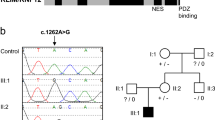
Abstract
Protein arginine N-methyltransferase 7 (PRMT7) encodes an arginine methyltransferase central to a number of fundamental biological processes, mutations in which result in an autosomal recessive developmental disorder characterized by short stature, brachydactyly, intellectual developmental disability and seizures (SBIDDS). To date, fewer than 15 patients with biallelic mutations in PRMT7 have been documented. Here we report brothers from a consanguineous Iraqi family presenting with a developmental disorder characterized by global developmental delay, shortened stature, facial dysmorphisms, brachydactyly, and kidney dysfunction. In both affected brothers, whole genome sequencing (WGS) identified a novel homozygous substitution in PRMT7 (ENST00000339507.5), c.1097 G > A (p.Cys366Tyr), considered to account for the majority of the phenotypic presentation. Rare compound heterozygous mutations in the dysplasia-associated perlecan-encoding HSPG2 gene (ENST00000374695.3) were also found (c.10721-2dupA, p.Ser71Asn and c.212 G > A), potentially accounting for the kidney dysfunction. In addition to expanding the known mutational spectrum of variably expressive PRMT7 mutations alongside potential digenic inheritance with HSPG2, this report underlines the diagnostic utility of a WGS-guided analysis in the detection of rare genetic disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Krause CD, Yang Z-H, Kim Y-S, Lee J-H, Cook JR, Pestka S. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharm Ther. 2007;113:50–87.
Carithers LJ, Moore HM. The Genotype-Tissue Expression (GTEx) Project. Biopreserv Biobank. 2015;13:307–8.
Migliori V, Müller J, Phalke S, Low D, Bezzi M, Mok WC, et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19:136–44.
Di Lorenzo A, Bedford MT. Histone arginine methylation. FEBS Lett. 2011;585:2024–31.
Szewczyk MM, Ishikawa Y, Organ S, Sakai N, Li F, Halabelian L, et al. Pharmacological inhibition of PRMT7 links arginine monomethylation to the cellular stress response. Nat Commun. 2020;11:2396.
Li WJ, He YH, Yang JJ, Hu GS, Lin YA, Ran T, et al. Profiling PRMT methylome reveals roles of hnRNPA1 arginine methylation in RNA splicing and cell growth. Nat Commun. 2021;12:1946.
Kernohan KD, McBride A, Xi Y, Martin N, Schwartzentruber J, Dyment DA, et al. Loss of the arginine methyltranserase PRMT7 causes syndromic intellectual disability with microcephaly and brachydactyly. Clin Genet. 2017;91:708–16.
Birnbaum R, Yosha-Orpaz N, Yanoov-Sharav M, Kidron D, Gur H, Yosovich K, et al. Prenatal and postnatal presentation of PRMT7 related syndrome: expanding the phenotypic manifestations. Am J Med Genet Part A. 2019;179:78–84.
Akawi N, McRae J, Ansari M, Balasubramanian M, Blyth M, Brady AF, et al. Discovery of four recessive developmental disorders using probabilistic genotype and phenotype matching among 4,125 families. Nat Genet. 2015;47:1363–9.
Jeong HJ, Lee HJ, Vuong TA, Choi KS, Choi D, Koo SH, et al. Prmt7 deficiency causes reduced skeletal muscle oxidative metabolism and age-related obesity. Diabetes. 2016;65:1868–82.
Blanc RS, Vogel G, Chen T, Crist C, Richard S. PRMT7 Preserves Satellite Cell Regenerative Capacity. Cell Rep. 2016;14:1528–39.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Broad Institute. Picard tools [Internet]. https://broadinstitute.github.io/picard/. 2016.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, et al. From FastQ Data to High-Confidence Variant Calls: the Genome Analysis Toolkit Best Practices Pipeline. In: Current Protocols in Bioinformatics. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2013. p.11.10.1–33.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, et al. From fastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinforma. 2013;43:11.10.1–33.
Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–81.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
1000 Genomes. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73.
Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92.
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinforma. 2012;13:134.
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122.
Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4.
Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;7:Unit7.20.
Lopes MC, Joyce C, Ritchie GRS, John SL, Cunningham F, Asimit J, et al. A combined functional annotation score for non-synonymous variants. Hum Hered. 2012;73:47–51.
Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AFA, Roskin KM, et al. Aligning Multiple Genomic Sequences With the Threaded Blockset Aligner. Genom Res. 2004;14:708–15.
Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–48.
Yeo G, Burge CB. Maximum Entropy Modeling of Short Sequence Motifs with Applications to RNA Splicing Signals. J Comput Biol. 2004;11:377–94.
Leman R, Gaildrat P, Gac GL, Ka C, Fichou Y, Audrezet MP, et al. Novel diagnostic tool for prediction of variant spliceogenicity derived from a set of 395 combined in silico/in vitro studies: an international collaborative effort. Nucleic Acids Res. 2018;46:7913–23.
Desmet FO, Hamroun D, Lalande M, Collod-Bëroud G, Claustres M, Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67.
Wang M, Marín A. Characterization and prediction of alternative splice sites. Gene. 2006;366:219–27.
Ferreira CR. The burden of rare diseases. Am J Med Genet A. 2019;179:885–92.
Boycott KM, Vanstone MR, Bulman DE, MacKenzie AE. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet. 2013;14:681–91.
Katsanis SH, Katsanis N. Molecular genetic testing and the future of clinical genomics. Nat Rev Genet. 2013;14:415–26.
Musante L, Ropers HH. Genetics of recessive cognitive disorders. Trends Genet. 2014;30:32–9.
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006.
Berry C, Thomas M, Langley B, Sharma M, Kambadur R. Single cysteine to tyrosine transition inactivates the growth inhibitory function of Piedmontese myostatin. Am J Physiol Cell Physiol. 2002;283:C135–41.
Kernohan KD, McBride A, Xi Y, Martin N, Schwartzentruber J, Dyment DA, et al. Loss of the arginine methyltranserase PRMT7 causes syndromic intellectual disability with microcephaly and brachydactyly. Clin Genet. 2017;91:708–16.
Agolini E, Dentici ML, Bellacchio E, Alesi V, Radio FC, Torella A, et al. Expanding the clinical and molecular spectrum of PRMT7 mutations: 3 additional patients and review. Clin Genet. 2018;93:675–81.
Valenzuela I, Segura-Puimedon M, Rodríguez-Santiago B, Fernández-Alvarez P, Vendrell T, Armengol L, et al. Further delineation of the phenotype caused by loss of function mutations in PRMT7. Eur J Med Genet. 2019;62:182–5.
Basalom S, Trakadis Y, Shear R, Azouz ME, De Bie I. Dyssegmental dysplasia, Silverman-Handmaker type: a challenging antenatal diagnosis in a dizygotic twin pregnancy. Mol Genet Genom Med. 2018;6:452–6.
Arikawa-Hirasawa E, Wilcox WR, Le AH, Silverman N, Govindraj P, Hassell JR, et al. Dyssegmental dysplasia, Silverman-Handmaker type, is caused by functional null mutations of the perlecan gene. Nat Genet. 2001;27:431–4.
Arikawa-Hirasawa E, Le AH, Nishino I, Nonaka I, Ho NC, Francomano CA, et al. Structural and Functional Mutations of the Perlecan Gene Cause Schwartz-Jampel Syndrome, with Myotonic Myopathy and Chondrodysplasia. Am J Hum Genet. 2002;70:1368–75.
Izzi B, Francois I, Labarque V, Thys C, Wittevrongel C, Devriendt K, et al. Methylation defect in imprinted genes detected in patients with an albright’s hereditary osteodystrophy like phenotype and platelet gs hypofunction. PLoS ONE. 2012;7:e38579.
Cianferotti L, Brandi ML. Pseudohypoparathyroidism. Minerva Endocrinol. 2018;43:156–67.
Mantovani G, De Sanctis L, Barbieri AM, Elli FM, Bollati V, Vaira V, et al. Pseudohypoparathyroidism and GNAS epigenetic defects: clinical evaluation of Albright hereditary osteodystrophy and molecular analysis in 40 patients. J Clin Endocrinol Metab. 2010;95:651–8.
Linglart A, Maupetit-Méhouas S, Silve C. GNAS-related loss-of-function disorders and the role of imprinting. Horm Res Paediatr. 2013;79:119–29.
Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–40.
Stavropoulos DJ, Merico D, Jobling R, Bowdin S, Monfared N, Thiruvahindrapuram B, et al. Whole-genome sequencing expands diagnostic utility and improves clinical management in paediatric medicine. NPJ Genom Med. 2016;1:15012.
Acknowledgements
The families are gratefully acknowledged for their participation in the study. The study was supported by the New Zealand eScience Infrastructure.
Funding
JCJ is supported by a government-funded Rutherford Discovery Fellowship administered by the Royal Society of New Zealand. The research was funded by the IHC Foundation.
Author information
Authors and Affiliations
Contributions
JCJ, KL and RGS conceived the experiments. JP and WW performed DNA-based laboratory experiments. JP, WW, KL, RGS and JCJ performed data and bioinformatic analysis. JT and SA conducted the clinical evaluation. JP and JCJ wrote the paper. All authors reviewed the paper.
Corresponding author
Ethics declarations
Ethical approval
Ethical approval was obtained by the Northern B Health and Disability Ethics Committee (12/NTB/59) prior to acquiring, sequencing, and analyzing all human genetic information. All procedures were performed in accordance with the ethical standards of the institutional and national responsible committees on human experimentation and with the 1975 Helsinki Declaration (as revised in 2000).
Patient consent
Consent was obtained from the patient’s family for publication of this report.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Poquérusse, J., Whitford, W., Taylor, J. et al. Novel PRMT7 mutation in a rare case of dysmorphism and intellectual disability. J Hum Genet 67, 19–26 (2022). https://doi.org/10.1038/s10038-021-00955-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-021-00955-5
This article is cited by
-
Short stature in PRMT7 Mutations: first evidence of response to growth hormone treatment
European Journal of Human Genetics (2023)
-
Critical Roles of Protein Arginine Methylation in the Central Nervous System
Molecular Neurobiology (2023)