Abstract
Natriuretic peptide type C (NPPC) secreted by mural granulosa cells (MGCs) maintains oocyte meiotic arrest via the activation of guanylyl cyclase-linked natriuretic peptide receptor 2 (NPR2). Here, we investigated the effect of transforming growth factor (TGF)-β on NPPC expression in MGCs and oocyte maturation. TGF-β ligands (TGFB1 and TGFB3, but not TGFB2) and receptors (TGFBR1 and TGFBR2) were predominantly expressed in MGCs. The activation of the follicle-stimulating hormone (FSH) receptor by FSH/equine chorionic gonadotropin (eCG) increased the levels of TGFB1, TGFBR2, and TGF-β downstream SMAD proteins in MGCs, which were decreased following the activation of the luteinizing hormone (LH) receptor by human chorionic gonadotropin (hCG). TGF-β significantly increased the gene and protein levels of NPPC in cultured MGCs through SMAD3 binding to Nppc promoter regions. In the presence of FSH, TGF-β further increased NPPC levels and inhibited oocyte meiotic resumption of cumulus-oocyte complexes (COCs). Moreover, Tgfbr2-specific depletion in granulosa cells using Fshr-Cre mice reduced NPPC mRNA and protein levels, resulting in the weak maintenance of oocyte meiotic arrest within large antral follicles. Tgfbr2 depletion also impaired follicle development, ovulation, and female fertility. Taken together, TGF-β-promoted NPPC in MGCs is involved in maintaining oocyte meiotic arrest. FSH and LH could regulate NPPC levels in MGCs via TGF-β and then control the process of oocyte meiosis.
Similar content being viewed by others
Introduction
Mammalian oocyte meiosis begins in the fetal period but is arrested at the diplotene stage of the first meiotic prophase around the time of birth for a prolonged period1,2. Oocytes within preantral follicles do not possess the competence to progress through meiosis due to low levels of cell cycle regulatory proteins inherent in the oocyte3. When a fluid-filled follicle antrum forms, the arrested oocytes reach their full size and have the ability to resume meiosis4,5. However, natriuretic peptide type C (NPPC, also known as CNP), secreted by mural granulosa cells (MGCs), maintains oocyte meiotic arrest via its cognate receptor natriuretic peptide receptor 2 (NPR2) producing cyclic guanosine monophosphate (cGMP)6,7,8,9.
The activation of the follicle-stimulating hormone (FSH) receptor by FSH/equine chorionic gonadotropin (eCG, a glycoprotein hormone that primarily has FSH activity) leads to the formation of early antral follicles that eventually develop into preovulatory follicles10. During follicle development, the levels of NPPC/NPR2 in granulosa cells are increased to ensure oocyte meiotic arrest. The activation of the luteinizing hormone (LH) receptor by LH/human chorionic gonadotropin (hCG, a pregnancy hormone that exhibits LH activity) leads to a decrease in NPPC/NPR2 levels11,12,13,14, and NPR2 activity by dephosphorylation and a reduction in NPPC-binding affinity15,16,17. All of these factors reduce the production of cGMP, thereby relieving cGMP-mediated inhibition of meiotic arrest7,8. In vitro evidences suggest that the regulation of the expression of NPPC/NPR2 in granulosa cells by FSH and LH could be an indirect effect that is mediated by autocrine factors produced by granulosa cells. For example, previous studies showed that estradiol (E2) promotes Nppc and Npr2 mRNA expression in cultured MGCs14,18. Given that FSH and LH affect E2 biosynthesis by specifically regulating the aromatizing enzyme system19,20, FSH and LH could regulate NPPC/NPR2 levels through the control of E2 production. However, aromatase deletion had no effect on oocyte maturation, and estrogen receptor (ER) knockout only had a partial effect on oocyte meiotic resumption21,22. Thus, other factors could also be involved in promoting the expression of NPPC/NPR2 for meiotic arrest23.
NPPC expression and secretion are increased by transforming growth factor β (TGF-β) in human and rat vascular smooth muscle cells and bovine aortic endothelial cells24,25,26. In mammals, the TGF-β family includes three ligands: TGFB1, TGFB2, and TGFB3. These ligands bind to their membrane receptors and trigger the serine/threonine protein kinase activity of type 2 receptor (TGFBR2) and type 1 receptor (TGFBR1), and then the activated receptor phosphorylates intracellular SMAD2 (Sma- and Mad-related protein 2) and SMAD3. Subsequently, phosphorylated SMAD2 and SMAD3 form a heteromeric complex with SMAD4 (co-SMAD, the central signaling component of the TGF-β superfamily)27,28. Ultimately, the complex translocates into the nucleus and regulates gene expression by binding to the target gene promoter region, termed an SMAD-binding element (SBE), and recruiting distinct transcription factors29.
The expression of TGF-β ligands and their receptors has been observed in mammalian ovaries from different species30,31,32. Accumulating evidence indicates that TGF-β participates in multiple ovary functions, including follicle growth33, granulosa cell proliferation34,35,36, and ovulation37. Moreover, TGFB1 had an inhibitory effect on the spontaneous meiotic resumption of porcine cumulus-oocyte complexes (COCs)38. Although Smad4 depletion in granulosa cells decreases the mRNA levels of Nppc and Npr2, resulting in the weak maintenance of oocyte meiosis arrest39, its specific upstream signaling molecule(s) is unclear. Thus, we investigated the expression pattern of TGF-β ligands and receptors in mouse ovaries and their effects on NPPC/NPR2 expression and oocyte maturation.
Results
The expression pattern of TGF-β ligands and receptors in mouse ovary
We first detected the localization of TGF-β ligands and receptors in eCG-primed mouse ovaries by immunofluorescence. TGFB1, TGFB3, TGFBR1, and TGFBR2 were clearly detected in the cytoplasm of MGCs and cumulus cells, and TGFB1 and TGFBR1 staining were also weak in the oocyte cytoplasm (Fig. 1a). In contrast, TGFB2 was mainly located in the cytoplasm of oocytes and was slightly stained in the cytoplasm of MGCs and cumulus cells (Fig. 1a). Theca cells had a slight expression of these ligands and receptors (Fig. 1a).
a Immunofluorescence analysis of TGFB1, TGFB2, TGFB3, TGFBR1, and TGFBR2 in the ovaries isolated from eCG-primed mice. The images are representative of three images captured. The small white boxes indicate the enlarged areas as shown in the following images. Scale bars: 200 μm. b Comparison of steady-state levels of Tgfb1, Tgfb2, Tgfb3, Tgfbr1, and Tgfbr2 mRNA in MGCs, cumulus cells (CCs) and oocytes isolated from eCG-primed mice. c Representative western blotting of TGFB1, TGFB2, TGFB3, TGFBR1, and TGFBR2 in MGCs, CCs and, oocytes isolated from eCG-primed mice. GAPDH was used as a loading control. Bars indicate the mean ± SEM of three independent replicates. Values not indicated by the same letter are significantly different (P < 0.05)
Next, we compared the expression levels of TGF-β ligands and receptors in MGCs, cumulus cells, and oocytes. The mRNA and protein levels of TGFB1 and TGFBR2 in MGCs were higher than in cumulus cells and oocytes (Fig. 1b, c). TGFB3 and TGFBR1 levels were higher in both MGCs and cumulus cells compared with those in oocytes (Fig. 1b, c). TGFB2 mRNA and protein levels were dramatically higher in oocytes than in MGCs and cumulus cells, which are consistent with the immunofluorescence results (Fig. 1b, c). Collectively, TGFB1, TGFB3, TGFBR1, and TGFBR2 are mainly expressed in MGCs, and TGFB2 is mainly expressed in oocytes. We speculate that TGF-β plays a physiological role in these cells.
Gonadotropins regulate the expression of TGF-β signaling molecules in MGCs in vivo
We studied the effect of eCG on the expression of TGF-β signaling molecules in MGCs during follicle development. Treatment with eCG significantly increased the mRNA levels of Tgfb1, Tgfbr1, and Tgfbr2 in MGCs (Fig. 2a). Consistent with this, western blotting results showed that the protein levels of TGFB1, TGFBR1, and TGFBR2 were significantly increased (Fig. 2b). Furthermore, eCG significantly increased the mRNA and protein levels of SMAD2, SMAD3, and SMAD4 (Fig. 2c, d). The phosphorylation levels of SMAD3, but not SMAD2, were also increased (Fig. 2d). We next examined the expression change in TGF-β signaling molecules in MGCs during hCG-induced oocyte maturation. After hCG treatment, the mRNA and/or protein levels of TGFB1, TGFBR2, SMAD3, SMAD4, and phosphorylated SMAD3 were decreased to pre-eCG levels (Fig. 2a–d). Gonadotropins had no effect on the expression of TGFB2 in oocytes (Fig. S2a) and TGFB3 in MGCs (Fig. 2a, b), indicating that other factor(s) regulates their expression.
MGCs were isolated from prepubertal mice, eCG-primed mice and eCG-primed mice followed by hCG as indicated in the figures. a Quantitative RT-PCR analysis of the gene expression of Tgfb1, Tgfb3, Tgfbr1, and Tgfbr2 in MGCs. b Western blotting analysis of the protein levels of TGFB1, TGFB3, TGFBR1, and TGFBR2 in MGCs. c Quantitative RT-PCR analysis of the gene expression of Smad2, Smad3, and Smad4 in MGCs. d Western blotting analysis of the protein levels of SMAD2/3, SMAD4, and phosphorylated SMAD2/3 (p-SMAD2/3) in MGCs. The blots shown are representative of three images captured in b, d. GAPDH was used as a loading control in b, d. Bars indicate the mean ± SEM of three independent replicates. Values not indicated by the same letter are significantly different (P < 0.05)
FSH upregulates TGF-β signaling molecules in MGCs in vitro
We studied the effect of FSH on the expression of TGF-β signaling molecules by culturing MGCs. FSH treatment significantly increased the mRNA levels of Tgfb1, Tgfbr2, Smad2, Smad3, and Smad4 (Fig. 3a). Consistent with the results of gene expression, western blotting analyses revealed that an FSH significantly increased the protein levels of TGFB1, TGFBR2, SMAD2, SMAD3, SMAD4, and phosphorylated SMAD3 (Fig. 3b). Immunofluorescence results also showed that FSH treatment obviously increased the cytoplasm accumulation of TGFB1 and TGFBR2 and the nuclear accumulation of SMAD4 and phosphorylated SMAD3 (Fig. 3c). Thus, FSH upregulates TGF-β signaling molecules in MGCs and activates the TGF-β signaling pathway directly.
MGCs collected from prepubertal mice were cultured for 24 h in cell culture medium without or with 10 ng/ml FSH. a Effects of FSH on Tgfb1, Tgfb3, Tgfbr1, Tgfbr2, Smad2, Smad3, and Smad4 mRNA levels in MGCs. b Effects of FSH on the protein levels of TGFB1, TGFBR2, SMAD2/3, SMAD4, and phosphorylated SMAD2/3 (p-SMAD2/3) in MGCs. The blots shown are representative of three images captured. GAPDH was used as a loading control. c Immunofluorescence analysis for TGFB1, TGFBR2, SMAD2/3, SMAD4, and phosphorylated SMAD3 (p-SMAD3) in MGCs. The images are representative of three images captured. Scale bars: 50 μm. Bars indicate the mean ± SEM of three independent replicates. Values not indicated by the same letter are significantly different (P < 0.05)
TGF-β promotes NPPC expression and inhibits oocyte meiotic resumption
We cultured MGCs to investigate the effect of TGF-β on NPPC expression. As a result, TGFB1, TGFB2, TGFB3, or the combination of these three TGF-β ligands (here as TGF-β) promoted the expression of Nppc mRNA (Fig. 4a). Western blotting and a luciferase immunoassay showed that TGF-β increased NPPC protein levels (Fig. 4b, c), which is consistent with qRT-PCR results. FSH, as reported previously18, had no effect on NPPC expression (Fig. 4a, b) and significantly enhanced TGF-β-induced NPPC expression (Fig. 4a–c). TGF-β-promoted NPPC was completely reversed by the TGF-β signaling inhibitor SD208 (Fig. 4a, b). These results indicate that TGF-β upregulates NPPC expression in MGCs. However, TGF-β had no effect on the NPR2 gene and protein expression in cultured MGCs (Fig. 4a, b).
a–c MGCs were collected from prepubertal mice and cultured for 24 h in cell culture medium supplemented with 10 ng/ml TGFB1, 50 ng/ml TGFB2, 10 ng/ml TGFB3, TGF-β (including 10 ng/ml TGFB1, 50 ng/ml TGFB2, 10 ng/ml TGFB3), 1 μmol/l SD208 (SD), and/or 10 ng/ml FSH. NPPC and/or NPR2 mRNA and protein levels were detected by qRT-PCR (a), western blotting (b) and a luciferase immunoassay (c), respectively. The blots shown are representative of three images captured. GAPDH was used as a loading control. d Effect of TGF-β on oocyte meiotic resumption of COCs. MGCs were cultured in the medium supplemented with TGF-β, SD208, and/or FSH for 24 h, and then COCs isolated from eCG-primed mice were cocultured with MGCs for 1, 2, 3, and 4 h. n = 30 for each treatment across all three replicates. **P ˂ 0.01 and ***P ˂ 0.001 compared with corresponding control. e Effect of TGF-β on the meiotic resumption of oocytes within follicles. Large antral follicles isolated from eCG-primed mice were cultured in the medium supplemented with TGF-β, and/or SD208 for 4 h. n = 30 for each treatment across all three replicates. Bars indicate the mean ± SEM of three independent replicates. Values not indicated by the same letter are significantly different (P < 0.05)
To study whether TGF-β had an inhibitory effect on oocyte meiotic resumption by promoting NPPC expression, MGCs were cultured in cell culture medium supplemented with TGF-β and/or FSH for 24 h (to induce NPPC production) before COCs were added. As shown in Fig. 4d, TGF-β, in the presence of FSH, significantly inhibited oocyte meiotic resumption when COCs were cocultured with MGCs for 1, 2, 3, and 4 h, which may be related to the ability of FSH to enhance TGF-β-induced NPPC expression by the increasing TGFBR2 as well as SMAD proteins (Fig. 3). FSH alone can inhibit oocyte maturation in a short period of time, possibly because FSH can transiently promote cAMP production40. TGF-β alone had no effect on oocyte maturation of COCs, possibly because TGF-β could not promote adequate expression of NPPC peptide due to the low levels of TGFBR2 and SMADs. During follicle culture, TGF-β also stimulated the expression of Nppc mRNA in MGCs (Fig. S3) and significantly inhibited the meiotic resumption of oocytes within large antral follicles (Fig. 4e). The inhibitory effect of TGF-β on meiotic resumption was blocked by SD208 (Fig. 4d, e). SD208 alone could promote the meiotic resumption of oocytes within the follicles (Fig. 4e), probably because SD208 inhibits TGF-β signaling present in the follicles. Therefore, TGF-β-promoted NPPC in MGCs has an inhibitory effect on oocyte meiotic resumption.
Previous reports indicate that E2 upregulates the expression of Nppc mRNA in MGCs and is involved in maintaining meiotic arrest18,21. Therefore, we investigated the relationship between TGF-β and E2 on NPPC expression in MGCs. The results showed that TGF-β treatment had no effect on the mRNA expression of ERs (Fig. S4a). In turn, E2 had no effect on the mRNA expression of TGF-β ligands and receptors (Fig. S4b). Moreover, the TGF-β signaling inhibitor SD208 did not inhibit the action of E2 on Nppc mRNA expression and oocyte meiotic arrest, and the ER inhibitor ICI182780 did not inhibit the action of TGF-β on Nppc mRNA expression and oocyte meiotic arrest (Fig. S4c, d). These results indicate that TGF-β and E2 independently promote NPPC expression in MGCs.
SMAD3 directly regulates Nppc gene transcription
After TGF-β stimulation, phosphorylated SMAD2/3 enters the nucleus by binding to SMAD4. In this process, activated SMAD3 binds to SBEs that contain AGAC motifs29. We analyzed the Nppc promoter 1–2000 base pairs (bp) before the 5′ untranslated region, which was chosen as the core promoter region. Multiple potential SMAD3-binding sites were identified in the Nppc promoter (Fig. 5a). Chromatin IP (ChIP) analysis was used to detect whether SMAD3 bound to the Nppc promoter to regulate gene transcription. The results showed that SMAD3 bound to the −1800 to −1600 bp (R2) and −600 to −400 bp (R8) regions of the Nppc promoter (Fig. 5b).
a Schematic diagram showing the putative SMAD3-binding elements (SBE, AGAC) in the mouse Nppc promoter. The Nppc promoter sequence was divided into ten regions (designated R1–R10). Each region denotes 200 bp. Red lines represent the potential SMAD3-binding sites in the Nppc promoter. The red lines below the axis represent the potential SMAD3-binding sites in the Nppc promoter complementary chain. b ChIP-qPCR analysis of the interaction between SMAD3 protein and the Nppc promoter in MGCs. c, d Effects of TGF-β on the levels of SMAD3 and phosphorylated SMAD3 (p-SMAD3) protein (c) and Nppc mRNA (d) in KK1 cells. KK1 cells were cultured in cell medium without or with TGF-β (including 10 ng/ml TGFB1, 50 ng/ml TGFB2, and 10 ng/ml TGFB3) for 24 h. The blots shown are representative of three images captured. GAPDH was used as a loading control. e Effects of Smad3 expression vectors (pSmad3) on the transiently transfected Nppc gene promoter enhancers (R2 and R8) fused to luciferase reporter vectors in KK1 cells. pNppc represents pGL3.0-basic plasmid containing R2 or R8 region of the Nppc promoter. KK1 cells were treated without or with TGF-β for 24 h. Bars indicate the mean ± SEM of three independent replicates. Values not indicated by the same letter are significantly different (P < 0.05)
To further confirm the interaction between SMAD3 protein and the Nppc promoter, a mouse granulosa cell line (KK1) responding to TGF-β stimulation was used for a dual-luciferase assay (Fig. 5c, d). We cloned R1–R10 regions and inserted them into the multiple cloning sites of a pGL3.0 vector, respectively. These Nppc promoter-driven luciferase reporter plasmids were then transfected with or without a Smad3 overexpression plasmid into KK1 cells for 24 h. In these assays, SMAD3 significantly enhanced the activities of the R2 and R8 regions, which were further enhanced in response to TGF-β treatment (Fig. 5e and S5). Thus, TGF-β promotes NPPC expression through SMAD3 binding to the Nppc promoter.
Selective Tgfbr2 depletion in granulosa cells impairs oocyte meiotic arrest, ovulation, and female fertility
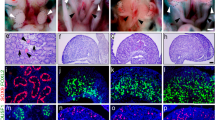
To study the physiological role of TGF-β in NPPC expression and oocyte meiotic arrest, we crossed Tgfbr2fl/fl mice with Fshr-Cre mice to knock out TGF-β-specific type II receptor (TGFBR2) in granulosa cells. In the resulting Tgfbr2gc−/− mice, TGFBR2 expression was dramatically decreased in granulosa cells of antral follicles (Fig. 6a). The reduction in TGFBR2 protein levels in granulosa cells was further confirmed by western blotting results (Fig. 6b). Then, we detected the NPPC level in MGCs. The results showed that the gene and protein levels of NPPC in MGCs from Tgfbr2gc−/− mice were significantly decreased compared with wild-type (WT) controls (Fig. 6c, d). Moreover, TGF-β did not promote the expression of Nppc mRNA in the culture of MGCs isolated from Tgfbr2gc−/− mice (Fig. 6e). Tgfbr2 depletion slightly decreased the mRNA levels of Fshr (encoding FSH receptor) but had no effect on the mRNA levels of Cyp19a1 (a well-established FSH target gene in granulosa cells) and Lhcgr (encoding LH receptor) in MGCs and the levels of E2 in serum (Fig. S6a and b). We next analyzed oocyte meiotic progression by examining serial sections of ovaries from eCG-primed mice. As expected, oocytes in large antral follicles in control ovaries were maintained at the germinal vesicle (GV) stage, and 26% of oocytes in the large antral follicles in Tgfbr2gc−/− mice had resumed meiosis (Fig. 6f, g). In addition, the number of large antral follicles was significantly reduced in Tgfbr2gc−/− mice (Fig. S6c). In the superovulation test, Tgfbr2gc−/− mice displayed a 50% ovulation rate compared with WT mice (Fig. 6i). Consistent with the decreased number of ovulations, fewer corpus lutea (CL) were observed in Tgfbr2gc−/− mice at 48 h after hCG treatment (Fig. 6h). A continuous breeding assay demonstrated that Tgfbr2gc−/− female mice had reduced fertility (Fig. 6j). Collectively, our results indicate that TGF-β promotes NPPC expression in MGCs, which is involved in maintaining oocyte meiotic arrest and plays an important role in normal female fertility.
a, b The detection of TGFBR2 knockout efficiency in MGCs from eCG-primed mice. The expression levels of TGFBR2 in MGCs were detected by immunofluorescence (a) and western blotting (b). c–e Effects of Tgfbr2 depletion on NPPC and NPR2 expressions. MGCs were isolated from eCG-primed mice. The gene and protein levels of NPPC and NPR2 were detected by qRT-PCR (c) and western blotting (d), respectively. MGCs isolated from Tgfbr2gc−/− mice were cultured for 24 h in cell medium without or with TGF-β (including 10 ng/ml TGFB1, 50 ng/ml TGFB2, and 10 ng/ml TGFB3) to detect the effect of TGF-β on Nppc mRNA levels (e). f, g Selective Tgfbr2 depletion in granulosa cells impairs oocyte meiotic arrest. A prophase-arrested oocyte (GV) within a large antral follicle of a WT ovary and an oocyte with metaphase I (MI) chromosomes within a large antral follicle of a Tgfbr2gc−/− ovary (f). Percentages of oocytes that had resumed meiosis, counted in serial sections of ovaries from eCG-primed mice (g). The small black boxes indicate the enlarged areas in the following images. n = 6. h–j Selective Tgfbr2 depletion in granulosa cells impairs ovulation and female fertility. Histology of ovarian sections from WT and Tgfbr2gc−/− ovaries at 48 h after hCG injection (h). CL: corpus luteum. The superovulation assay shows decreased ovulation in Tgfbr2gc−/− mice at 16 h after hCG injection (i). Comparison of the cumulative number of progeny per female WT and Tgfbr2gc−/− female mice over a 6-month period (j). n = 6. *P ˂ 0.05, **P ˂ 0.01, and ***P ˂ 0.001 compared with corresponding control. The images and blots shown are representative of three images captured in a, b, d, f, h. Scale bars: 200 μm in a, f, h. GAPDH was used as a loading control in b, d. Bars indicate the mean ± SEM of three independent replicates. Values not indicated by the same letter are significantly different (P < 0.05)
Discussion
MGCs-secreted NPPC maintains oocyte meiotic arrest by NPR2 producing cGMP6,7,8. In the present study, we found that TGF-β ligands and receptors were mainly expressed in MGCs of mouse ovaries. TGF-β promoted the expression of NPPC in MGCs and inhibited oocyte meiotic resumption in vitro. Moreover, specifically depleting the Tgfbr2 gene in granulosa cells showed a decrease in NPPC levels in MGCs, resulting in the weak maintenance of oocyte meiosis arrest within large antral follicles. Tgfbr2 depletion also decreased ovulation number and litter size. Therefore, TGF-β promotes the expression of NPPC in MGCs and participates in maintaining oocyte meiotic arrest.
The immunofluorescence, western blotting, and qRT-PCR analyses revealed that TGFB1, TGFB3, and their receptors were mainly expressed in mouse MGCs, and TGFB2 was primarily expressed in oocytes, which is consistent with previous immunohistochemical results in mouse and other species41,42. TGFB1 promotes NPPC expression and secretion in vascular smooth muscle cells24,25. In the present study, the addition of TGF-β increased the gene and protein levels of NPPC in the cultured MGCs by SMAD3 binding to the NPPC promoter region and inhibited oocyte meiotic resumption. Furthermore, in Tgfbr2gc−/− mice, the NPPC levels were decreased, and 26% of oocytes resumed meiosis in the large antral follicles. Thus, TGF-β is involved in maintaining oocyte meiotic arrest by promoting the expression of NPPC in MGCs. However, knocking out either Tgfb1 in granulosa cells or Tgfb2 in oocytes had no effect on NPPC expression and oocyte maturation (data not shown), indicating that there is a compensatory effect between ligands as reported in mouse embryonic gonadal development43. Tgfbr2 depletion also decreased the number of large antral follicles, ovulation, and little size. According to previous reports, both TGF-β and NPPC could promote follicle development33,44. Thus, TGF-β may promote follicle development through the upregulation of NPPC. In Tgfbr2-depleted mice, the possible reason for the fertility defect is that the decreased number of large antral follicles, the weakened ability to maintain oocyte meiotic arrest, the reduced oocyte developmental competence45,46, and the ovulation defect. E2 upregulates the expression of NPPC in MGCs and maintains oocyte meiotic arrest18,21. In our study, TGF-β, independent of the E2 pathway, promotes NPPC expression in MGCs. Knocking out either Tgfbr2 or ER could only partially cause oocyte precocious meiotic resumption. These results indicate that both TGF-β and E2-promoted NPPC expression in MGCs are required for the maintenance of oocyte meiosis arrest.
The injection of eCG increases NPPC expression in MGCs6, but FSH treatment did not increase the NPPC levels in cultured MGCs in our study or another study18. This indicates that eCG/FSH promotes NPPC expression indirectly. In vivo, eCG promoted the mRNA and protein levels of TGFB1, TGFBRs, SMAD2, SMAD3, and SMAD4 in MGCs. In vitro, FSH also increased the levels of TGFB1, TGFBR2, SMAD2, SMAD3, and SMAD4, which is consistent with the previous study where FSH upregulated the expression of Tgfb1 mRNA in cultured caprine follicles31. Furthermore, the phosphorylation levels of SMAD3 were also significantly increased by eCG/FSH, suggesting that TGF-β signaling is activated. There was no significant increase in the phosphorylation level of SMAD2, probably due to its lower sensitivity to hormone stimulation during TGF-β signaling transduction47. Therefore, the activation of the FSH receptor could promote NPPC expression in MGCs by the upregulation of TGF-β ligands and receptors and TGF-β downstream SMAD proteins to maintain oocyte meiotic arrest during follicular development. However, FSH could not stimulate the expression of NPPC in cultured MGCs. The possible reason is that FSH-induced TGFB1 diffuses into the culture medium, causing a reduction in concentrations to levels unable to act in an autocrine manner. Interestingly, FSH increased TGF-β-promoted NPPC expression in cultured MGCs. This may be because FSH increases the levels of TGFBR2 as well as SMAD proteins. The activation of the LH receptor decreases ovarian NPPC levels and NPR2 activity, resulting in oocyte meiotic resumption13,15,17. In a previous study, LH inhibits TGFB1 secretion in theca-interstitial cells48. In the present study, hCG treatment significantly decreased the levels of TGFB1, TGFBR2, and SMAD proteins in MGCs, which may result in the decrease in NPPC to induce meiotic resumption.
TGFBR2 was mainly expressed in MGCs. In contrast, bone morphogenetic protein type II receptor (BMPR2), the oocyte-derived paracrine factors (BMP15, growth differentiation factor 9) binding receptor49, was mostly expressed in cumulus cells (Fig. S2b). Thus, TGF-β mainly acts on MGCs to promote NPPC expression but had no effect on NPR2 expression. Oocyte-derived paracrine factors act on cumulus cells adjacent to oocytes to promote NPR2 expression6,50. This may be the reason for the high levels of NPPC in MGCs and NPR2 in cumulus cells6. The depletion of Smad4 reduces not only NPPC expression but also NPR2 expression in granulosa cells39, possibly because both TGF-β and oocyte-derived paracrine factors pathways are blocked in Smad4 depletion mice.
E2 promotes NPPC/NPR2 expression in MGCs. Here, we show that TGF-β upregulates NPPC expression in MGCs. Both E2 and TGF-β are required for maintaining oocyte meiotic arrest. Moreover, there is a compensatory effect between E2 and TGF-β to ensure normal follicle development and female fertility. Our findings not only contribute to a better understanding of the molecular mechanisms responsible for the regulation of NPPC in the follicles but also provide new therapeutic targets for the treatment of infertility.
Materials and methods
Animals and chemicals
WT C57/BL6 female mice and ICR (CD1) female mice were purchased from the Laboratory Animal Center of the Institute of Genetics and Developmental Biology (Beijing, China). Tgfbr2fl/fl mice (stock number 012603) were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). Mice with granulosa cell-specific knockout of Tgfbr2 (Tgfbr2gc−/−) were generated by crossing Tgfbr2fl/fl mice with previously reported Fshr-Cre mice51,52. Female mice (21–23 days old) were injected with 5 IU of eCG 48 h before use to stimulate follicle development. In some experiments, the female mice were treated with 5 IU eCG followed by 5 IU hCG to stimulate ovulation. All animal procedures were approved by the Institutional Animal Care and Use Committee of China Agricultural University and maintained according to the Guide for the Care and Use of Laboratory Animals (Institute for Learning and Animal Research at China Agricultural University). The reagents used in this study, unless otherwise stated, were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Isolation of MGCs, cumulus cells, and oocytes
MGCs were collected through gentle puncture of antral follicles from 21–23 days old mice with a 25-gauge syringe needle. In some experiments, MGCs were collected from eCG-primed mice or eCG-primed mice followed by 5 IU hCG. COCs with the oocytes at the GV stage were collected from eCG-primed mice, and the cumulus cells were stripped from the oocytes by repeatedly drawing the oocytes in and out of a glass pipet slightly smaller in diameter than the oocytes. The collected MGCs, cumulus cells and oocytes were immediately frozen in liquid nitrogen and stored at −80 °C until analysis for gene or protein levels.
MGCs and COCs cultures
MGCs isolated from 21–23 days old mice were washed and then cultured at a density of 1 × 106 cells. The cell culture medium was Dulbecco’s Modified Eagle Medium (DMEM)/F12 with 2.2 mg/ml NaHCO3 and 5% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA, USA). The culture was carried out at 37 °C in a controlled atmosphere of 5% O2, 5% CO2, and 90% N2. After the overnight culture, the unattached, nonviable MGCs were then removed by washing with this medium. The attached cells were then cultured with FSH (10 ng/ml), E2 (100 nmol/l), TGFB1 (10 ng/ml), TGFB2 (50 ng/ml), TGFB3 (10 ng/ml), and/or SD208 (1 μmol/l) for 24 h. Then, the MGCs were either collected using trypsin-EDTA (0.25%) digestion for gene expression and protein analysis or continuously cocultured with COCs for varying times. At the end of culture, oocyte meiotic resumption was assessed by scoring released oocytes for GV breakdown (GVB). GVB was scored by the absence of an obvious GV in the oocyte.
Isolation and culture of follicles
Large antral follicles (350–400 μm) from eCG-primed mice were separated by gentle dissection with a 25-gauge syringe needle. The follicle culture medium was bicarbonate-buffered minimum Eagle medium (MEM)-alpha with Earle balanced salts supplemented with 2.5 mg/ml insulin, 5 µg/ml transferrin, 75 µg/ml penicillin G, 50 µg/ml streptomycin sulfate, 0.23 mM pyruvate, and 3 mg/ml bovine serum albumin. The follicles were cultured on Millicell culture inserts (Millipore, Billerica, MA, USA) supplemented with SD208 (1 μmol/l), ICI182780 (10 μmol/l), E2, TGFB1, TGFB2, and/or TGFB3. The culture was carried out at 37 °C in a controlled atmosphere (5% O2, 5% CO2, and 90% N2) for 4 h. At the end of culture, the MGCs and oocytes were isolated from the follicles for the analysis of gene expression and meiotic resumption, respectively.
Immunofluorescence and histologic analysis
For immunofluorescence, the ovaries from eCG-primed mice were fixed with 4% paraformaldehyde for 12 h, dehydrated, embedded in paraffin and sectioned at 5 μm. The sections were deparaffinized and rehydrated through a graded ethanol series and subjected to antigen retrieval using 0.01% sodium citrate buffer (pH 6.0). The nonspecific binding was blocked with 10% normal donkey serum for 1 h at room temperature. Then, the sections were incubated with primary antibodies overnight at 4 °C, followed by Alexa Fluor 488-conjugated secondary antibodies (1:100, Thermo Fisher Scientific, Waltham, MA, USA) at room temperature for 2 h. Subsequently, the sections were washed with PBS and stained with DAPI for 5 min. The primary antibodies used were as follows: rabbit anti-TGFB1 (ab92486; Abcam, Cambridge, UK) at 1:200, mouse anti-TGFB2 (ab36495; Abcam) at 1:50, rabbit anti-TGFB3 (ab15537; Abcam) at 1:50, rabbit anti-TGFBR1 (ab31013; Abcam) at 1:100, rabbit anti-TGFBR2 (ab186838; Abcam) at 1:200, rabbit anti-SMAD2/3 (8685; Cell Signaling Technology, Danvers, MA, USA) at 1:100, rabbit anti-SMAD4 (46535; Cell Signaling Technology) at 1:100, and rabbit anti-phospho-SMAD3 (9520; Cell Signaling Technology) at 1:100. In some experiments, the cultured MGCs were fixed with 4% paraformaldehyde for 20 min, permeabilized with PBS containing 0.3% Triton X-100 (PBST) for 30 min, and blocked with 5% BSA in PBST at room temperature. Then, cells were used for immunofluorescence staining with the antibodies as described above.
For histologic analysis, paraffin-embedded ovarian samples prepared from eCG-primed mice or eCG-primed mice followed by 5 IU hCG were serially sectioned at 5 μm and stained with periodic acid/Schiff reagent and hematoxylin. The number of large antral follicles and oocytes with meiotic resumption was counted by examining serial sections through the entire ovary. The oocyte count method was determined as previously reported6. If the GV was no longer present and condensed chromosomes were visible, oocytes were scored as having resumed meiosis.
Western blotting
Total proteins were extracted in a tissue and cell lysis solution (CellChip Biotechnology, Beijing, China), and protein concentration was quantified using the BCA Protein Assay Kit (Beyotime, Shanghai, China). Then, the protein lysates (30 μg total protein per lane) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrically transferred to polyvinylidene fluoride membranes (Millipore). Membranes were first blocked with 5% nonfat milk in Tris-buffered saline (pH = 7.6) for 2 h at room temperature, followed by incubation overnight at 4 °C with primary antibodies (each diluted 1:1000): TGFB1, TGFB2, TGFB3, TGFBR1, TGFBR2, SMAD2/3, SMAD4, phospho-SMAD3, rabbit anti-SMAD3 (9523; Cell Signaling Technology), rabbit anti-phospho-SMAD2/3 (8828; Cell Signaling Technology), rabbit anti-CNP (sc-374043; Santa Cruz Biotechnology, CA, USA. It can be used to detect CNP precursor and all active peptides by western blotting), rabbit anti-NPR2 (ab14357; Abcam), or rabbit anti-GAPDH (5174, Cell Signaling Technology). Then, the membranes were washed and further incubated for 1 h at room temperature with horseradish peroxidase-conjugated secondary antibodies (each diluted 1:5000) (Zhongshan Golden Bridge Biotechnology, Beijing, China). Protein visualization was performed with a Tanon 5200 chemiluminescent imaging system (Tanon, Shanghai, China). The levels of GAPDH were detected as a loading control.
RNA extraction and quantitative RT-PCR analysis
Total RNA was extracted and purified from cells using the RNeasy micro-RNA Isolation Kit (Qiagen, Valencia, CA, USA), and reverse transcription into cDNA was conducted using the QuantiTect Reverse Transcription System (Qiagen) according to the manufacturer’s instructions. The relative amount of target gene expression for each sample was determined using an ABI 7500 real-time PCR instrument (Applied Biosystems, Foster City, CA, USA). The relative gene expression was quantified on the basis of the threshold cycle value and normalized using a housekeeping gene, ribosomal protein L19 (Rpl19). The relative transcript level of the control group was set as 1, and relative transcript levels of other samples were compared with the control. qRT-PCR primers for Nppc, Npr2, and Rpl19 were reported previously5. Other primer sequences used for qRT-PCR are shown in Supplementary Table S1.
Measurement of NPPC levels
The cultured MGCs with different treatments were collected and transferred to a 1.5 ml centrifuge tube. Then, the samples were treated according to a previous study53. Briefly, MGCs were boiled in 100 μl of 1.0 M acetic acid for 5 min and homogenized on ice using a tissue homogenizer (T10 basic, IKA, Germany) and were lysed with an ultrasonic cell pulverizer (Scientz-IID, Ningbo, China). We added 500 μl of MeOH to dissolve the lipids in the sample. After centrifugation at 20,000 × g at 4 °C for 30 min, the supernatant containing 2.0–5.0 mg of protein was frozen in liquid nitrogen. Lyophilized sample extracts were assayed for NPPC levels using a luciferase immunoassay kit (Phoenix Pharmaceuticals, Belmont, CA, USA) according to the manufacturer’s instructions.
Chromatin IP (ChIP) assay
ChIP assays were performed using a ChIP kit (Active Motif, Carlsbad, CA, USA) according to the manufacturer’s instructions. The MGCs harvested from 21–23 days old mice were washed three times with ice-cold PBS. Samples were fixed, cross-linked and lysed. The lysate was centrifuged and resuspended in 350 μl of digestion buffer. Then, chromatin was enzymatically sheared until the average DNA length was ~200–300 bp, as evaluated by agarose gel electrophoresis. The sample containing 25 μg of sheared chromatin was used for immunoprecipitation with 3 μg of SMAD3 antibody or normal rabbit IgG (sc-2774; Santa Cruz Biotechnology, Santa Cruz, CA, USA). Following elution, cross-link reversal and proteinase K digestion, the DNA fragments were purified by phenol/chloroform extraction and ethanol precipitation from the protein/DNA complexes. Immunoprecipitated chromatin fragments and input chromatin were detected by qRT-PCR. The primers used for the ChIP assay are indicated in Supplementary Table S2.
Plasmids constructs and a dual-luciferase reporter assay
For the luciferase assay, the DNA fragments containing the R1–R10 regions of the NPPC promoter (according to the results of the ChIP assay) were amplified by PCR from mouse genomic DNA using specific primers and inserted into the pGL3.0 luciferase reporter vector (E1910; Promega, Madison, WI, USA). The forward primer contained a restriction enzyme site of MluI, and the reverse primer contained a restriction enzyme site of XhoI. The cDNA-derived mouse Smad3 sequence was amplified using PCR with the primer containing the MluI–KpnI restriction sites and inserted into the pcDNA3.1 vector (1332; Addgene, Cambridge, MA, USA). Primer sequences were listed in Supplementary Table S3. KK1 cells were maintained in cell culture medium at 37 °C with 5% CO2. This cells were transiently transfected with Smad3 expression vector, Nppc luciferase reporter vector, and pTK-Renilla vector using Lipofectamine 2000 reagent (Thermo Fisher Scientific). An empty luciferase reporter vector was used as a control. After 24 h of transfection, the cells were harvested, and luciferase activities were measured using the Dual-Luc Assay Kit (E1960; Promega). Values shown by the fluc to rluc ratio were normalized to an empty luciferase reporter control.
E2 level assays
The 21–23 days old mice were placed in the supine position, and blood was collected by cardiac puncture. Serum and blood cells were separated by centrifugation, and the serum E2 level was determined by Beijing North Biotechnology Institute (Beijing, China).
Superovulation and fertility
To investigate ovulation, 21–23 days old female Tgfbr2gc−/− mice and WT female mice were injected with 5 IU eCG followed by 5 IU hCG 48 h later to induce ovulation. Then, we counted the number of oocytes by separating COCs from the ampulla at 16 h after hCG treatment. To evaluate reproductive performance, 6-week-old Tgfbr2gc−/− female mice and WT female mice were continually mated to WT males of known fertility. The number of litters was recorded over a 6-month period.
Statistical analysis
All experiments were repeated at least three times, and values were expressed as the mean ± SEM. The proportional data were subjected to an arcsine transformation. A t-test was used to analyze the significant difference between the treatment and control groups. ANOVA (SAS Institute, Inc., Cary, NC, USA) was used to compare data from multiple groups. When a significant F ratio was detected by the ANOVA, the groups were compared using the Holm–Šidák test. Values of P < 0.05 were considered significant.
References
Sorensen, R. A. & Wassarman, P. M. Relationship between growth and meiotic maturation of the mouse oocyte. Dev. Biol. 50, 531–536 (1976).
Erickson, G. F. & Sorensen, R. A. In vitro maturation of mouse oocytes isolated from late, middle, and pre-antral graafian follicles. J. Exp. Zool. 190, 123–127 (1974).
Conti, M., Hsieh, M., Zamah, A. M. & Oh, J. S. Novel signaling mechanisms in the ovary during oocyte maturation and ovulation. Mol. Cell. Endocrinol. 356, 65–73 (2012).
Solc, P., Schultz, R. M. & Motlik, J. Prophase I arrest and progression to metaphase I in mouse oocytes: comparison of resumption of meiosis and recovery from G2-arrest in somatic cells. Mol. Hum. Reprod. 16, 654–664 (2010).
Szybek, K. In-vitro maturation of oocytes from sexually immature mice. J. Endocrinol. 54, 527–528 (1972).
Zhang, M., Su, Y. Q., Sugiura, K., Xia, G. & Eppig, J. J. Granulosa cell ligand NPPC and its receptor NPR2 maintain meiotic arrest in mouse oocytes. Science 330, 366–369 (2010).
Zhang, T., Zhang, C., Fan, X., Li, R. & Zhang, J. Effect of C-type natriuretic peptide pretreatment on in vitro bovine oocyte maturation. Vitr. Cell. Dev. Biol. Anim. 53, 199–206 (2017).
Celik, O., Celik, N., Gungor, S., Haberal, E. T. & Aydin, S. Selective regulation of oocyte meiotic events enhances progress in fertility preservation methods. Biochem. Insights 8, 11–21 (2015).
Norris, R. P. et al. Cyclic GMP from the surrounding somatic cells regulates cyclic AMP and meiosis in the mouse oocyte. Development 136, 1869–1878 (2009).
Zeleznik, A. J. The physiology of follicle selection. Reprod. Biol. Endocrinol. 2, 31 (2004).
Tsuji, T., Kiyosu, C., Akiyama, K. & Kunieda, T. CNP/NPR2 signaling maintains oocyte meiotic arrest in early antral follicles and is suppressed by EGFR-mediated signaling in preovulatory follicles. Mol. Reprod. Dev. 79, 795–802 (2012).
Kawamura, K. et al. Pre-ovulatory LH/hCG surge decreases C-type natriuretic peptide secretion by ovarian granulosa cells to promote meiotic resumption of pre-ovulatory oocytes. Hum. Reprod. 26, 3094–3101 (2011).
Zamah, A. M., Xie, F., Xiong, L. & Conti, M. Gonadotropin regulation of natriuretic peptide precursor type C as a trigger to oocyte maturation. Fertil. Steril. 98, S152 (2012).
Zhang, M. et al. Estradiol promotes and maintains cumulus cell expression of natriuretic peptide receptor 2 (NPR2) and meiotic arrest in mouse oocytes in vitro. Endocrinology 152, 4377–4385 (2011).
Egbert, J. R. et al. Dephosphorylation and inactivation of NPR2 guanylyl cyclase in granulosa cells contributes to the LH-induced decrease in cGMP that causes resumption of meiosis in rat oocytes. Development 141, 3594–3604 (2014).
Liu, X., Xie, F., Zamah, A. M., Cao, B. & Conti, M. Multiple pathways mediate luteinizing hormone regulation of cGMP signaling in the mouse ovarian follicle. Biol. Reprod. 91, 9 (2014).
Hao, X. et al. Epidermal growth factor-mobilized intracellular calcium of cumulus cells decreases natriuretic peptide receptor 2 affinity for natriuretic peptide type C and induces oocyte meiotic resumption in the mouse. Biol. Reprod. 95, 45 (2016).
Lee, K. B. et al. Hormonal coordination of natriuretic peptide type C and natriuretic peptide receptor 3 expression in mouse granulosa cells. Biol. Reprod. 88, 42 (2013).
Emmen, J. M. et al. In vitro growth and ovulation of follicles from ovaries of estrogen receptor (ER) α and ERβ null mice indicate a role for ERβ in follicular maturation. Endocrinology 146, 2817–2826 (2005).
Palermo, R. Differential actions of FSH and LH during folliculogenesis. Reprod. Biomed. Online 15, 326–337 (2007).
Liu, W. et al. Estrogen receptors in granulosa cells govern meiotic resumption of pre-ovulatory oocytes in mammals. Cell Death Dis. 8, e2662 (2017).
Britt, K. L. et al. The ovarian phenotype of the aromatase knockout (ArKO) mouse. J. Steroid Biochem. Mol. Biol. 79, 181–185 (2001).
Babadagli, M. E., Tezcan, B., Yilmaz, S. T. & Tufan, A. C. Matrilin-3 as a putative effector of C-type natriuretic peptide signaling during TGF-β induced chondrogenic differentiation of mesenchymal stem cells. Mol. Biol. Rep. 41, 5549–5555 (2014).
Mendonca, M. C., Koles, N., Doi, S. Q. & Sellitti, D. F. Transforming growth factor-beta1 regulation of C-type natriuretic peptide expression in human vascular smooth muscle cell: dependence on TSC22D1. Am J Physiol. Heart Circ. Physiol. 299, H2018–H2027 (2010).
Vlachopoulos, C. et al. Amino-terminal pro-C-type natriuretic peptide is associated with the presence, severity, and duration of vasculogenic erectile dysfunction. Eur. Urol. 56, 552–558 (2009).
Ohta, S., Shimekake, Y. & Nagata, K. Molecular cloning and characterization of a transcription factor for the C-type natriuretic peptide gene promoter. Eur. J. Biochem. 242, 460–466 (1996).
Fang, X., Gao, Y. & Li, Q. SMAD3 activation: a converging point of dysregulated TGF-Beta superfamily signaling and genetic aberrations in granulosa cell tumor development? Biol. Reprod. 95, 105–105 (2016).
Schmierer, B. & Hill, C. S. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 8, 970–982 (2007).
Subramaniam, N. et al. Cross-talk between 1, 25-dihydroxyvitamin D3 and transforming growth factor-beta signaling requires binding of VDR and Smad3 proteins to their cognate DNA recognition elements. J. Biol. Chem. 276, 15741–15746 (2001).
Sriperumbudur, R., Zorrilla, L. & Gadsby, J. E. Transforming growth factor-β (TGF-β) and its signaling components in peri-ovulatory pig follicles. Anim. Reprod. Sci. 120, 84–94 (2010).
Rodrigues, G. Q. et al. Relative mRNA expression and immunolocalization for transforming growth factor-beta (TGF-β) and their effect on in vitro development of caprine preantral follicles. Vitr. Cell Dev. Biol. Anim. 50, 688–699 (2014).
Chegini, N. & Flanders, K. C. Presence of transforming growth factor-beta and their selective cellular localization in human ovarian tissue of various reproductive stages. Endocrinology 130, 1707–1715 (1992).
Fortune, J. E., Yang, M. Y. & Muruvi, W. The earliest stages of follicular development: follicle formation and activation. Soc. Reprod. Fertil. Suppl. 67, 203–216 (2010).
Li, Y. et al. SMAD3 regulates the diverse functions of rat granulosa cells relating to the FSHR/PKA signaling pathway. Reproduction 146, 169–179 (2013).
Du, X. et al. TGF-β signaling controls FSHR signaling-reduced ovarian granulosa cell apoptosis through the SMAD4/miR-143 axis. Cell Death Dis. 7, e2476 (2016).
Chen, Y. C. et al. Transforming growth factor-β1 up-regulates connexin43 expression in human granulosa cells. Hum. Reprod. 30, 2190–2201 (2015).
Fang, L., Chang, H. M., Cheng, J. C., Leung, P. C. & Sun, Y. P. TGF-β1 induces COX-2 expression and PGE2 production in human granulosa cells through Smad signaling pathways. J. Clin. Endocrinol. Metab. 99, E1217–E1226 (2014).
Coskun, S. & Lin, Y. C. Effects of transforming growth factors and activin-A on in vitro porcine oocyte maturation. Mol. Reprod. Dev. 38, 153–159 (1994).
Yu, C., Zhang, Y. L. & Fan, H. Y. Selective Smad4 knockout in ovarian preovulatory follicles results in multiple defects in ovulation. Mol. Endocrinol. 27, 966–978 (2013).
Hamberger, L., Nordenstrom, K., Rosberg, S. & Sjogren, A. Acute influence of LH and FSH on cyclic AMP formation in isolated granulosa cells of the rat. Acta Endocrinol. 88, 567–579 (1978).
Gueripel, X., Benahmed, M. & Gougeon, A. Sequential gonadotropin treatment of immature mice leads to amplification of transforming growth factor beta action, via upregulation of receptor-type 1, Smad 2 and 4, and downregulation of Smad 6. Biol. Reprod. 70, 640–648 (2004).
Gilchrist, R. B., Morrissey, M. P., Ritter, L. J. & Armstrong, D. T. Comparison of oocyte factors and transforming growth factor-beta in the regulation of DNA synthesis in bovine granulosa cells. Mol. Cell. Endocrinol. 201, 87–95 (2003).
Memon, M. A., Anway, M. D., Covert, T. R., Uzumcu, M. & Skinner, M. K. Transforming growth factor beta (TGFβ1, TGFβ2 and TGFβ3) null-mutant phenotypes in embryonic gonadal development. Mol. Cell. Endocrinol. 294, 70–80 (2008).
Sato, Y., Cheng, Y., Kawamura, K., Takae, S. & Hsueh, A. J. C-type natriuretic peptide stimulates ovarian follicle development. Mol. Endocrinol. 26, 1158–1166 (2012).
Santiquet, N. W., Greene, A. F., Becker, J., Schoolcraft, W. B. & Krisher, R. L. A pre-in vitro maturation medium containing cumulus oocyte complex ligand-receptor signaling molecules maintains meiotic arrest, supports the cumulus oocyte complex and improves oocyte developmental competence. Mol. Hum. Reprod. 23, 594–606 (2017).
Sánchez, F. et al. An improved IVM method for cumulus-oocyte complexes from small follicles in polycystic ovary syndrome patients enhances oocyte competence and embryo yield. Hum. Reprod. 32, 2056–2068 (2017).
Xu, J., Oakley, J. & McGee, E. A. Stage-specific expression of Smad2 and Smad3 during folliculogenesis. Biol. Reprod. 66, 1571–1578 (2002).
Magoffin, D. A., Hubertleslie, D. & Zachow, R. J. Estradiol-17 beta, insulin-like growth factor-I, and luteinizing hormone inhibit secretion of transforming growth factor beta by rat ovarian theca-interstitial cells. Biol. Reprod. 53, 627–635 (1995).
Liu, C. et al. Effects of mir-375-BMPR2 as a key factor downstream of BMP15/GDF9 on the Smad1/5/8 and Smad2/3 Signaling Pathways. Cell. Physiol. Biochem. 46, 213–225 (2018).
Wigglesworth, K. et al. Bidirectional communication between oocytes and ovarian follicular somatic cells is required for meiotic arrest of mammalian oocytes. Proc. Natl Acad. Sci. USA 110, E3723–E3729 (2013).
Yu, C., Zhou, J. J. & Fan, H. Y. Studying the functions of TGF-β signaling in the ovary. Methods Mol. Biol. 1344, 301–311 (2016).
Chodankar, R. et al. Cell-nonautonomous induction of ovarian and uterine serous cystadenomas in mice lacking a functional Brca1 in ovarian granulosa cells. Curr. Biol. 15, 561–565 (2005).
Kong, N. et al. Natriuretic peptide type C induces sperm attraction for fertilization in mouse. Sci. Rep. 7, 39711 (2017).
Acknowledgements
We thank Professor Louis Dubeau (University of Southern California) for providing the Fshr-Cre mice. This work was supported by the National Key Research and Development Program of China (2017YFC1002002 and 2018YFC1003801 to M.Z.), the National Science Fund for Distinguished Young Scholars of China (31425024 to M.Z.), National Natural Science Foundation of China (31771658 to M.Z.), the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (81521002 to J.Q.), and the Project of State Key Laboratory of Agrobiotechnology (grant number 2019KLAB6–8 to Y.S.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by D. Aberdam
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, J., Zhang, Y., Xu, X. et al. Transforming growth factor-β is involved in maintaining oocyte meiotic arrest by promoting natriuretic peptide type C expression in mouse granulosa cells. Cell Death Dis 10, 558 (2019). https://doi.org/10.1038/s41419-019-1797-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-019-1797-5
This article is cited by
-
The molecular regulatory mechanisms of meiotic arrest and resumption in Oocyte development and maturation
Reproductive Biology and Endocrinology (2023)
-
Transcriptomic profiles reveal the characteristics of oocytes and cumulus cells at GV, MI, and MII in follicles before ovulation
Journal of Ovarian Research (2023)
-
Expression and Clinical Significance of HIF-1α in Follicular Fluid and Granulosa Cells in Infertile PCOS Patients
Reproductive Sciences (2023)
-
Identification, characterization and functional analysis of gonadal long noncoding RNAs in a protogynous hermaphroditic teleost fish, the ricefield eel (Monopterus albus)
BMC Genomics (2022)
-
SphK-produced S1P in somatic cells is indispensable for LH-EGFR signaling-induced mouse oocyte maturation
Cell Death & Disease (2022)