Abstract
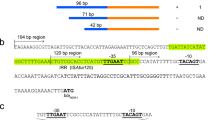
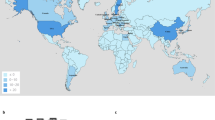
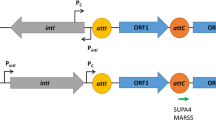
The first report of transmissible carbapenem resistance encoded by blaIMP-1 was discovered in Pseudomonas aeruginosa GN17203 in 1988, and blaIMP-1 has since been detected in other bacteria, including Enterobacterales. Currently, many variants of blaIMPs exist, and point mutations in the blaIMP promoter have been shown to alter promoter strength. For example, the promoter (Pc) of blaIMP-1, first reported in P. aeruginosa GN17203, was a weak promoter (PcW) with low-level expression intensity. This study investigates whether point mutations in the promoter region have helped to create strong promoters under antimicrobial selection pressure. Using bioinformatic approaches, we retrieved 115 blaIMPs from 14,529 genome data of Pseudomonadota and performed multiple alignment analyses. The results of promoter analysis of the 115 retrieved blaIMPs showed that most of them used the Pc located in class 1 integrons (n = 112, 97.4%). The promoter analysis by year revealed that the blaIMP population with the strong promoter, PcS, was transient. In contrast, the PcW-TG population, which had acquired a TGn-extended −10 motif in PcW and had an intermediate promoter strength, gradually spread throughout the world. An inverse correlation between Pc promoter strength and Intl1 integrase excision efficiency has been reported previously [1]. Because of this trade-off, it is unlikely that blaIMPs with strong promoters will increase rapidly, but the possibility that promoter strength will increase with the use of other integrons cannot be ruled out. Monitoring of the blaIMP genes, including promoter analysis, is necessary for global surveillance of carbapenem-resistant bacteria.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Jové T, Re SD, Denis F, Mazel D, Ploy M-C. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet. 2010;6:e1000793.
Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35:147–51.
Osano E, et al. Molecular characterization of an enterobacterial metallo β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob Agents Chemother. 1994;38:71–8.
Minami S, et al. Imipenem and cephem resistant Pseudomonas aeruginosa carrying plasmids coding for class B β-lactamase. J Antimicrob Chemother. 1996;37:433–44.
Iyobe S, Yamada H, Minami S. Insertion of a carbapenemase gene cassette into an integron of a Pseudomonas aeruginosa plasmid. J. Antimicrob Chemother. 1996;38:1114–5.
Arakawa Y, et al. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother. 1995;39:1612–5.
Collis CM, Kim M-J, Partridge SR, Stokes HW, Hall RM. Characterization of the class 3 integron and the site-specific recombination system it determines. J Bacteriol. 2002;184:3017–26.
Dixon N, et al. IMP-27, a unique metallo-β-lactamase identified in geographically distinct isolates of Proteus mirabilis. Antimicrob Agents Chemother. 2016;60:6418–21.
Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site‐specific gene‐integration functions: integrons. Mol Microbiol. 1989;3:1669–83.
Ghaly TM, et al. The natural history of integrons. Microorganisms. 2021;9:2212.
Partridge SR, Tsafnat G, Coiera E, Iredell JR. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009;33:757–84.
Lévesque C, Brassard S, Lapointe J, Roy PH. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integron. Gene. 1994;142:49–54.
Bunny KL, Hall RM, Stokes HW. New mobile gene cassettes containing an aminoglycoside resistance gene, aacA7, and a chloramphenicol resistance gene, catB3, in an integron in pBWH301. Antimicrob Agents Chemother. 1995;39:686–93.
Papagiannitsis CC, Tzouvelekis LS, Miriagou V. Relative strengths of the class 1 integron promoter hybrid 2 and the combinations of strong and hybrid 1 with an active P2 promoter. Antimicrob. Agents Chemother. 2009;53:277–80.
Yamamoto M, et al. Molecular analysis of a blaIMP-1-harboring class 3 integron in multidrug-resistant Pseudomonas fulva. Antimicrob Agents Chemother. 2018;62:e00701–18.
Wang T, Zhu Y, Zhu W, Cao M, Wei Q. Molecular characterization of class 1 integrons in carbapenem-resistant Enterobacterales isolates. Microb Pathog. 2023;177:106051.
Wei Q, et al. Diversity of gene cassette promoter variants of class 1 integrons in uropathogenic Escherichia coli. Curr Microbiol. 2013;67:543–9.
Vinué L, Jové T, Torres C, Ploy M-C. Diversity of class 1 integron gene cassette Pc promoter variants in clinical Escherichia coli strains and description of a new P2 promoter variant. Int J Antimicrob Agents. 2011;38:526–9.
Nešvera J, Hochmannová J, Pátek M. An integron of class 1 is present on the plasmid pCG4 from Gram‐positive bacterium Corynebacterium glutamicum. FEM Microbiol Lett. 1998;169:391–5.
Kumar A, et al. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J Mol Biol. 1993;232:406–18.
Barne KA, Bown JA, Busby SJ, Minchin SD. Region 2.5 of the Escherichia coli RNA polymerase σ70 subunit is responsible for the recognition of the “extended-10” motif at promoters. EMBO J. 1997;16:4034–40.
Burr T, Mitchell J, Kolb A, Minchin S, Busby S. DNA sequence elements located immediately upstream of the –10 hexamer in Escherichia coli promoters: a systematic study. Nucleic Acids Res. 2000;28:1864–70.
Wei Q, et al. Transcription of integron‐harboured gene cassette impacts integration efficiency in class 1 integron. Mol Microbiol. 2011;80:1326–36.
Chapman B, Chang J. Biopython: Python tools for computational biology. ACM SIGBIO Newsl. 2000;20:15–19.
Robinson O, Dylus D, Dessimoz C. Phylo.io: Interactive viewing and comparison of large phylogenetic trees on the web. Mol Bio Evol. 2016;33:2163–6.
Kuraku S, Zmasek CM, Nishimura O, Katoh K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013;41:W22–W28.
South A. rworldmap: a new R package for mapping global data. R J. 2011;3:35.
Chen F-J, et al. Molecular epidemiology of emerging carbapenem resistance in Acinetobacter nosocomialis and Acinetobacter pittii in Taiwan, 2010 to 2014. Antimicrob Agents Chemother. 2019;63:e02007–18.
Li L-H, et al. Clinical and molecular characterization of Acinetobacter seifertii in Taiwan. J. Antimicrob Chemother. 2020;76:312–21.
Chen F, et al. Uncovering the hidden threat: The widespread presence of chromosome-borne accessory genetic elements and novel antibiotic resistance genetic environments in Aeromonas. Virulence. 2023;14:2271688.
Yuan S, Wu G, Zheng B. Complete genome sequence of an IMP-8, CTX-M-14, CTX-M-3 and QnrS1 co-producing Enterobacter asburiae isolate from a patient with wound infection. J Glob Antimicrob Resist. 2019;18:52–54.
Wyrsch ER, et al. Urban Wildlife Crisis: Australian silver gull is a bystander host to widespread clinical antibiotic resistance. mSystems. 2022;7:e00158–22.
Boyd DA, et al. Emergence of Morganellaceae harboring blaIMP-27 metalloenzyme in Canada. mSphere. 2021;6:e00048–21.
Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-β-Lactamases: the quiet before the storm? Clin Microbiol Rev. 2005;18:306–25.
Turton JF, et al. IncN3 and IncHI2 plasmids with an In1763 integron carrying blaIMP-1 in carbapenem-resistant Enterobacterales clinical isolates from the UK. J Méd Microbiol. 2020;69:739–47.
Chu Y-W, et al. IMP-4, a novel metallo-β-lactamase from nosocomial Acinetobacter spp. Collected in Hong Kong between 1994 and 1998. Antimicrob Agents Chemother. 2001;45:710–4.
Acknowledgements
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology, KAKENHI grants from the Japan Society for the Promotion of Science (nos. 19K07542 to A.A. and 20K07485, 23K06531 to A.K.) and AMED grants (nos. JP20nk0101552 to A.A. and JP22nk0101587 to A.A.). The funders had no role in the study design, data collection or analysis, the decision to publish, or the preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kikuchi, Y., Yoshida, M., Kuwae, A. et al. Correlation between the spread of IMP-producing bacteria and the promoter strength of blaIMP genes. J Antibiot 77, 315–323 (2024). https://doi.org/10.1038/s41429-024-00715-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-024-00715-5