Abstract
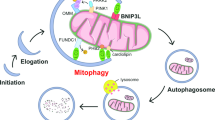
Mitophagy plays an important role in the maintenance of mitochondrial homeostasis and can be categorized into two types: ubiquitin-mediated and receptor-mediated pathways. During receptor-mediated mitophagy, mitophagy receptors facilitate mitophagy by tethering the isolation membrane to mitochondria. Although at least five outer mitochondrial membrane proteins have been identified as mitophagy receptors, their individual contribution and interrelationship remain unclear. Here, we show that HeLa cells lacking BNIP3 and NIX, two of the five receptors, exhibit a complete loss of mitophagy in various conditions. Conversely, cells deficient in the other three receptors show normal mitophagy. Using BNIP3/NIX double knockout (DKO) cells as a model, we reveal that mitophagy deficiency elevates mitochondrial reactive oxygen species (mtROS), which leads to activation of the Nrf2 antioxidant pathway. Notably, BNIP3/NIX DKO cells are highly sensitive to ferroptosis when Nrf2-driven antioxidant enzymes are compromised. Moreover, the sensitivity of BNIP3/NIX DKO cells is fully rescued upon the introduction of wild-type BNIP3 and NIX, but not the mutant forms incapable of facilitating mitophagy. Consequently, our results demonstrate that BNIP3 and NIX-mediated mitophagy plays a role in regulating mtROS levels and protects cells from ferroptosis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All relevant data are available from the authors. Original immunoblot data are provided in the Supplemental Material.
References
Gambert S, Ricquier D. Mitochondrial thermogenesis and obesity. Curr Opin Clin Nutr Metab Care. 2007;10:664–70.
Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407.
Wanders RJ, Ruiter JP, IJLst L, Waterham HR, Houten SM. The enzymology of mitochondrial fatty acid beta-oxidation and its application to follow-up analysis of positive neonatal screening results. J Inherit Metab Dis. 2010;33:479–94.
Palikaras K, Lionaki E, Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20:1013–22.
Pickles S, Vigié P, Youle RJ. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol. 2018;28:R170–85.
Ganley IG, Simonsen A. Diversity of mitophagy pathways at a glance. J Cell Sci. 2022;135:23.
Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803.
Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298.
Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–21.
Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–14.
Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51.
Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson Å. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012;287:19094–104.
Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–85.
Murakawa T, Yamaguchi O, Hashimoto A, Hikoso S, Takeda T, Oka T, et al. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat Commun. 2015;6:7527.
Bhujabal Z, Birgisdottir Å, Sjøttem E, Brenne HB, Øvervatn A, Habisov S, et al. FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 2017;18:947–61.
Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–95.
Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13.
Oh SJ, Ikeda M, Ide T, Hur KY, Lee MS. Mitochondrial event as an ultimate step in ferroptosis. Cell Death Discov. 2022;8:414.
Chourasia AH, Tracy K, Frankenberger C, Boland ML, Sharifi MN, Drake LE, et al. Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 2015;16:1145–63.
O’Sullivan TE, Johnson LR, Kang HH, Sun JC. BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity. 2015;43:331–42.
Yamashita SI, Kanki T. Detection of iron depletion- and hypoxia-induced mitophagy in mammalian cells. Methods Mol Biol. 2018;1782:315–24.
Igarashi R, Yamashita SI, Yamashita T, Inoue K, Fukuda T, Fukuchi T, et al. Gemcitabine induces Parkin-independent mitophagy through mitochondrial-resident E3 ligase MUL1-mediated stabilization of PINK1. Sci Rep. 2020;10:1465.
Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26.
Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91.
Park SJ, Shin JH, Kim ES, Jo YK, Kim JH, Hwang JJ, et al. Mitochondrial fragmentation caused by phenanthroline promotes mitophagy. FEBS Lett. 2012;586:4303–10.
Allen GF, Toth R, James J, Ganley IG. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127–35.
Yamashita SI, Jin X, Furukawa K, Hamasaki M, Nezu A, Otera H, et al. Mitochondrial division occurs concurrently with autophagosome formation but independently of Drp1 during mitophagy. J Cell Biol. 2016;215:649–65.
Ryu D, Mouchiroud L, Andreux PA, Katsyuba E, Moullan N, Nicolet-Dit-Félix AA, et al. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat Med. 2016;22:879–88.
Rojansky R, Cha MY, Chan DC. Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. Elife. 2016;5:e17896
Denison SR, Wang F, Becker NA, Schüle B, Kock N, Phillips LA, et al. Alterations in the common fragile site gene Parkin in ovarian and other cancers. Oncogene. 2003;22:8370–8.
Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–903.
Xia M, Huang R, Sun Y, Semenza GL, Aldred SF, Witt KL, et al. Identification of chemical compounds that induce HIF-1alpha activity. Toxicol Sci. 2009;112:153–63.
Sulkshane P, Ram J, Thakur A, Reis N, Kleifeld O, Glickman MH. Ubiquitination and receptor-mediated mitophagy converge to eliminate oxidation-damaged mitochondria during hypoxia. Redox Biol. 2021;45:102047.
Hayes JD, Dinkova-Kostova AT, Tew KD. Oxidative stress in cancer. Cancer Cell. 2020;38:167–97.
Tonelli C, Chio IIC, Tuveson DA. Transcriptional regulation by Nrf2. Antioxid Redox Signal. 2018;29:1727–45.
Pap EH, Drummen GP, Winter VJ, Kooij TW, Rijken P, Wirtz KW, et al. Ratio-fluorescence microscopy of lipid oxidation in living cells using C11-BODIPY(581/591). FEBS Lett. 1999;453:278–82.
Hirata Y, Cai R, Volchuk A, Steinberg BE, Saito Y, Matsuzawa A, et al. Lipid peroxidation increases membrane tension, Piezo1 gating, and cation permeability to execute ferroptosis. Curr Biol. 2023;33:1282–94.e1285.
Zhao JF, Rodger CE, Allen GFG, Weidlich S, Ganley IG. HIF1α-dependent mitophagy facilitates cardiomyoblast differentiation. Cell Stress. 2020;4:99–113.
Wilhelm LP, Zapata-Muñoz J, Villarejo-Zori B, Pellegrin S, Freire CM, Toye AM, et al. BNIP3L/NIX regulates both mitophagy and pexophagy. EMBO J. 2022;41:e111115.
Cao Y, Zheng J, Wan H, Sun Y, Fu S, Liu S, et al. A mitochondrial SCF-FBXL4 ubiquitin E3 ligase complex degrades BNIP3 and NIX to restrain mitophagy and prevent mitochondrial disease. EMBO J. 2023;42:e113033.
Elcocks H, Brazel AJ, McCarron KR, Kaulich M, Husnjak K, Mortiboys H, et al. FBXL4 ubiquitin ligase deficiency promotes mitophagy by elevating NIX levels. EMBO J. 2023;42:e112799.
Nguyen-Dien GT, Kozul KL, Cui Y, Townsend B, Kulkarni PG, Ooi SS, et al. FBXL4 suppresses mitophagy by restricting the accumulation of NIX and BNIP3 mitophagy receptors. EMBO J. 2023;42:e112767.
Niemi NM, Serrano LR, Muehlbauer LK, Balnis CE, Wei L, Smith AJ, et al. PPTC7 maintains mitochondrial protein content by suppressing receptor-mediated mitophagy. Nat Commun. 2023;14:6431.
Sun Y, Cao Y, Wan H, Memetimin A, Li L, Wu C, et al. A mitophagy sensor PPTC7 controls BNIP3 and NIX degradation to regulate mitochondrial mass. Mol Cell. 2024;84:327–344.e329.
Giorgi C, Marchi S, Simoes ICM, Ren Z, Morciano G, Perrone M, et al. Mitochondria and reactive oxygen species in aging and age-related diseases. Int Rev Cell Mol Biol. 2018;340:209–344.
Wang Y, Yen FS, Zhu XG, Timson RC, Weber R, Xing C, et al. SLC25A39 is necessary for mitochondrial glutathione import in mammalian cells. Nature. 2021;599:136–40.
Hosoi KI, Miyata N, Mukai S, Furuki S, Okumoto K, Cheng EH, et al. The VDAC2-BAK axis regulates peroxisomal membrane permeability. J Cell Biol. 2017;216:709–22.
Dubreuil MM, Morgens DW, Okumoto K, Honsho M, Contrepois K, Lee-McMullen B, et al. Systematic identification of regulators of oxidative stress reveals non-canonical roles for peroxisomal import and the pentose phosphate pathway. Cell Rep. 2020;30:1417–33.e1417.
Okumoto K, El Shermely M, Natsui M, Kosako H, Natsuyama R, Marutani T, et al. The peroxisome counteracts oxidative stresses by suppressing catalase import via Pex14 phosphorylation. Elife. 2020;9:e55896
Park H, He A, Tan M, Johnson JM, Dean JM, Pietka TA, et al. Peroxisome-derived lipids regulate adipose thermogenesis by mediating cold-induced mitochondrial fission. J Clin Investig. 2019;129:694–711.
Harris IS, Treloar AE, Inoue S, Sasaki M, Gorrini C, Lee KC, et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell. 2015;27:211–22.
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72.
Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–85.
Li Y, Zheng N, Ding X. Mitophagy disequilibrium, a prominent pathological mechanism in metabolic heart diseases. Diabetes Metab Syndr Obes. 2021;14:4631–40.
Gan B. Mitochondrial regulation of ferroptosis. J Cell Biol. 2021:220:e202105043
Liang KX, Kristiansen CK, Mostafavi S, Vatne GH, Zantingh GA, Kianian A, et al. Disease-specific phenotypes in iPSC-derived neural stem cells with POLG mutations. EMBO Mol Med. 2020;12:e12146.
Vara-Pérez M, Rossi M, Van den Haute C, Maes H, Sassano ML, Venkataramani V, et al. BNIP3 promotes HIF-1α-driven melanoma growth by curbing intracellular iron homeostasis. EMBO J. 2021;40:e106214.
Simpson CL, Tokito MK, Uppala R, Sarkar MK, Gudjonsson JE, Holzbaur ELF. NIX initiates mitochondrial fragmentation via DRP1 to drive epidermal differentiation. Cell Rep. 2021;34:108689.
Schmid ET, Pyo JH, Walker DW. Neuronal induction of BNIP3-mediated mitophagy slows systemic aging in. Nat Aging. 2022;2:494–507.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23.
Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol. 2011;18:1042–52.
Acknowledgements
We thank Feng Zhang (Broad Institute of MIT and Harvard) and Eric Campeau (University of Massachusetts Medical School) for providing plasmids and CCRF (Niigata University) for supporting FACS analysis.
Funding
This work was supported in part by the Japan Society for the Promotion of Science KAKENHI grants (16KK0162 and 22K07207 to SY and 19H05712, 18H04858, and 18H04691, to TK), AMED grant (JP21gm6110013, JP23ek0109647 and JP23gm1710006 to TK), and the Takeda Science Foundation (to SY).
Author information
Authors and Affiliations
Contributions
SY, YS, and TK designed the experiments. SY generated receptor KO cell lines using the CRIPSR/Cas9 genome editing system. SY performed all cellular analyses. YS, YM, and RM performed the metabolomic analysis. SY, KI, KF, TF, DCC, and TK wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yamashita, Si., Sugiura, Y., Matsuoka, Y. et al. Mitophagy mediated by BNIP3 and NIX protects against ferroptosis by downregulating mitochondrial reactive oxygen species. Cell Death Differ (2024). https://doi.org/10.1038/s41418-024-01280-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41418-024-01280-y