Abstract
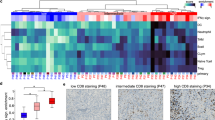
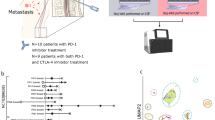
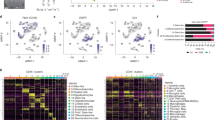
Patients diagnosed with glioblastoma (GBM) have the most aggressive tumor progression and lethal recurrence. Research on the immune microenvironment landscape of tumor and cerebrospinal fluid (CSF) is limited. At the single-cell level, we aim to reveal the recurrent immune microenvironment of GBM and the potential CSF biomarkers and compare tumor locations. We collected four clinical samples from two patients: malignant samples from one recurrent GBM patient and non-malignant samples from a patient with brain tumor. We performed single-cell RNA sequencing (scRNA-seq) to reveal the immune landscape of recurrent GBM and CSF. T cells were enriched in the malignant tumors, while Treg cells were predominately found in malignant CSF, which indicated an inhibitory microenvironment in recurrent GBM. Moreover, macrophages and neutrophils were significantly enriched in malignant CSF. This indicates that they an important role in GBM progression. S100A9, extensively expressed in malignant CSF, is a promising biomarker for GBM diagnosis and recurrence. Our study reveals GBM’s recurrent immune microenvironment after chemoradiotherapy and compares malignant and non-malignant CSF samples. We provide novel targets and confirm the promise of liquid CSF biopsy for patients with GBM.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The expressional and survival validation data of S100A9 were downloaded from The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/) and the Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org.cn/) database. The scRNA-seq datasets generated during and analyzed during the current study have been uploaded in SRA with ID: PRJNA970244 and are available from Hengzhu Zhang and Yizhi Ge on reasonable request.
References
Xiao Y, Wang Z, Zhao M, Deng Y, Yang M, Su G, et al. Single-cell transcriptomics revealed subtype-specific tumor immune microenvironments in human glioblastomas. Front Immunol. 2022;13:914236.
Sareen H, Garrett C, Lynch D, Powter B, Brungs D, Cooper A, et al. The role of liquid biopsies in detecting molecular tumor biomarkers in brain cancer patients. Cancers. 2020;12:1831.
Yeo AT, Rawal S, Delcuze B, Christofides A, Atayde A, Strauss L, et al. Single-cell RNA sequencing reveals evolution of immune landscape during glioblastoma progression. Nat Immunol. 2022;23:971–84.
Hu H, Mu Q, Bao Z, Chen Y, Liu Y, Chen J, et al. Mutational landscape of secondary glioblastoma guides MET-targeted trial in brain tumor. Cell. 2018;175:1665–78.e18.
Abdelfattah N, Kumar P, Wang C, Leu JS, Flynn WF, Gao R, et al. Single-cell analysis of human glioma and immune cells identifies S100A4 as an immunotherapy target. Nat Commun. 2022;13:767.
Chen Z, Hambardzumyan D. Immune microenvironment in glioblastoma subtypes. Front Immunol. 2018;9:1004.
Watermann C, Pasternack H, Idel C, Ribbat-Idel J, Brägelmann J, Kuppler P, et al. Recurrent HNSCC harbor an immunosuppressive tumor immune microenvironment suggesting successful tumor immune evasion. Clin Cancer Res. 2021;27:632–44.
Fecci PE, Mitchell DA, Whitesides JF, Xie W, Friedman AH, Archer GE, et al. Increased regulatory T-cell fraction amidst a diminished CD4 compartment explains cellular immune defects in patients with malignant glioma. Cancer Res. 2006;66:3294–302.
Piccirillo CA, Shevach EM. Cutting edge: control of CD8+ T cell activation by CD4+CD25+ immunoregulatory cells. J Immunol. 2001;167:1137–40.
Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–9.
Kryczek I, Wu K, Zhao E, Wei S, Vatan L, Szeliga W, et al. IL-17+ regulatory T cells in the microenvironments of chronic inflammation and cancer. J Immunol. 2011;186:4388–95.
Ronvaux L, Riva M, Coosemans A, Herzog M, Rommelaere G, Donis N, et al. Liquid biopsy in glioblastoma. Cancers. 2022;14:3394.
Jung M, Klotzek S, Lewandowski M, Fleischhacker M, Jung K. Changes in concentration of DNA in serum and plasma during storage of blood samples. Clin Chem. 2003;49:1028–9.
Sarkaria JN, Hu LS, Parney IF, Pafundi DH, Brinkmann DH, Laack NN, et al. Is the blood-brain barrier really disrupted in all glioblastomas? A critical assessment of existing clinical data. Neuro Oncol. 2018;20:184–91.
Miller AM, Shah RH, Pentsova EI, Pourmaleki M, Briggs S, Distefano N, et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565:654–8.
Mikolajewicz N, Khan S, Trifoi M, Skakdoub A, Ignatchenko V, Mansouri S, et al. Leveraging the CSF proteome toward minimally-invasive diagnostics surveillance of brain malignancies. Neuro-Oncol Adv. 2022;4:vdac161.
Mabbott NA, Baillie JK, Brown H, Freeman TC, Hume DA. An expression atlas of human primary cells: inference of gene function from coexpression networks. BMC Genomics. 2013;14:632.
Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14:979–82.
Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624.e24.
Wu Y, Yang S, Ma J, Chen Z, Song G, Rao D, et al. Spatiotemporal immune landscape of colorectal cancer liver metastasis at single-cell level. Cancer Discov. 2022;12:134–53.
Ma R, Kang X, Zhang G, Fang F, Du Y, Lv H. High expression of UBE2C is associated with the aggressive progression and poor outcome of malignant glioma. Oncol Lett. 2016;11:2300–4.
Hyman G, Manglik V, Rousch JM, Verma M, Kinkebiel D, Banerjee HN. Epigenetic approaches in glioblastoma multiforme and their implication in screening and diagnosis. Methods Mol Biol. 2015;1238:511–21.
Zhou J, Guo H, Liu L, Hao S, Guo Z, Zhang F, et al. Construction of co-expression modules related to survival by WGCNA and identification of potential prognostic biomarkers in glioblastoma. J Cell Mol Med. 2021;25:1633–44.
Xia S, Lal B, Tung B, Wang S, Goodwin CR, Laterra J. Tumor microenvironment tenascin-C promotes glioblastoma invasion and negatively regulates tumor proliferation. Neuro Oncol. 2016;18:507–17.
Monteiro C, Miarka L, Perea-García M, Priego N, García-Gómez P, Álvaro-Espinosa L, et al. Stratification of radiosensitive brain metastases based on an actionable S100A9/RAGE resistance mechanism. Nat Med. 2022;28:752–65.
Biswas AK, Han S, Tai Y, Ma W, Coker C, Quinn SA, et al. Targeting S100A9-ALDH1A1-retinoic acid signaling to suppress brain relapse in EGFR-mutant lung cancer. Cancer Discov. 2022;12:1002–21.
Chen Y, Ouyang Y, Li Z, Wang X, Ma J. S100A8 and S100A9 in cancer. Biochim Biophysica Acta Rev Cancer. 2023;1878:188891.
Wang H, Mao X, Ye L, Cheng H, Dai X. The role of the S100 protein family in glioma. J Cancer. 2022;13:3022–30.
Zhao MJ, Lu T, Ma C, Wang ZF, Li ZQ. A narrative review on the management of glioblastoma in China. Chin Clin Oncol. 2022;11:29.
Wen PY, Chang SM, Lamborn KR, Kuhn JG, Norden AD, Cloughesy TF, et al. Phase I/II study of erlotinib and temsirolimus for patients with recurrent malignant gliomas: North American Brain Tumor Consortium trial 04-02. Neuro Oncol. 2014;16:567–78.
Reardon DA, Brandes AA, Omuro A, Mulholland P, Lim M, Wick A, et al. Effect of nivolumab vs bevacizumab in patients with recurrent glioblastoma: the CheckMate 143 phase 3 randomized clinical trial. JAMA Oncol. 2020;6:1003–10.
O’Rourke DM, Nasrallah MP, Desai A, Melenhorst JJ, Mansfield K, Morrissette JJD, et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Transl Med. 2017;9:eaaa0984.
Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013;110:4009–14.
Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178:835–849.e21.
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–401.
Chen S, Xie Y, Cai Y, Hu H, He M, Liu L, et al. Multiomic analysis reveals comprehensive tumor heterogeneity and distinct immune subtypes in multifocal intrahepatic cholangiocarcinoma. Clin Cancer Res. 2022;28:1896–910.
Yekula A, Tracz J, Rincon-Torroella J, Azad T, Bettegowda C. Single-cell RNA sequencing of cerebrospinal fluid as an advanced form of liquid biopsy for neurological disorders. Brain Sci. 2022;12:812.
Wu L, Wu W, Zhang J, Zhao Z, Li L, Zhu M, et al. Natural coevolution of tumor and immunoenvironment in glioblastoma. Cancer Discov. 2022;12:2820–37.
Kim J, Lee IH, Cho HJ, Park CK, Jung YS, Kim Y, et al. Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell. 2015;28:318–28.
Valiente M, Sepúlveda JM, Pérez A. Emerging targets for cancer treatment: S100A9/RAGE. ESMO Open. 2023;8:100751.
Cai Q, Zhu J, Cui X, Xia Y, Gao H, Wang X, et al. S100A9 promotes inflammatory response in diabetic nonalcoholic fatty liver disease. Biochem Biophys Res Commun. 2022;618:127–32.
Fang S, Cheng X, Shen T, Dong J, Li Y, Li Z, et al. CXCL8 up-regulated LSECtin through AKT signal and correlates with the immune microenvironment modulation in colon cancer. Cancers. 2022;14:5300.
Ribeiro-Dias F, Oliveira IBN. A critical overview of interleukin 32 in Leishmaniases. Front Immunol. 2022;13:849340.
Pereira AC, De Pascale J, Resende R, Cardoso S, Ferreira I, Neves BM, et al. ER-mitochondria communication is involved in NLRP3 inflammasome activation under stress conditions in the innate immune system. Cell Mol Life Sci. 2022;79:213.
Fanelli GN, Grassini D, Ortenzi V, Pasqualetti F, Montemurro N, Perrini P, et al. Decipher the glioblastoma microenvironment: the first milestone for new groundbreaking therapeutic strategies. Genes. 2021;12:445.
Jacobs JFM, Idema AJ, Bol KF, Grotenhuis JA, de Vries IJM, Wesseling P, et al. Prognostic significance and mechanism of Treg infiltration in human brain tumors. J Neuroimmunol. 2010;225:195–9.
Acknowledgements
We deeply acknowledged the great assistant from OE Biotech Co., Ltd. (Shanghai, China) and their technicist XiaoHua Yao. We also thank the data upload service from NCBI SRA Submissions.
Funding
This study was supported by the Postdoctoral Fund of the People’s Government of Jiangsu Province (200730000107) and the Postdoctoral Fund of Jiangsu Cancer Hospital (SZL202013).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Xingdong Wang, Yizhi Ge, and Yuting Hou were involved in the conceptualization, design, writing, and methodology of this study. Xiaodong Wang, Zhengcun Yan, and Yuping Li polish language. Lun Dong, Lei She, and Can Tang participated in the patients’ selection. Hengzhu Zhang and Min Wei performed conceptualization, design, data analysis, and critically revised the manuscript. All authors have read and approved the final version of the manuscript and consent to its publication.
Corresponding authors
Ethics declarations
Ethics approval
Approval of the research protocol by an Institutional Reviewer Board: Subei People Hospital Institutional Review Board. Informed Consent: Informed consent was obtained from all patients. Registry and the Registration No. of the study/trial: N/A. Animal Studies: write N/A.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, X., Ge, Y., Hou, Y. et al. Single-cell atlas reveals the immunosuppressive microenvironment and Treg cells landscapes in recurrent Glioblastoma. Cancer Gene Ther (2024). https://doi.org/10.1038/s41417-024-00740-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41417-024-00740-4