Abstract
Biallelic pathogenic variants in ALDH1A3 are responsible for approximately 11% of recessively inherited cases of severe developmental eye anomalies. Some individuals can display variable neurodevelopmental features, but the relationship to the ALDH1A3 variants remains unclear. Here, we describe seven unrelated families with biallelic pathogenic ALDH1A3 variants: four compound heterozygous and three homozygous. All affected individuals had bilateral anophthalmia/microphthalmia (A/M), three with additional intellectual or developmental delay, one with autism and seizures and three with facial dysmorphic features. This study confirms that individuals with biallelic pathogenic ALDH1A3 variants consistently manifest A/M, but additionally display neurodevelopmental features with significant intra- and interfamilial variability. Furthermore, we describe the first case with cataract and highlight the importance of screening ALDH1A3 variants in nonconsanguineous families with A/M.
Similar content being viewed by others
Introduction
Anophthalmia (absence of visible ocular tissue) and microphthalmia (reduced ocular size) (A/M) are developmental eye anomalies affecting around 11.9 per 100,000 live births [1]. More than half of affected individuals exhibit variable extraocular features [2]. Pathogenic variants in at least 120 genes are known to underlie A/M, including several in the retinoic acid pathway: STRA6, RBP4, RARB and ALDH1A3 [3]. ALDH1A3 (Aldehyde dehydrogenase 1 family member A3) catalyses retinoic acid formation, playing a key role in embryonic eye development [4]. Pathogenic biallelic variants in ALDH1A3 are responsible for ∼11% of cases in consanguineous families [5,6,7,8]. To date, the majority of ALDH1A3 variants are reported in individuals from consanguineous families and are consistently associated with bilateral A/M, with additional systemic features described in some cases [6, 8, 9]. However, genotype-phenotype correlations are unclear, with particular uncertainty surrounding whether the neurodevelopmental manifestations are solely linked to these variants.
Herein, we report nine cases with biallelic ALDH1A3 variants from seven families. All affected individuals display bilateral A/M, with variable additional neurodevelopmental anomalies in some cases, providing further insights into the phenotypic spectrum.
Materials and methods
We identified seven families from a cohort of 202 undiagnosed UK, French and Spanish families with A/M. Families 1, 2, 4 and 7 are from research studies: UK ‘Genetics of Eye and Brain anomalies’ (Cambridge East Ethics Committee (04/Q0104/12)), Deciphering Developmental Disorders (DDD) Study (Cambridge South Research Ethics Committee (10/H0305/83), Republic of Ireland (GEN/284/12)) and Genetics of Congenital Ocular Disorders, Fundación Jimenez Díaz University Hospital (Ethics Research Committee FJD (PIC015-18)), respectively. Families 3, 5 and 6 were identified through diagnostic testing. Informed consent was obtained from all individuals in accordance with the Declaration of Helsinki.
ALDH1A3 (NM_000693.4) variants were identified from the cohort (n = 202) using whole genome (n = 20)/exome (n = 88) sequencing (WGS/WES), customized NGS panels (n = 91) and Sanger sequencing (n = 3). WGS/WES was performed using TruSeq Nano DNA Sample Prep (Illumina Inc., San Diego, CA, USA) and Agilent SureSelect Human All Exon V6 (Agilent Technologies, Santa Clara, CA, USA) kits, respectively. The majority of individuals (n = 181) received copy number variant screening using SNP-Array or array-CGH. WGS/WES data were annotated and filtered using an in-house pipeline. We used SIFT [10], Polyphen-2 [11] and CADD [12] in silico tools to predict pathogenicity, and Human Splicing Finder [13] to identify splicing effects. Variants were classified according to the ACMG guidelines [14], and confirmed by Sanger sequencing with segregation analysis when samples were available. Variants were submitted to the ClinVar database (SCV002761233, SCV002761243). Details of in silico predictions and ACMG/AMP classifications are described in Supplementary Table S1.
Results
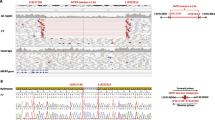
We identified nine affected individuals with eleven biallelic ALDH1A3 variants from seven unrelated families (Table 1 and Fig. 1). Unless stated, no other likely pathogenic variants relevant to the individuals’ phenotypes were identified.
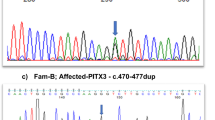
A–G Pedigrees, genotypes and consented clinical images in families. H Schematic of ALDH1A3, its domains and variant locations reported to date. The variants identified in this study are listed above and previously reported variants below the domain structure. Compound heterozygous variant pairs are indicated in purple, orange, green, yellow and blue circles, homozygous variants in red circles. The previously reported variants also identified in our cohort are listed above and below the domain structure. The number of the reports are indicated in brackets.
Compound heterozygous variants
Family 1
The proband (II:2) had bilateral anophthalmia, a right lower lid cyst and normal development. His brother (II:4) presented with bilateral anophthalmia, and mildly delayed development until 2 years-of-age after which it gradually slowed. He was diagnosed with nonverbal autism at 3–4 years-of-age. Both brothers have a large sandal gap between the first and second toe. They carried compound heterozygous missense ALDH1A3 variants (c.874G>T, p.(Asp292Tyr), maternal; c.1393A>T, p.(Ile465Phe), paternal). Both variants were absent in gnomAD and reported in DECIPHER. The proband had a normal SNP-Array and his brother had a normal array-CGH. A similar family has been published by Patel et al. [15], however it is unclear if this is the same family as described here.
Family 2
The proband (II:1) presented with bilateral anophthalmia with lower lid cysts. He had developmental delay and severe learning difficulties, delayed speech, autistic features, and tonic-clonic seizures (onset 13 years-of-age). He carries compound heterozygous missense variants (rs547918064, c.845G>C, p.(Gly282Ala), maternal, gnomAD MAF: 0.000019; c.1459A>G, p.(Arg487Gly), paternal, absent in gnomAD). The p.(Gly282Ala) variant was reported in the homozygous state by Alabdullatif et al. [16]. The parents are healthy with no family history of seizures.
Family 3
The proband (II:1) presented with bilateral microphthalmia and coloboma, bilateral retinal detachments with microcalcifications and vascularization at 1 month-of-age and normal development. She carries compound heterozygous variants: an inframe deletion of a highly conserved amino acid (c.847_849del, p.(Gly283del), maternal) and a missense (c.953C>A, p.(Ser318Tyr), paternal) variant. Both variants are absent in gnomAD. She had a normal array-CGH.
Family 4
The proband (II:1) presented with bilateral microphthalmia and normal development. We identified compound heterozygous nonsense (c.566G>A, p.(Trp189*), maternal, absent in gnomAD) and splice (rs1422193527, c.100-2A>G, paternal, gnomAD MAF: 0.00003183) variants.
Homozygous variants
Family 5
The proband (II:1), born to consanguineous parents, presented with bilateral microphthalmia (extreme on the left) and additional coloboma and cataract of the right eye. He had normal development. He carries a homozygous splice variant (c.1233 + 2T>C, absent in gnomAD). His parents are heterozygous carriers. A paternal aunt had bilateral anophthalmia, but was unavailable for testing.
Family 6
The proband (II:2), born to consanguineous parents, presented with bilateral anophthalmia and mild intellectual delay. Her sister (II:4) had bilateral anophthalmia and normal development. Both carry a homozygous missense variant (c.1144G>A, p.(Gly382Arg), absent in gnomAD). Both parents were heterozygous carriers.
Family 7
The proband (II:1) presented with bilateral microphthalmia, iris and chorioretinal coloboma, abnormal anterior segment morphology and facial dysmorphic features, including high forehead, telecanthus, epicanthic folds, ptosis, full cheeks, everted upper lip, and micrognathia. He carries a homozygous missense variant (c.434C>T, p.(Ala145Val), gnomAD MAF: 0.000004). His parents are heterozygous carriers. There was no history of parental consanguinity, but the parents come from the same small town. Array-CGH was normal.
Discussion
We report nine individuals from seven families with biallelic ALDH1A3 variants (Fig. 1). In each case the variants were predicted disease-causing, with no other variants detected in genes associated with developmental eye disorders by WES/WGS/panel/CNV analysis. This study describes further cases with compound heterozygous ALDH1A3 variants in A/M and highlights inter- and intrafamilial phenotypic variability.
Since many developmental eye genes are critical in the development of other organ systems, extraocular features are often observed in individuals with variants in developmental eye genes, such as SOX2, OTX2 and STRA6 [17]. Individuals with biallelic ALDH1A3 variants have been previously reported with additional variable systemic features, including severe neurodevelopmental delay and autism, in addition to bilateral A/M [6, 9, 18]. Similarly, our nine cases consistently exhibited bilateral A/M; two with facial dysmorphic features (Families 6 and 7), and three also manifesting neurodevelopmental anomalies, including intellectual disability (Families 1, 2 and 6), autism (Families 1 and 2) and seizures (Family 2). Importantly, while additional ocular features are frequently reported in ALDH1A3 cases, our study represents the first report of the presence of cataract (Family 5). The facial dysmorphic features described in individuals with ALDH1A3 variants include bilateral small palpebral fissures or blepharophimosis [8, 9, 19, 20], which is often seen in individuals with small eye sockets secondary to severe A/M, irrespective of the genetic cause. However, broad eyebrows, synophrys and high arched palate are also reported in some cases [8]. Interestingly, one of our cases (Family 7, II:1) displayed multiple additional dysmorphic features, although it remains unclear if these are related to the ALDH1A3 variants.
ALDH1A3 is a member of the retinoic acid pathway, encoding aldehyde dehydrogenase involved in oxidation of retinaldehyde to retinoic acid (RA). RA levels are tightly regulated during embryonic development and are essential for normal eye morphogenesis [4]. Including this study, 32 pathogenic ALDH1A3 variants have been reported: 18 missense, 7 splicing, 4 nonsense, 2 frameshift and 1 inframe deletion. Some of the missense (p.(Arg89Cys), p.(Ala493Pro), p.(Arg96His), p.(Gly237Arg)) and nonsense (p.(Lys190*), p.(Lys389*)) variants have been shown to impair protein production and cause loss of function and were reported in patients with variable additional neurodevelopmental phenotypes in the A/M spectrum [6, 19, 21].
To date, even with the growing evidence from cases, there is no consistent genotype-phenotype correlation. The majority of variants (16) are located in the catalytic domain, followed by the NAD binding domains (13) and the oligomerization domains (3) (Fig. 1). However, the location of variants does not appear to correlate with distinct phenotypic features or differences in severity. Furthermore, there is also striking inter- and intrafamilial variation even for the same variants. For example, Roos et al. [9] described neurodevelopmental intrafamilial variability in a large consanguineous family with microphthalmia/coloboma and a homozygous ALDH1A3 variant (p.(Cys174Tyr)). Similarly, the affected brothers in Family 1, carrying the same compound heterozygous variants, had variable phenotypes: the older brother had isolated bilateral anophthalmia with normal intellect while the younger had severe neurodevelopmental delay. Furthermore, of the two affected sisters presenting with bilateral anophthalmia and facial dysmorphic features in Family 6 (p.(Gly382Arg)) the proband has mild intellectual delay whereas her younger sister has normal cognition. Interestingly, the same variant has been previously reported in four affected members of a family who presented with bilateral anophthalmia and facial dysmorphism, but normal psychomotor development [8], bringing into question the link between ALDH1A3 variants and intellectual and developmental delay. In addition, the proband in Family 7 has the same variant (p.(Ala145Val)) that had been previously described in individuals from 3 independent families and a simplex case, all with isolated bilateral microphthalmia [5, 22], whereas he had some additional facial dysmorphic features. Therefore, the presence of inter- and intrafamilial variability suggests a more complex interplay between ALDH1A3 variants and genetic and/or environmental factors, as might be expected for such a fundamentally important gene.
In conclusion, we present the clinical and genetic analysis of nine cases with biallelic ALDH1A3 variants from seven families. Our data confirms that pathogenic ALDH1A3 variants are consistently associated with bilateral A/M and highlights additional susceptibility to neurodevelopmental manifestations, with significant intra- and interfamilial variability. Moreover, one individual displayed microphthalmia with coloboma and cataract, broadening the ocular phenotype, and suggesting that it would be important to include this gene on cataract gene panels. Finally, the identification of four families with compound heterozygous variants underscores the importance of ALDH1A3 screening in nonconsanguineous families.
Data availability
Data will be made available upon reasonable request. Variants were submitted to ClinVAR database (SCV002761233, SCV002761243).
Change history
28 April 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41431-023-01363-3
References
Shah SP, Taylor AE, Sowden JC, Ragge NK, Russell-Eggitt I, Rahi JS, et al. Anophthalmos, microphthalmos, and typical coloboma in the United Kingdom: a prospective study of incidence and risk. Investig Ophthalmol Vis Sci. 2011;52:558–64.
Shah SP, Taylor AE, Sowden JC, Ragge N, Russell-Eggitt I, Rahi JS, et al. Anophthalmos, microphthalmos, and coloboma in the United Kingdom: clinical features, results of investigations, and early management. Ophthalmology. 2011;119:362–8.
Martin AR, Williams E, Foulger RE, Leigh S, Daugherty LC, Niblock O, et al. PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nat Genet. 2019;51:1560–5.
Metzler MA, Sandell LL. Enzymatic Metabolism of Vitamin A in Developing Vertebrate Embryos. Nutrients 2016;8:812.
Patel N, Khan AO, Alsahli S, Abdel-Salam G, Nowilaty SR, Mansour AM, et al. Genetic investigation of 93 families with microphthalmia or posterior microphthalmos. Clin Genet. 2018;93:1210–22.
Fares-Taie L, Gerber S, Chassaing N, Clayton-Smith J, Hanein S, Silva E, et al. ALDH1A3 mutations cause recessive anophthalmia and microphthalmia. Am J Hum Genet. 2013;92:265–70.
Lin S, Harlalka GV, Hameed A, Moattar Reham H, Yasin M, Muhammad N, et al. Novel mutations in ALDH1A3 associated with autosomal recessive anophthalmia/microphthalmia, and review of the literature. BMC Med Genet. 2018;19:160.
Abouzeid H, Favez T, Schmid A, Agosti C, Youssef M, Marzouk I, et al. Mutations in ALDH1A3 represent a frequent cause of microphthalmia/anophthalmia in consanguineous families. Hum Mutat. 2014;35:949–53.
Roos L, Fang M, Dali C, Jensen H, Christoffersen N, Wu B, et al. A homozygous mutation in a consanguineous family consolidates the role of ALDH1A3 in autosomal recessive microphthalmia. Clin Genet. 2014;86:276–81.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81.
Adzhubei I, Jordan DM, Sunyaev SR Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013. https://doi.org/10.1002/0471142905.hg0720s76.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:67.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Patel A, Hayward JD, Tailor V, Nyanhete R, Ahlfors H, Gabriel C, et al. The Oculome Panel Test: Next-Generation Sequencing to Diagnose a Diverse Range of Genetic Developmental Eye Disorders. Ophthalmology. 2019;126:888–907.
Alabdullatif MA, Al Dhaibani MA, Khassawneh MY, El-Hattab AW. Chromosomal microarray in a highly consanguineous population: diagnostic yield, utility of regions of homozygosity, and novel mutations. Clin Genet. 2017;91:616–22.
Slavotinek A. Genetics of anophthalmia and microphthalmia. Part 2: Syndromes associated with anophthalmia-microphthalmia. Hum Mutat. 2019;138:831–6.
Semerci CN, Kalay E, Yildirim C, Dincer T, Olmez A, Toraman B, et al. Novel splice-site and missense mutations in the ALDH1A3 gene underlying autosomal recessive anophthalmia/microphthalmia. Br J Ophthalmol. 2014;98:832–40.
Liu Y, Lu Y, Liu S, Liao S. Novel compound heterozygous mutations of ALDH1A3 contribute to anophthalmia in a non-consanguineous Chinese family. Genet Mol Biol. 2017;40:430–5.
Plaisancie J, Bremond-Gignac D, Demeer B, Gaston V, Verloes A, Fares-Taie L, et al. Incomplete penetrance of biallelic ALDH1A3 mutations. Eur J Med Genet. 2016;59:215–8.
Yahyavi M, Abouzeid H, Gawdat G, de Preux AS, Xiao T, Bardakjian T, et al. ALDH1A3 loss of function causes bilateral anophthalmia/microphthalmia and hypoplasia of the optic nerve and optic chiasm. Hum Mol Genet. 2013;22:3250–8.
Aldahmesh MA, Khan AO, Hijazi H, Alkuraya FS. Mutations in ALDH1A3 cause microphthalmia. Clin Genet. 2013;84:128–31.
Acknowledgements
We would like to thank the patients and their families for their participation to this study. We would like to thank Dr. Francesca Forzano, Dr. Jane Hurst and Dr. Louise Wilson for their clinical support and North East Thames Regional Genetics Service for parental testing of the ALDH1A3 variants in Family 2. We would like to thank Dr. Richard Holt for his support in the preparation of the manuscript.
Funding
This work was supported by Grants from Baillie Gifford, MACS (Microphthalmia, Anophthalmia, Coloboma Support) and Instituto de Salud Carlos III (ISCIII) of the Spanish Ministry of Health (FIS; PI17/00164 and PI20/00851), Comunidad de Madrid (CAM, RAREGenomics Project, B2017/BMD-3721) and ONCE (Spanish National Organization of the Blind). AD is supported by a PhD fellowship from ISCIII (FI18/00123), MC is supported by a Miguel Servet program contract from ISCIII (CPII17/00006). The DDD study presents independent research commissioned by the Health Innovation Challenge Fund [grant number HICF-1009-003]. This study makes use of DECIPHER (http://www.deciphergenomics.org), which is funded by Wellcome. See Nature PMID: 25533962 or www.ddduk.org/access.html for full acknowledgement.
Author information
Authors and Affiliations
Contributions
YK performed data analysis and interpretation, validation, and prepared the paper. FC performed data annotation. AD, FBK, CA, MC, NC, PC performed clinical examinations and data analysis. DB performed research coordination. KW performed data analysis. VP, CM, CR performed clinical examinations of the families. JP performed validation of the findings. NR supervised the project, performed clinical examinations of the families, reviewed and discussed the results and critically reviewed the paper. All authors provided critical feedback to paper preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Families 1, 2, 4 and 7 are from research studies: UK ‘Genetics of Eye and Brain anomalies’ (Cambridge East Ethics Committee (04/Q0104/12)), Deciphering Developmental Disorders (DDD) Study (Cambridge South Research Ethics Committee (10/H0305/83), Republic of Ireland (GEN/284/12)) and Genetics of Congenital Ocular Disorders, Fundación Jimenez Díaz University Hospital (Ethics Research Committee FJD (PIC015-18)), respectively. Families 3, 5 and 6 were identified through diagnostic testing. Written informed consent for the scientific use and publication of the clinical data and images presented was obtained from all individuals in accordance with the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: In this article the layout of table 1 was wrong and it appeared incorrectly.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kesim, Y., Ceroni, F., Damián, A. et al. Clinical and genetic analysis further delineates the phenotypic spectrum of ALDH1A3-related anophthalmia and microphthalmia. Eur J Hum Genet 31, 1175–1180 (2023). https://doi.org/10.1038/s41431-023-01342-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-023-01342-8
This article is cited by
-
Expanding what we know about rare genetic diseases
European Journal of Human Genetics (2023)