Abstract
Recently, variants in DONSON have been reported to cause different disorders of the microcephalic primordial dwarfism spectrum. Using whole-exome sequencing, we identified two novel, compound heterozygous DONSON variants in a pair of siblings, one of whom was previously diagnosed with Fanconi anemia. This occurred because the present cases exhibited clinical findings in addition to those of the microcephalic primordial dwarfism disorder, including severe limb malformations. These findings suggest that the DONSON and Fanconi anemia proteins could have supplementary roles in developmental processes as they have in the maintenance of genomic integrity, resulting in related disease phenotypes.
Similar content being viewed by others
Introduction
Reynolds et al. [1] recently provided evidence that the previously uncharacterized gene designated as “downstream neighbor of SON (DONSON)” is implicated in autosomal recessive human disease. These authors identified DONSON as a novel replication fork protection factor that is also required for S-phase checkpoint activity. They reported biallelic DONSON variants in 29 individuals with microcephalic primordial dwarfism (MPD). Of note, the patients of one of their families (P13-1, 2, and 3) presented with forearm and thumb dysplasia, and were earlier suspected to have a Fanconi anemia-like disorder, although none of these individuals had hematological evidence of bone marrow failure.
More recently, Evrony et al. [2] set out to identify the genetic cause of the microcephaly-micromelia syndrome (MMS) using a combined RNA-seq plus genome sequencing approach. The syndrome’s main clinical features include intrauterine growth restriction, severe microcephaly with distinctive craniofacial dysmorphic features, marked limb malformations, and almost uniform perinatal lethality such that most of the affected individuals are either stillborn or die within the first week of life due to respiratory failure [3]. In 12 affected individuals of nine families of Cree descent from the First Nations population of Northern Saskatchewan in Canada, Evrony et al. [2] detected homozygosity for the c.1047-9A>G noncoding variant resulting in retention of intron 6 (exons and introns are numbered as with Reynolds et al. [1] and Evrony et al. [2]). Retention of intron 6 translates into a premature stop codon after 52 bp of the 109-bp intron, which is predicted to cause nonsense-mediated decay of the mutant transcript or to result in a truncated protein that terminates with 17 aberrant amino acids prior to the premature stop [2].
Of note, all DONSON variants thus far reported to affect protein function are hypomorphic, i.e., they represent incomplete loss-of-function rather than null alleles, consistent with the fact that knockout of mouse Donson leads to developmental lethality [2]. Most of the MPD phenotypes Reynolds et al. [1] reported are nonlethal and milder than MMS, implying that their variants had milder effects on DONSON functions than the MMS variant of Evrony et al. [2]. However, Reynolds et al. [1] also reported two patients of a Saudi Arabian family (P21-1 and 2) who resemble the MMS phenotype described by Evrony et al. [2], and who, remarkably, featured the same homozygous splice variant c.1047-9A>G [1, 2].
DONSON has been recognized as a component of the replisome [1]. This protein localization is consistent with an important task in timely genome replication as can be inferred from examples of other genes encoding factors at the replication fork that are required at damaged and/or stalled forks in response to replication stress, and which also cause MPD disorders when mutated [4]. Deficiency or defects of proteins that protect replication forks, or repair and restart damaged forks, can compromise cell proliferation kinetics. The current notion is that primordial dwarfism gene variants may impair the efficiency of cell cycle progression and act to reduce the total cell number generated during development, leading to reduced tissue and organism size [5]. This way, impaired genome replication due to DONSON deficiency is conceptually founded to lead to impaired cellular proliferation and decreased total growth, in particular of organs such as the brain whose development depends on a high number of progenitor cell mitoses [2].
Here, we describe two further individuals with DONSON gene defects, one of whom was previously suspected to suffer from a Fanconi anemia-like disorder because of cell cycle delay and clinical overlap with Fanconi anemia-associated symptoms such as radial ray malformation, short stature, and microcephaly.
Patients and Methods
The parents of the present cases are of German descent, not consanguineous and both healthy. The mother sought medical advice as a 33-year-old fifth gravida (two healthy children and two abortions). Fetal growth retardation, an abnormal facial profile and limb malformations were noted on routine and high-resolution ultrasound screens at 20 weeks of gestation. Subsequent chromosome studies in amniocytes revealed a normal female karyotype, 46,XX. Array-comparative genomic hybridization (CGH) analysis using a 180K oligonucleotide microarray (Agilent Technologies) was inconspicuous. Based on the morphological findings, differential diagnoses were considered. The thrombocytopenia-absent radius syndrome, characterized by bilateral absence of the radii, presence of the thumbs and thrombocytopenia, caused by chromosome 1q21.1 microdeletion, was excluded by flourescence in situ hybridization (FISH) analysis. Roberts syndrome (RBS), a rare autosomal recessive disorder with key features including limb reduction, pre- and post-natal growth retardation, and craniofacial abnormalities became unlikely by exclusion of variants affecting the function of the ESCO2 gene that could cause RBS in the fetal DNA sample. Fanconi anemia was suspected and amniotic cells were exposed to mitomycin C. The results demonstrated increased levels of G2-phase arrest compared to controls, compatible with but not specific for Fanconi anemia. However, multiplex ligation-dependent probe amplification analysis did neither reveal large deletions nor duplications of FANCA, nor did targeted enrichment and MiSeq NGS of 17 Fanconi anemia genes (FANC-A,-B,-C,-D1,-D2,-E,-F,-G,-I,-J,-L,-M,-N,-O,-P,-Q,-S) detect any variants that could affect functioning of the FANC genes.
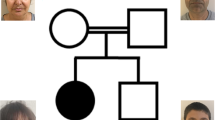
The pregnancy was terminated at 23 gestational weeks because of severe fetal malformations. Pathological examination of the female fetus showed marked growth retardation (weight 230 g; delayed for about 4 weeks), dysmorphic facial features with hypertelorism, flat midface, microtia, dysplastic low set ears, and microcephaly (Fig. 1). Malformations of the upper and lower limbs displayed overall dysplasia, including shortening of both ulnae and the left radius, complete absence of the right radius, and bilateral brachydactyly. Moreover, hypoplastic left thumb, ulnar deviation of the left hand, radial deviation of the right hand, flexion contractures of both elbows and wrists, syndactyly II–V at both feet, bilateral hypoplasia of the first digitus with shortening of the halluces, and contractures of the ankles were observed (Fig. 1a, c, e). Regarding the brain, severe hypotrophy of the cerebrum, the telencephalon without any suggestion of lobar division, and lissencephaly resulting in a lack of development of primary gyri and sulci were noted (Fig. 1b). Apart from this pathology, we encountered an age-appropriate morphology of the di-, meso-, and rhombencephalon. The cranial nerves were unremarkable. Clitoromegaly was also noticed (Fig. 1d).
Microcephalic dwarfism phenotype and pedigree. (Upper panel), pedigree of the family, black symbols represent affected female siblings. (Lower left panel), II/3 female fetus at 23 weeks of gestation. (Lower right panel), II/4 female patient at the age of 9 months. a Hypoplastic appendage-like proximally placed thumb with missing radius. b Lissencephaly of the brain. c Fetus displaying microcephaly, facial dysmorphism, and limb anomalies. d Enlargement of the clitoris. e Syndactyly at both feet and hypoplastic first toe. f Microtia and retrognatism, hypoplasia of the mandibula. g Axial brain MRI with pachygyria especially of the frontal lobes. h Sagittal brain MRI with small but completely development. i, j Radiographs of the right and left arm with hypoplastic radius and bilateral fifth finger clinodactyly. k Syndactyly, keeping the feet in a position of pes equinovarus
During the next pregnancy a few months later, ultrasonography again revealed severe growth retardation of the fetus. In the 19th week of gestation, microcephaly and limb malformations were documented. Fetal MRI showed a flat sulcus centralis and failed to detect the corpus callosum. Microtia and dysplasia of the upper and lower limbs were apparent. Mesomelia of the upper limbs included shortening of both radii and ulnae; bilateral brachydactyly was also present. Syndactyly II–V at both feet was noted. Striking shortening of the halluces qualified as bilateral hypoplasia of the first digit. Ankle flexion contractures of the feet were also observed in the second affected sib.
Cytogenetic analysis of amniocytes revealed a normal female karyotype in 21 metaphases examined. Two different structural chromosome aberrations, 46,XX,del(14)(q2?4) and 46,XX,del(15)(q1?5), were observed in single metaphases and considered artifacts since array-CGH analysis was inconspicuous, arr(1-22,X)x2, as in the previous fetus. This time MMC-induced G2-phase accumulation in amniocyte cultures was below the limits of Fanconi anemia.
The girl was born after 38 + 2 gestational weeks with a birth weight of 2000 g (−2.9 SD), a birth length of 40 cm (−4.67 SD), and a head circumference of 29 cm (−4.14 SD). The malformations of the face and limbs were confirmed, including bilateral microtia grade II, retrognathia with hypoplasia of the mandibula, moderate hypertelorism, mesomelia of the upper limbs, bilateral clinodactyly of the fifth finger, and bilateral syndactyly of toes II–V. Club feet-like deformity was also present bilaterally (Fig. 1f, i, j, k).
During the first days of life, inspiratory stridor was noted, which worsened over the next months. Bronchoscopy revealed subglottic stenosis while MRI showed a narrowed diameter of the mainstem bronchiae. Continuous positive airway pressure became necessary during sleep. At the age of 9 months, the subglottic stenosis became life threatening, thus tracheotomy was performed.
Cerebral MRI in the first month of life showed pachygyria with an emphasis on both frontal lobes, whereas the basal ganglia, brain stem, and infratentorial structures demonstrated age-appropriate development. The corpus callosum, however, was small (Fig. 1g, h).
During the first weeks, oral food intake was adequate. Later on, the child did not adapt to the increasing nutritional needs such that naso-gastric tube-feeding became necessary. At 9 months of age, a percutaneous gastric tube was placed. At this time, the diagnosis of MPD was confirmed. Her weight was 4.85 kg (−4.43 SD), her length 54 cm (−6.74 SD), and her fronto-occipital head circumference 36 cm (−8.69 SD). She revealed gross motor developmental delay comparable to a 3-month-old child, whereas her social skills and hand functions were significantly better and almost appropriate for age.
Given the microcephalic, “bird-face-like” aspect and limb defects, a thorough investigation for immunodeficiencies was carried out. Indeed, the girl presented with leukopenia (3600 per µl). T-lymphocyte subsets showed decreased numbers including CD3+ (97 per µl), CD8+ (60 per µl), and CD4+ (44 per µl), and NK cell counts were also low (CD56+, 96 per µl), whereas B-cell counts were normal (CD19+, 316 per µl) resembling a T−, B+, NK− severe combined immunodeficiency (SCID) phenotype. T-cell receptor excision circles were only slightly reduced while kappa-deleting recombination excision circle counts were normal consistent with regular B-cell function. Immunoglobulins G, A, and M showed normal levels. No remaining maternal T cells were detected. SCID screens yielded normal results. Alpha-1-fetoprotein determined at post-natal day 42 (421.9 ng/ml) was within the normal range for age excluding ataxia telangiectasia. DiGeorge syndrome was ruled out by array-CGH analysis and Nijmegen breakage syndrome by absent cellular hyperradiosensitivity and failure to detect variants in the NBN gene affecting its function. A cellular MMC sensitivity study was repeated and excluded Fanconi anemia again. Because of neutropenia and lymphocytopenia, prophylactic cotrimoxazole therapy was initiated. No severe infections were observed. Live vaccinations were postponed awaiting an increase in T-cell numbers.
Results
Assuming the same genetic cause of disease in both siblings, whole-exome sequencing of the index patient, one unaffected sibling, and both parents was performed. Included into analysis was the workflow described by Zemojtel et al. [6] to identify a function affecting variant in the disease-associated genome. Because the pedigree suggested a recessive trait in a non-consanguineous family, sequence data were filtered for variants that are compound heterozygous or homozygous in the index, heterozygous in the parents, and neither compound heterozygous nor homozygous in the unaffected sibling using GeneTalk [7, 8]. Additional positive filtering criteria were an allele frequency of <0.0001, a sequence variant affecting protein quality or level or both (missense, stopgain, stoploss, and splice variants, and frameshift or non-frameshift small indels), a quality score >10, and a coverage >30 in at least one sample. Evaluation of the 13 single nucleotide variants in eight genes that passed filtering suggested two compound heterozygous variants in DONSON (NM_017613.3: [c.1433C>T];[1047-2A>G]) as the most likely cause of the patient’s disorder. One is likely to result in an amino acid substitution (NM_017613.3: c.1433C>T; p.(Pro478Leu) that is NC_000021.8:g.34951786G>A) while the other abrogates a canonical splice acceptor (NM_017613.3: c.1047-2A>G that is NC_000021.8:g.34954363T>C) in DONSON (all variants described in this paper refer to the DONSON transcript NM_017613.3 positioned at NC_000021.8:g.34949859-34961267). Of 121,404 alleles listed in the database of Exome Aggregation Consortium (ExAC) [9], only three feature the missense variant (reflecting an allele frequency of 0.000025) that is consistent with c.1433C>T being a missense variant having a deleterious effect, while the splice site variant is not listed at all on the ExAC browser. Neither of the two variants is listed in the 1000 Genomes Project’s database [10]. Both affect highly conserved positions of the genome with a phyloP score [11] of 5.5 and 2.52, respectively. Both were predicted to be disease-causing by MutationTaster [12]. Sanger sequencing confirmed maternal segregation of the missense and paternal segregation of the splice site variant. Both affected siblings proved to be compound heterozygous for both variants, while of the two unaffected siblings, one was heterozygous for the missense variant and the other featured neither of the two variants. Variant and phenotypic data can be found in the Leiden Open Variation Database (www.LOVD.nl/DONSON) individual IDs 00133271 and 00134048.
Discussion
In the present report, we describe siblings with microcephaly, short stature, and limb malformations, one of them initially misdiagnosed with Fanconi anemia as this has been the case in three previous siblings from a consanguineous Palestinian family [13], later included as P13-1, P13-2, and P13-3 into the cohort of individuals with microcephaly, short stature, and limb abnormalities and DONSON variants of Reynolds et al. [1] because they were homozygous for the missense variant c.1337T>C, p.(Met446Thr). Figure 2 depicts all DONSON variants predicted to have a deleterious effect on protein function. Whole-exome sequencing in the present cases identified compound heterozygous variants in DONSON. The missense variant c.1433C>T; p.(Pro478Leu) had been identified in neither of the two previous studies that linked DONSON variants with microcephaly syndromes [1, 2]. Nevertheless, it should be noted that DONSON Pro478 is highly conserved among vertebrates (Fig. 3). The splice site variant c.1047-2A>G has likewise not been reported before as a DONSON variant. It is very close and similar to the confirmed splice site variant c.1047-9A>G that was identified as disease-causing in a large consanguineous pedigree of a First Nations family in Saskatchewan, first described by Ives and Houston [3], whereas the other patients in the report of Evrony et al. [2] belonged to the same population and were known to be descendants of the founders of this pedigree, although the exact relationships were unknown. This same splice variant was independently detected in the two Saudi Arabian siblings, P21-1 and P21-2 [1]. It is located only 7 bp upstream of the splice variant identified in the present report. The variant c.1047-2A>G of the present siblings is located within the canonical splice acceptor dinucleotide of DONSON intron 6. The almost invariant AG sequence that terminates an intron at the 3′ end is essential for canonical splicing, termed the lariat pathway, and exon definition. Intron retention is the most common effect of base substitutions in canonical splice dinucleotides [14].
DONSON variants predicted to have a deleterious effect on protein function. Blocks represent coding exons of DONSON (NM_017613.3 exon numbering according to Reynolds et al. [1]). Variants are described using cDNA nomenclature. Variants beneath the exon cartoon detected by Reynolds et al. [1] and Evrony et al. [2]. Underlined variants belong to one haplotype (co-segregating). Variants above the exon cartoon are described in the present study. Note the close proximity of c.1047-2A>G to c.1047-9A>G, and of c.1433C>T to other missense variants in exon 9
Reynolds et al. [1] showed that c.1047-9A>G had an effect on splicing and resulted in lower expression levels of DONSON. Furthermore, Evrony et al. [2] presented compelling evidence that c.1047-9A>G causes intron 6 retention, premature termination of translation, and largely downscaled levels of functional DONSON protein. They also demonstrated that the splice acceptor of DONSON intron 6 is weak and even wild-type intron 6 is already spliced imperfectly in normal cells. They postulated that variants around this splice junction in general could compromise intron 6 splicing efficiency [2]. It is conceivable that if c.1047-9A>G already is a splice variant, this is all the more true for c.1047-2A>G.
Absent phenotypes of confirmed heterozygous carriers of DONSON variants in our present study, including the parents and one of the unaffected brothers of the present cases, are consistent with previous observations in DONSON variant carriers and are also consistent with autosomal recessive inheritance of MPD and MMS due to DONSON variants.
Variants in DONSON have been reported to result either in MPD, in the microcephaly, short stature, and limb abnormality disorder, or in the MMS [1, 2]. The current studies by us and others convey the idea that the boundaries between these entities are fluid. We think the most appropriate diagnosis for the present siblings would be the microcephaly, short stature, and limb abnormality disorder as the second affected sib did not show the perinatal lethal phenotype of the MMS although she revealed a severe-type splice variant similar to that of Evrony et al. [2]. Radial defects in MPD have attracted little attention although they are not an uncommon symptom in microcephaly cases, but their underlying genetic basis is still poorly understood. We add more evidence that radial/thumb defects are recurrent phenotypes of microcephaly syndromes. This provides a link to Fanconi anemia, a genetically heterogeneous disorder whose underlying genes are involved in DNA repair as is DONSON. Almost 2/3 of individuals affected by Fanconi anemia have physical abnormalities that can include short stature, congenital malformations of the heart, eyes, kidneys, and skeletal system (especially thumbs and forearms), hearing loss, endocrine problems, and developmental delay that may manifest themselves similar to DONSON variants [15]. The Fanconi anemia pathway repairs DNA interstrand crosslinks in the genome and shares components with other DNA repair processes. Moreover, Fanconi anemia proteins have other functions in genome maintenance including stabilization of the replication fork. DONSON, similar to the Fanconi anemia proteins, is emerging as an important regulator of genomic integrity implicated in key processes of DNA replication and repair.
References
Reynolds JJ, Bicknell LS, Carroll P, et al. Mutations in DONSON disrupt replication fork stability and cause microcephalic dwarfism. Nat Genet. 2017;49:537–49.
Evrony GD, Cordero DR, Shen J, et al. Integrated genome and transcriptome sequencing identifies a noncoding mutation in the genome replication factor DONSON as the cause of microcephaly-micromelia syndrome. Genome Res. 2017;27:1323–35.
Ives EJ, Houston CS. Autosomal recessive microcephaly and micromelia in Cree Indians. Am J Med Genet. 1980;7:351–60.
Harley ME, Murina O, Leitch A, et al. TRAIP promotes DNA damage response during genome replication and is mutated in primordial dwarfism. Nat Genet. 2016;48:36–43.
Klingseisen A, Jackson AP. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev. 2011;25:2011–24.
Zemojtel T, Kohler S, Mackenroth L, et al. Effective diagnosis of genetic disease by computational phenotype analysis of the disease-associated genome. Sci Transl Med. 2014;6:252ra123.
Kamphans T, Krawitz PM. GeneTalk: an expert exchange platform for assessing rare sequence variants in personal genomes. Bioinformatics. 2012;28:2515–6.
Kamphans T, Sabri P, Zhu N, et al. Filtering for compound heterozygous sequence variants in non-consanguineous pedigrees. PLoS ONE. 2013;8:e70151.
Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74.
Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–21.
Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–6.
Milner RD, Khallouf KA, Gibson R, Hajianpour A, Mathew CG. A new autosomal recessive anomaly mimicking Fanconi’s anemia phenotype. Arch Dis Child. 1993;68:101–3.
Krawczak M, Thomas NS, Hundrieser B, et al. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum Mutat. 2007;28:150–8.
Shimamura A, Alter BP. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev. 2010;24:101–22.
Acknowledgements
We thank the family for their cooperation. M.A.M. is participant in the BIH Charité Junior Clinician Scientist Program funded by the Charité—Universitätsmedizin Berlin and the Berlin Institute of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Schulz, S., Mensah, M.A., de Vries, H. et al. Microcephaly, short stature, and limb abnormality disorder due to novel autosomal biallelic DONSON mutations in two German siblings. Eur J Hum Genet 26, 1282–1287 (2018). https://doi.org/10.1038/s41431-018-0128-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-018-0128-0
This article is cited by
-
The expanding genetic and clinical landscape associated with Meier-Gorlin syndrome
European Journal of Human Genetics (2023)
-
Differentiation of MISSLA and Fanconi anaemia by computer-aided image analysis and presentation of two novel MISSLA siblings
European Journal of Human Genetics (2019)