold of 0.77 was trained to generate the lowest density networks as described in the Materials and Methods section (Fig. 6a). Finally, two independent modules were revealed for the rose MIKCC genes (Fig. 6b). The first module of the network, which represented vegetative tissue development and flower transition, contained seven MIKCC genes (nodes), i.e., RcAGL19.3, RcAGL24.1, RcAGL24.2, RcSOC1.1, RcSOC1.4, RcSOC1.5, and RcAGL42.2, and 559 other adjacent genes (nodes) and 1894 co-expression events (edges) (Fig. 6b, upper panel). All of the detected MIKCC genes can be considered as hubs due to their high connectivity in the network. The frequency of co-expression events for these genes ranged from 22 to 770, and in which the edge numbers of RcSOC1.1 (770 edges), RcSOC1.4 (318), RcAGL24.2 (275), and RcAGL19.3 (261) were remarkably high, suggesting their prominent roles in vegetative tissue development and flower transition in rose. Furthermore, out of 559 other adjacent genes in the network, 234 (41.9%) had at least two connections with detected MIKCC genes. Among them, 22 genes were identified as having the highest connectivity to MIKCC genes with at least five edges, suggesting their important biological functions in regulating flower transition in rose.
The second module of the network, which contained eight MIKCC genes (nodes), 297 other adjacent genes (nodes), and 1039 co-expression events (edges), represented reproductive tissues and flower development due to the high expression of member genes in reproductive tissues (Fig. 6b, lower panel). Interestingly, only genes from the AP3/PI and SEP clades were detected in the network, i.e., the RcAP3.2, RcAP3.3, RcPI3, RcPI4, RcPI5, RcSEP2.1, RcSEP3.1, and RcSEP4.1 genes, all of which were considered to be hubs due to their high connectivity in the network. Among them, the edge number of the AP3/PI clade genes, including RcPI3 (109 edges), RcPI5 (99), RcAP3.2 (94), and RcPI4 (86), was much higher than that of the SEP clade genes, suggesting that the AP3/PI clade genes may have critical roles in regulating floral organogenesis and development in rose. Furthermore, out of 297 other adjacent genes, 158 (53.2%) had at least 2 connections with detected MIKCC genes and 55 (18.5%) of them had 4 connections with detected MIKCC genes, suggesting their potential roles in regulating floral development via the ABCDE model in rose.
In addition, Gene Ontology (GO) term annotations were surveyed for all the adjacent genes (859 genes) in the networks to gain more insight into their biological function bias. Among these, 260 (30.3%) genes were found to be enriched in one biological process (BP) network (Fig. 6c). The following GO categorizations were significantly enriched: biological regulation, metabolism, cellular process, and single-organism processes. More importantly, 19 genes were enriched in developmental processes and reproductive process categorizations (Fig. 6c), suggesting their potential roles in regulating floral organogenesis and development in rose. Furthermore, 118 adjacent genes were identified to be vegetative or reproductive tissue specific (Supplementary File S4).
Discussion
The evolution of reproductive organ morphologies in seed plants, especially of floral transition and floral organogenesis in angiosperms, is considered to be a major evolutionary innovation in plant evolution and likely had a key role in Darwin’s ‘abominable mystery’3. MIKCC genes, which were initially discovered as floral organ identity genes, originated in ancestral seed plants and have central regulatory roles in reproductive organ morphologies and flowering transition. Although MADS-box genes have been studied for approximately three decades, primarily in model plants such as Arabidopsis2,3,11, the biological functions and regulatory mechanisms of MIKCC genes during flowering transition and floral organogenesis are still not fully understood, especially in Rosaceae. Comprehensive analysis of MIKCC genes in rose will facilitate the understanding of plant floral organogenesis and development.
The expansion of ABCDE model genes, and FLC and AGL17 clade genes was not detected in R. chinensis
In our study, 58 non-redundant MIKCC unigenes were identified from rose transcriptomes based on the presence of the highly conserved MADS-box domain (Supplementary File S2). Phylogenetic analysis then divided the MIKCC genes into 12 major gene clades along with their counterparts in Arabidopsis and strawberry1,53 (Fig. 1a). Interestingly, the ABCDE model and SOC1 and AGL6 clades in rose (Fig. 1b, II cluster) were found to be remarkably expanded relative to the corresponding clades in Arabidopsis, suggesting that enhanced and/or novel roles of the associated genes might be required for specific aspects of floral development in rose. In contrast to these expanded clades, no rose MIKCC proteins are classified into the AGL17 clade observed in Arabidopsis and strawberry, and neither rose nor strawberry has genes in the FLC clade (Fig. 1b, I cluster), suggesting that significant gene loss might have happened in these clades after the split of Arabidopsis and rosaceae during their evolution. Furthermore, it is worth mentioning that the FLOWERING LOCUS C (FLC) gene and the MADS AFFECTING FLOWERING (MAFs) genes in the FLC clade act as key regulators in plant vernalization, the process that represses flowering and ensures that plants survive winter54,55,56,57. The non-detection of FLC clade genes in rose or strawberry could potentially indicate a re-modulation of the plant flowering mechanisms in these species23 and may even be associated with CF traits.
Comparative expression analysis of AP1/FUL, AP3/PI, AG, and SEP clade genes between Old Blush and Viridiflora reveals their roles in rose floral organogenesis
Dubois et al.43 constructed a high-throughput gene expression atlas for rose using RNA-seq, which covered almost all the typical organs (six vegetative and seven reproductive organs) of rose plants. In total, 52 annotated unigenes were identified as MIKCC genes from the unigene datasets in our study (Fig. 3a). Most of the genes from the AP1/FUL, AP3/PI, AG, and SEP clades, as well as from the AGL6 clade, were specifically expressed in the reproductive organs of rose, which is consistent with their particular roles in floral organogenesis and development2,3,11. Furthermore, the increase in members of the AP3/PI, AG, and SEP clades reflects the remarkable expansion of ABCDE model genes and supports their robust roles in rose floral organogenesis and development.
Furthermore, the expression profiles of genes from the AP1/FUL, AP3/PI, AG and SEP clades were extensively surveyed in multiple floral organ initiation stages in Old Blush (Fig. 4a) and the homeotic mutant Viridiflora (Fig. 4b) using qRT-PCR. The results showed that almost all of the examined genes were significantly up-regulated in response to floral organ initiation in both Old Blush and Viridiflora, suggesting roles for these genes in floral organogenesis and development in rose. The cluster analyses showed that AG and AP1 genes were always separated into cluster II and cluster III, respectively (Fig. 4c,d), corresponding with the fact that A- class genes are regionally antagonistic to the expression of C-class genes in different whorls of flowers9,58. In addition, three genes (RcAP3.2, RcAP3.4, and RcPI3) from the AP3/PI clade and one gene (RcSEP2.1) from the SEP clade were found to have shifted cluster positions from cluster III to II, suggesting their potential roles in contributing to the floral organ mutations of Viridiflora as well as their critical roles in rose floral organogenesis and development. Subsequently, the fold changes of all of the examined genes were calculated between two adjacent floral organ differentiation stages for sepal, petal, stamen, and carpel organogenesis in rose buds (Fig. 4e). The fold changes for specific floral organ initiation of all genes from the AP3/PI clade (B- class), two genes (RcAP1.1 and RcAP1.1) from the AP1/FUL clade, and three genes (RcSEP4.1, RcSEP4.3, and RcSEP4.4) from the SEP clade (E-class) were dramatically different between Old Blush and Viridiflora, further revealing their possible role in floral organogenesis and development.
Regional expression in different whorls of flowers is a key aspect of A-, B-, C-, D-, and E-class genes that contribute to floral organogenesis11,12. In our study, the regional expression restrictions of genes from the AP1/FUL, AP3/PI, AG and SEP clades were found to be dramatically changed in Viridiflora, with the expression peak of many of the genes shifted from whorl 2 and whorl 3 to whorl 4 in Viridiflora relative to Old Blush (Fig. 5). Furthermore, the expressions of RcAP1.1, RcAP1.2, RcFUL1, RcSEP4.1, and RcSOC1.1 were extended in whorls 2 and 3 in Viridiflora, suggesting their possible important roles in rose floral organogenesis.
Co-expression analysis further positions specific MIKCC genes as hubs of the regulatory network of rose flowering development and organogenesis
Even though MIKCC genes have been extensively studied in plants for almost 30 years2,3,11, the full nature of their position in the regulatory networks controlling flowering time and floral organogenesis remains unclear. In addition, other types of genes and factors (e.g., small RNAs, long non-coding RNA, and histone modification) have also been found to be involved in the control of floral organ structure and development11,59,60,61,62,63,64,65. In our study, co-expression networks of MIKCC genes were constructed from transcriptome data using public RNA-seq data43. Fifteen (24.2%) out of the 58 detected MIKCC genes were found to be network hubs, with 856 other adjacent genes co-expressed with them (Fig. 6b), suggesting their potential regulatory roles in controlling flowering time and floral organogenesis in rose. Interestingly, RcAGL19, RcAGL24, and RcSOC1 were found in the vegetative module of the networks—this is surprising given work in Arabidopsis establishing these genes as key components of the vernalization pathway23, suggesting their crucial roles in regulating floral transition in rose. Interestingly, the FLC genes along with their homologs (MAFs) in the FLC clade that were characterized as the prolonged repressors of the vernalization pathway23,66 have been exhaustively lost in rose, leading us to hypothesize that the loss of genes from the FLC clade may be one of the factors contributing to CF in rose. In addition, only genes from the AP3/PI clade (RcAP3.2, RcAP3.3, RcPI3, RcPI4, and RcPI) and SEP clade (RcSEP3.1 and RcSEP4.1) were found in the reproductive module of the networks (Fig. 6b), suggesting prominent roles for AP3/PI clade genes in regulating floral organogenesis and development in rose. This is consistent with the results from the analyses of the MADS-box domain divergence and expression patterns we described above (Fig. 2a, c, 4e and 5c, d).
In conclusion, we identified 58 non-redundant MIKCC unigenes from the rose transcriptome. Subsequent analysis revealed that ABCDE model genes (corresponding to AP1/FUL, AP3/PI, AG and SEP clades), and SOC1 and AGL6 clade genes were remarkably expanded in rose. Comparative expression analysis of MIKCC genes, especially the ABCDE model genes, between R. chinensis cv. Old Blush and the closely related homeotic mutant genotype R. chinensis cv. Viridiflora, confirmed their role in regulating floral organogenesis and development in rose. Furthermore, the co-expression networks provided an overview of the regulatory networks of MIKCC genes and shed further light on the prominent roles of AP3/PI clade genes in floral organogenesis and the roles of RcAGL19, RcAGL24, and RcSOC1 in regulating floral transition in rose.
Materials and methods
Data retrieval for the rose transcriptome
Rose transcriptome data were retrieved from public RNA-seq databases, including NCBI/SRA and GEO DataSets (https://www.ncbi.nlm.nih.gov), the DDBJ/sequence Read Archive (http://trace.ddbj.nig.ac.jp/dra/index_e.htm), GDR (https://www.rosaceae.org/), and LIPM (https://lipm-browsers.toulouse.inra.fr/plants/R.chinensis), as well as from published papers43,48,49,50,67,68. In addition, two high-quality RNA-seq datasets of R. chinensis cv. Old Blush and R. chinensis cv. Viridiflora (including four vegetative samples and nine floral organ samples) were recently generated by our own laboratory (Illumina HiSeq X-ten; Pair-end; N50 > 1463; Q30: 99.9%), and the unigene datasets were integrated into the database (unpublished). To produce a more accurate transcriptome database for rose, only three high-quality RNA-seq datasets from public databases43,48,49 and the two datasets produced previously by our group were selected to construct a rose transcriptome database (composed of 368,514 unigenes) for further analysis in our study (Supplementary File S5). This database included 36 samples and covered all the typical organs and their developmental stages of the rose plant.
Transcriptomic identification of MIKCC genes in rose
All sequences from our rose transcriptome database were translated in all six reading frames by a Perl script (Supplementary File S6). A hidden Markov model (HMM) search was then carried out in the six-frame protein database using two different HMM profiles. One profile was constructed by Gramzow and Theißen32 (Supplementary File S7). The second HMM profile used was the SRF-TF domain HMM profile (PF00319) obtained from the Pfam database (http://pfam.xfam.org/). HMM searches were executed using the HMMER3.0 software package with default thresholds (http://hmmer.org/)69. The protein sequences of the output unigenes were subsequently verified through public databases including SMART (http://smart.embl-heidelberg.de/) and Pfam to confirm the integrity of the MADS-box domains. Finally, the corresponding nucleotide sequences of all candidate genes were submitted to the CD-HIT Suite server, with 90% sequence identity cut-off (http://weizhong-lab.ucsd.edu/cdhit_suite/cgi-bin/index.cgi?cmd=Server%20home) to remove redundant sequences along with manual checking (Supplementary File S8). Finally, MIKCC genes were separated from all identified MADS-box genes by further phylogenetic analysis. The same protocols were performed for the identification of MADS-box genes in the strawberry (Fragaria vesca) genome (Supplementary File S9).
Sequence alignment and phylogenetic analysis
Multiple sequence alignment was performed via the alignment software MAFFT v7.037b70 using the accurate E-INS-I alignment strategy, which is stated to be suitable for sequences containing large unalignable regions. For the phylogenetic analysis, accurate maximum-likelihood trees were constructed using FastTree software with default parameters (http://www.microbesonline.org/fasttree/)71. The phylogenetic tree was visualized using MEGA7 (http://www.megasoftware.net/home)72. The tertiary structures and homo-/heterodimeric states of MADS-domain proteins were predicted using the SWISS-MODEL online program (https://swissmodel.expasy.org/interactive)73.
Plant materials and sample collection
Two-year-old cutting seedlings of R. chinensis cultivars, Viridiflora, and Old Blush (both are Chinese old roses), were grown in a greenhouse. Flower buds were collected with stems carrying two leaves and were dipped directly into RNase-free water. The flower buds were then vertically dissected into two equal parts under a stereo microscope (Fluorescent Stereo Microscope Leica M165 FC, Wetzlar, Germany) to confirm accurate determination of the stages of floral organ differentiation. The validated samples were immediately placed into liquid nitrogen and stored at − 80 ℃ until RNA extractions. This procedure was completed for each sample in < 2 min.
RNA isolation and qRT-PCR determination
Total RNAs of samples were extracted using the BioTeke Quick RNA Isolation Kit (Catalog number: RP3301, BioTeke Corporation, Beijing, China) according to the manufacturer’s instructions. High-quality total RNA (1 μg) was reverse transcribed using the PrimeScriptTM RT Reagent Kit with gDNA Eraser (Perfect Real Time) (catalog number: RR047A, TaKaRa, Dalian, China) according to the manufacturer’s instructions. Gene-specific primers for MIKCC genes were designed using GenScript (https://www.genscript.com/) and the IDT (http://sg.idtdna.com/scitools/Applications/RealTimePCR/) online server. qRT-PCR assays were carried out by the QuantStudio 6 Real-Time PCR System (Thermo Fisher Scientific, California, USA) using SYBR® Premix Ex TaqTM (Tli RNaseH Plus) (catalog number: RR420A, TaKaRa) according to the manufacturer’s instructions. Expression levels were normalized with the RcTUBULIN, RcGAPDH, and RcTCTP reference genes, and the 2–ΔCT method was used for calculating the relative expression of rose MIKCC genes. Three biological replicates with three qPCR technical replicates were performed for each experiment. Heatmaps were generated using Mev v4.9.0 software (http://www.tm4.org/), and the Pearson’s correlation distance was used for cluster analysis. The sequences of primers are available in Supplementary File S10 and their melting curve analyses are presented in Supplementary File S11.
Co-expression networks
Co-expression networks were generated by using the RNA-seq datasets of rose downloaded from LIPM (https://lipm-browsers.toulouse.inra.fr/plants/R.chinensis)43, which covered 13 tissues (6 vegetative and 7 reproductive samples) of rose plants. First, t-tests were performed to calculate the significance level of all unigenes between vegetative and reproductive samples based on gene expression (FPKM values). A total of 3014 genes differed significantly between vegetative and reproductive samples, with a p-value of < 0.05 (Supplementary File S12). Then, the R package was employed to determine the Pearson’s correlation coefficient (PCC) values between all selected unigenes. An effective threshold (0.77) of the PCC value was trained to generate the lowest density networks using R script (Supplementary File S13). Finally, the MIKCC genes (nodes) with their adjacent connections (edges) were extracted for our analysis (Supplementary File S14). The constructed co-expression networks were visualized using the Cytoscape v. 3.5.1 program (http://www.cytoscape.org/)74. GO term enrichment was performed using Blast2GO software (https://www.blast2go.com/) and was visualized using REVIGO (http://revigo.irb.hr/)75.
References
Becker, A. & Theissen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29, 464–489 (2003).
Gramzow, L. & Theissen, G. A hitchhiker’s guide to the MADS world of plants. Genome Biol. 11, 214 (2010).
Smaczniak, C., Immink, R. G. H., Angenent, G. C. & Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139, 3081–3098 (2012).
Yanofsky, M. F. et al. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39 (1990).
Weigel, D. & Meyerowitz, E. M. The ABCs of floral homeotic genes. Cell 78, 203–209 (1994).
Li, C. et al. Genome-wide characterization of the MADS-Box gene family in radish (Raphanus sativus L.) and assessment of its roles in flowering and floral organogenesis. Front. Plant Sci. 7, 1390 (2016).
Parenicova, L. et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15, 1538–1551 (2003).
Theissen, G. & Saedler, H. Plant biology. Flor. Quartets Nat. 409, 469–471 (2001).
Theissen, G. Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant. Biol. 4, 75–85 (2001).
Angenent, G. C. & Colombo, L. Molecular control of ovule development. Trends Plant. Sci. 1, 228–232 (1996).
Causier, B., Schwarz-Sommer, Z. & Davies, B. Floral organ identity: 20 years of ABCs. Semin. Cell. Dev. Biol. 21, 73–79 (2010).
Coen, E. S. & Meyerowitz, E. M. The war of the whorls - Genetic interactions controlling flower development. Nature 353, 31–37 (1991).
Goodrich, J. et al. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51 (1997).
Sridhar, V. V., Surendrarao, A., Gonzalez, D., Conlan, R. S. & Liu, Z. Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc. Natl Acad. Sci. USA 101, 11494–11499 (2004).
Zhao, D., Yu, Q., Chen, M. & Ma, H. The ASK1 gene regulates B function gene expression in cooperation with UFO and LEAFY. Arab. Dev. 128, 2735–2746 (2001).
Ditta, G., Pinyopich, A., Robles, P., Pelaz, S. & Yanofsky, M. F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol.: CB 14, 1935–1940 (2004).
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E. & Yanofsky, M. F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203 (2000).
Andres, F. et al. SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc. Natl Acad. Sci. USA 111, E2760–E2769 (2014).
Marin-Gonzalez, E. et al. SHORT VEGETATIVE PHASE up-regulates TEMPRANILLO2 floral repressor at low ambient temperatures. Plant. Physiol. 169, 1214–1224 (2015).
Hu, J. Y. et al. miR824-regulated AGAMOUS-LIKE16 contributes to flowering time repression in Arabidopsis. Plant Cell 26, 2024–2037 (2014).
Ratcliffe, O. J., Kumimoto, R. W., Wong, B. J. & Riechmann, J. L. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15, 1159–1169 (2003).
Adamczyk, B. J., Lehti-Shiu, M. D. & Fernandez, D. E. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 50, 1007–1019 (2007).
Alexandre, C. M. & Hennig, L. FLC or not FLC: the other side of vernalization. J. Exp. Bot. 59, 1127–1135 (2008).
Gramzow, L., Ritz, M. S. & Theissen, G. On the origin of MADS-domain transcription factors. Trends Genet. 26, 149–153 (2010).
De Bodt, S. & Maere, S. & Van de Peer, Y. Genome duplication and the origin of angiosperms. Trends Ecol. Evol. 20, 591–597 (2005).
Arora, R. et al. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8, 242 (2007).
Leseberg, C. H., Li, A., Kang, H., Duvall, M. & Mao, L. Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene 378, 84–94 (2006).
Kaufmann, K., Melzer, R. & Theissen, G. MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347, 183–198 (2005).
Kramer, E. M. & Hall, J. C. Evolutionary dynamics of genes controlling floral development. Curr. Opin. Plant. Biol. 8, 13–18 (2005).
Becker, A., Winter, K. U., Meyer, B., Saedler, H. & Theissen, G. MADS-Box gene diversity in seed plants 300 million years ago. Mol. Biol. Evol. 17, 1425–1434 (2000).
Theissen, G. et al. A short history of MADS-box genes in plants. Plant. Mol. Biol. 42, 115–149 (2000).
Gramzow, L. & Theißen, G. Phylogenomics of MADS-Box genes in plants - two opposing life styles in one gene family. Biology 2, 1150–1164 (2013).
Zahn, L. M. et al. The evolution of the SEPALLATA subfamily of MADS-Box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169, 2209–2223 (2005).
Jimenez, S., Lawton-Rauh, A. L., Reighard, G. L., Abbott, A. G. & Bielenberg, D. G. Phylogenetic analysis and molecular evolution of the dormancy associated MADS-box genes from peach. BMC Plant. Biol. 9, 81 (2009).
Gramzow, L. & Theissen, G. Phylogenomics reveals surprising sets of essential and dispensable clades of MIKCc-group MADS-box genes in flowering plants. J. Exp. Zool. B 324, 353–362 (2015).
Bendahmane, M., Dubois, A., Raymond, O. & Bris, M. L. Genetics and genomics of flower initiation and development in roses. J. Exp. Bot. 64, 847–857 (2013).
Magnard, J. L. et al. Biosynthesis of monoterpene scent compounds in roses. Science 349, 81–83 (2015).
Kitahara, K. & Matsumoto, S. Rose MADS-box genes ‘MASAKO C1 and D1’ homologous to class C floral identity genes. Plant Sci. 151, 121–134 (2000).
Kitahara, K., Hibino, Y., Aida, R. & Matsumoto, S. Ectopic expression of the rose AGAMOUS-like MADS-box genes ‘MASAKO C1 and D1’ causes similar homeotic transformation of sepal and petal in Arabidopsis and sepal in Torenia. Plant Sci. 166, 1245–1252 (2004).
Kitahara, K., Hirai, S., Fukui, H. & Matsumoto, S. Rose MADS-box genes ‘MASAKO BP and B3’ homologous to class B floral identity genes. Plant Sci. 161, 549–557 (2001).
Hibino, Y., Kitahara, K., Hirai, S. & Matsumoto, S. Structural and functional analysis of rose class B MADS-box genes ‘MASAKO BP, euB3, and B3: Paleo-type AP3 homologue ‘MASAKO B3’ association with petal development. Plant Sci. 170, 778–785 (2006).
Remay, A. et al. A survey of flowering genes reveals the role of gibberellins in floral control in rose. Theor. Appl. Genet. 119, 767–781 (2009).
Dubois, A. et al. Transcriptome database resource and gene expression atlas for the rose. BMC Genomics 13, 638 (2012).
Chmelnitsky, I., Azizbekova, N., Khayat, E. & Zieslin, N. Morphological development of normal and phyllody expressing Rosa hybrida cv. Motrea flowers. Plant. Growth Regul. 37, 215–221 (2002).
Chmelnitsky, I., Colauzzi, M., Algom, R. & Zieslin, N. Effects of temperature on phyllody expression and cytokinin content in floral organs of rose flowers. Plant. Growth Regul. 35, 207–214 (2001).
Sugio, A. & Hogenhout, S. A. The genome biology of phytoplasma: modulators of plants and insects. Curr. Opin. Microbiol. 15, 247–254 (2012).
Hogenhout, S. A. et al. Phytoplasmas: bacteria that manipulate plants and insects. Mol. Plant Pathol. 9, 403–423 (2008).
Yan, H. J. et al. The Rosa chinensis cv. Viridiflora phyllody phenotype is associated with misexpression of flower organ identity genes. Front. Plant Sci. 7, 996 (2016).
Koning-Boucoiran, C. F. S. et al. Using RNA-Seq to assemble a rose transcriptome with more than 13,000 full-length expressed genes and to develop the WagRhSNP 68k Axiom SNP array for rose (Rosa L.). Front. Plant Sci. 6, 249 (2015).
Gao, Y. R. et al. Transcriptome profiling of petal abscission zone and functional analysis of an Aux/IAA FamilyGene RhIAA16 involved in petal shedding in Rose. Front. Plant Sci. 7, 1375 (2016).
Nakamura, N. et al. Genome structure of Rosa multiflora, a wild ancestor of cultivated roses. DNA Res., https://doi.org/10.1093/dnares/dsx042 (2017).
Dubois, A. et al. Genomic approach to study floral development genes in Rosa sp. PLoS ONE 6, e28455 (2011).
Diaz-Riquelme, J., Lijavetzky, D., Martinez-Zapater, J. M. & Carmona, M. J. Genome-wide analysis of MIKCC-type MADS box genes in grapevine. Plant. Physiol. 149, 354–369 (2009).
Aikawa, S., Kobayashi, M. J., Satake, A., Shimizu, K. K. & Kudoh, H. Robust control of the seasonal expression of the Arabidopsis FLC gene in a fluctuating environment. Proc. Natl Acad. Sci. USA 107, 11632–11637 (2010).
Michaels, S. D. & Amasino, R. M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956 (1999).
Sheldon, C. C. et al. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458 (1999).
Sheldon, C. C., Rouse, D. T., Finnegan, E. J., Peacock, W. J. & Dennis, E. S. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl Acad. Sci. USA 97, 3753–3758 (2000).
Bowman, J. L., Smyth, D. R. & Meyerowitz, E. M. The ABC model of flower development: then and now. Development 139, 4095–4098 (2012).
Clark, S. E., Running, M. P. & Meyerowitz, E. M. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418 (1993).
Clark, S. E., Running, M. P. & Meyerowitz, E. M. Clavata3 is a specific regulator of shoot and floral meristem development affecting the same processes as Clavata1. Development 121, 2057–2067 (1995).
Talbert, P. B., Adler, H. T., Parks, D. W. & Comai, L. The revoluta gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121, 2723–2735 (1995).
Running, M. P. & Meyerowitz, E. M. Mutations in the PERIANTHIA gene of Arabidopsis specifically alter floral organ number and initiation pattern. Development 122, 1261–1269 (1996).
Griffith, M. E., Conceicao, A. D. & Smyth, D. R. PETAL LOSS gene regulates initiation and orientation of second whorl organs in the Arabidopsis flower. Development 126, 5635–5644 (1999).
Brewer, P. B. et al. PETAL LOSS, a trihelix transcription factor gene, regulates perianth architecture in the Arabidopsis flower. Development 131, 4035–4045 (2004).
Takeda, S., Matsumoto, N. & Okada, K. RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 131, 425–434 (2004).
Helliwell, C. A., Wood, C. C., Robertson, M., James Peacock, W. & Dennis, E. S. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 46, 183–192 (2006).
Kim, J. et al. Small RNA and transcriptome deep sequencing proffers insight into floral gene regulation in Rosa cultivars. BMC Genomics 13, 657 (2012).
Zhang, X. Y. et al. Transcriptome sequencing and de novo analysis of Rosa multiflora under cold stress. Acta Physiol. Plant 38, https://doi.org/10.1007/s11738-016-2184-9 (2016).
Eddy, S. R. Profile hidden Markov models. Bioinformatics 14, 755–763 (1998).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650 (2009).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Biasini, M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Supek, F., Bosnjak, M., Skunca, N. & Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800 (2011).
Acknowledgements
This research was funded by the Fundamental Research Funds for the Central Universities (KYYJ201706), the Key Research and Development Project of Jiangsu Province (BE2016377), and the China Postdoctoral Science Foundation (2016M600425). We thank Dr. Geoff Benn from UC Davis for providing language editing for this paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
41438_2018_31_MOESM1_ESM.tif
Supplementary File S1 The flower phenotypes of Rosa chinensis cv. Old Blush (a, c) and Viridiflora (b, d). (e and f) present the almost identical leaves of Old Blush (e) and Viridiflora (f), respectivel(TIF 1075 kb)
41438_2018_31_MOESM3_ESM.xlsx
Supplementary File S3. The homo-/heterodimeric state of the MADS-box domain proteins form Arabidopsis, strawberry and rose(XLSX 13 kb)
41438_2018_31_MOESM6_ESM.txt
Supplementary File S6. Perl script for translating the nucleotide sequences of rose RNA-seq unigenes into six reading frames proteins(TXT 3 kb)
41438_2018_31_MOESM9_ESM.txt
Supplementary File S9. The protein sequences of MADS-box genes identified from strawberry (Fragaria vesca) genome(TXT 69 kb)
41438_2018_31_MOESM12_ESM.xlsx
Supplementary File S12. Total 3014 genes that varied significant between vegetative and reproductive tissues in rose transcriptome(XLSX 317 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, J., Fu, X., Dong, Y. et al. MIKCC-type MADS-box genes in Rosa chinensis: the remarkable expansion of ABCDE model genes and their roles in floral organogenesis. Hortic Res 5, 25 (2018). https://doi.org/10.1038/s41438-018-0031-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-018-0031-4
This article is cited by
-
Genome-Wide Characterization of the MADS-Box Gene Family and Expression Pattern in Different Tissues and Stresses in Zanthoxylum armatum
Journal of Plant Growth Regulation (2024)
-
Characterization of the MIKCC-type MADS-box gene family in blueberry and its possible mechanism for regulating flowering in response to the chilling requirement
Planta (2024)
-
Resequencing of Rosa rugosa accessions revealed the history of population dynamics, breed origin, and domestication pathways
BMC Plant Biology (2023)
-
Genome-wide identification of the MADS-box transcription factor family in autotetraploid cultivated alfalfa (Medicago sativa L.) and expression analysis under abiotic stress
BMC Genomics (2021)
-
The SOC1-like gene BoMADS50 is associated with the flowering of Bambusa oldhamii
Horticulture Research (2021)