Abstract
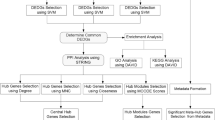
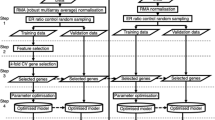
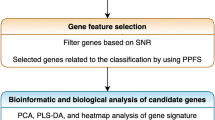
Triple-negative breast cancer (TNBC), colon adenocarcinoma (COAD), ovarian cancer (OV), and glioblastoma multiforme (GBM) are common malignant tumors, in which significant challenges are still faced in early diagnosis, treatment, and prognosis. Therefore, further identification of genes related to those malignant tumors is of great significance for the improvement of management of the diseases. The database of the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) repository was used as the data source of gene expression profiles in this study. Malignant tumors genes were selected using a feature selection algorithm of maximal relevance and minimal redundancy (mRMR) and the protein–protein interaction (PPI) network. And finally selected 20 genes as potential related genes. Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed on the potential related genes, and different tumor-specific genes and similarities and differences between network modules and pathways were analyzed. Further, using the potential cancer-related genes found above in this study as features, a support vector machine (SVM) model was developed to predict high-risk malignant tumors. As a result, the prediction accuracy reached more than 85%, indicating that such a model can effectively predict the four types of malignant tumors. It is demonstrated that such genes found above in this study indeed play important roles in the differentiation of the four types of malignant tumors, providing basis for future experimental biological validation and shedding some light on the understanding of new molecular mechanisms related to the four types of tumors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Liu G, Wong L, Chua HN. Complex discovery from weighted ppi networks. Bioinformatics. 2009;25:1891–7.
Wolfe CJ, Kohane IS, Butte AJ. Systematic survey reveals general applicability of “guilt-by-association” within gene coexpression networks. BMC Bioinf. 2005;6:227–0.
Li M, Guo Y, Feng Y-M, Zhang N. Identification of Triple-negative Breast Cancer Genes And A Novel High-risk Breast Cancer Prediction Model Development Based On Ppi Data And Support Vector Machines. Front Genet. 2019;10:180.
Cao HH, Zhang YH, Zhao J, Zhu L, Wang Y, Li JR. et al. Prediction of the ebola virus infection related human genes using protein-protein interaction network. Comb Chem High Throughput Screen. 2017;20:999.
Zou Q, Zeng J, Cao L, Ji R. A novel features ranking metric with application to scalable visual and bioinformatics data classification. Neurocomputing. 2015;173:346–54.
Peng H, Long F, Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell. 2005;27:1226–38.
Li BQ, Zhang J, Huang T, Zhang L, Cai YD. Identification of retinoblastoma related genes with shortest path in a protein–protein interaction network. Biochimie. 2012;94:1910–7.
Csardi G, Nepusz T. The igraph software package for complex network research. Inter J Complex Syst. 2006;1695:1–9.
George GVS, Raj VC. Review on feature selection techniques and the impact of svm for cancer classification using gene expression profile. Int J Computer Sci Eng Surv. 2011;2:16–27.
Nguyen HN, Ohn SY, Park J, Park KS. Combined kernel function approach in SVM for diagnosis of cancer. Int Conf Adv Nat Comput. 2005;3610:1017–26.
Junker M, Hoch R, & Dengel A. On the evaluation of document analysis components by recall, precision, and accuracy. International Conference on Document Analysis & Recognition. IEEE. 1999;713–6.
Burks HE, Elliott S, Hoang VT, Matossian MD, Burow BC, Burow ME. Abstract 5079: inhibition of hdac2, but not hdac1, abrogates triple negative breast cancer progression and metastasis. Cancer Res. 2017;77:5079–5079.
D'Souza-Schorey C, Li G, Colombo M, Stahl P. A regulatory role for arf6 in receptor-mediated endocytosis. Science. 1995;267:1175–8.
Uchihara T, Giasson BI. Propagation of alpha-synuclein pathology: hypotheses, discoveries, and yet unresolved questions from experimental and human brain studies. Acta Neuropathol. 2016;131:49–73.
Deaderick & William P. Positive regulation of localization of cell division proteins in escherichia coli. Chancellor’s Honors Program Projects. 2015.
Rodenburg CJ, Cornelisse CJ, Hermans J, Fleuren GJ. Dna flow cytometry and morphometry as prognostic indicators in advanced ovarian cancer: a step forward in predicting the clinical outcome. Gynecologic Oncol. 1988;29:176–87.
Pustjens MF. α-catenin in the adherens junction complex and the possible implications in cancer. Master’s thesis. 2012 (Open Access).
Bajenova O, Chaika N, Tolkunova E, Davydov-Sinitsyn A, Gapon S, Thomas P, et al. Carcinoembryonic antigen promotes colorectal cancer progression by targeting adherens junction complexes. Exp Cell Res. 2014;324:115–23.
Zhang L, Feng T, Spicer L. The role of tight junction proteins in ovarian follicular development and ovarian cancer. Reproduction. 2018;155:REP-17-0503.
Davies EJ. Distribution and clinical significance of heparan sulfate proteoglycans in ovarian cancer. Clin Cancer Res. 2004;10:5178–86.
Mcsherry EA, Brennan K, Hudson L, Hill AD, & Hopkins AM. Breast cancer cell migration is regulated through junctional adhesion molecule-a-mediated activation of rap1 gtpase. Breast Cancer Res. 2011;13:13.
Jiang P, Enomoto A, Takahashi M. Cell biology of the movement of breast cancer cells: intracellular signalling and the actin cytoskeleton. Cancer Lett. 2009;284:0–130.
Chung L, Orberg ET, Geis AL, et al. Bacteroides fragilis, toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells[J]. Cell Host Microbe. 2018;23:421.
Zheng Y, Rodrik V, Toschi A, Shi M, Hui L, Shen Y, et al. Phospholipase d couples survival and migration signals in stress response of human cancer cells. J Biol Chem. 2006;281:15862–8.
Yoshioka S, King ML, Ran S, Okuda H, Hayashi K. Wnt7a regulates tumor growth and progression in ovarian cancer through the wnt/beta-catenin pathway. Mol Cancer Res. 2012;10:469–82.
Aloisi A, Gregorio SD, Stagno F, Guglielmo P, Mannino F, Sormani MP, et al. Bcr-abl nuclear entrapment kills human cml cells: ex vivo study on 35 patients with the combination of imatinib mesylate and leptomycin b. Blood. 2006;107:1591–8.
Tang W, Wan S, Yang Z, Andrew ET, Zou Q. Tumor origin detection with tissue-specific miRNA and DNA methylation markers. Bioinformatics. 2018;34:398–406.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, M., Wang, P., Zhang, N. et al. Identification of genes of four malignant tumors and a novel prediction model development based on PPI data and support vector machines. Cancer Gene Ther 27, 715–725 (2020). https://doi.org/10.1038/s41417-019-0143-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-019-0143-5