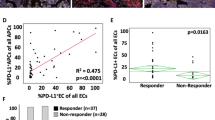
Abstract
The past decade has witnessed a revolution in cancer treatment by the shift from conventional drugs (chemotherapies) towards targeted molecular therapies and immune-based therapies, in particular the immune-checkpoint inhibitors (ICIs). These immunotherapies selectively release the host immune system against the tumour and have shown unprecedented durable remission for patients with cancers that were thought incurable such as advanced non-small cell lung cancer (aNSCLC). The prediction of therapy response is based since the first anti-PD-1/PD-L1 molecules FDA and EMA approvals on the level of PD-L1 tumour cells expression evaluated by immunohistochemistry, and recently more or less on tumour mutation burden in the USA. However, not all aNSCLC patients benefit from immunotherapy equally, since only around 30% of them received ICIs and among them 30% have an initial response to these treatments. Conversely, a few aNSCLC patients could have an efficacy ICIs response despite low PD-L1 tumour cells expression. In this context, there is an urgent need to look for additional robust predictive markers for ICIs efficacy in thoracic oncology. Understanding of the mechanisms that enable cancer cells to adapt to and eventually overcome therapy and identifying such mechanisms can help circumvent resistance and improve treatment. However, more than a unique universal marker, the evaluation of several molecules in the tumour at the same time, particularly by using multiplex immunostaining is a promising open room to optimise the selection of patients who benefit from ICIs. Therefore, urgent further efforts are needed to optimise to individualise immunotherapy based on both patient-specific and tumour-specific characteristics. This review aims to rethink the role of multiplex immunostaining in immuno-thoracic oncology, with the current advantages and limitations in the near-daily practice use.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Not applicable.
References
Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64.
Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707–23.
Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol. 2018;18:153–67.
Marcus L, Fashoyin-Aje LA, Donoghue M, Yuan M, Rodriguez L, Gallagher PS, et al. FDA approval summary: pembrolizumab for the treatment of tumor mutational burden-high solid tumors. Clin Cancer Res. 2021;27:4685–9.
Fincham REA, Bashiri H, Lau MC, Yeong J. Editorial: multiplex immunohistochemistry/immunofluorescence technique: the potential and promise for clinical application. Front Mol Biosci. 2022;9:831383.
Lu S, Stein JE, Rimm DL, Wang DW, Bell JM, Johnson DB, et al. Comparison of biomarker modalities for predicting response to PD-1/PD-L1 checkpoint blockade: a systematic review and meta-analysis. JAMA Oncol. 2019;5:1195–204.
Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, et al. Updated analysis of KEYNOTE-024: pembrolizumab versus platinum-based chemotherapy for advanced non-small-cell lung cancer with PD-L1 tumor proportion score of 50% or greater. J Clin Oncol. 2019;37:537–46.
Paz-Ares L, Luft A, Vicente D, Tafreshi A, Gumus M, Mazieres J, et al. Pembrolizumab plus chemotherapy for squamous non-small-cell lung cancer. N Engl J Med. 2018;379:2040–51.
Ilie M, Hofman V, Dietel M, Soria JC, Hofman P. Assessment of the PD-L1 status by immunohistochemistry: challenges and perspectives for therapeutic strategies in lung cancer patients. Virchows Arch. 2016;468:511–25.
Doroshow DB, Bhalla S, Beasley MB, Sholl LM, Kerr KM, Gnjatic S, et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol. 2021;18:345–62.
Mino-Kenudson M, Schalper K, Cooper W, Dacic S, Hirsch FR, Jain D, et al. Predictive biomarkers for immunotherapy in lung cancer: perspective from the International Association for the Study of Lung Cancer Pathology Committee. J Thorac Oncol. 2022;17:1335–54.
Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19:133–50.
Berland L, Heeke S, Humbert O, Macocco A, Long-Mira E, Lassalle S, et al. Current views on tumor mutational burden in patients with non-small cell lung cancer treated by immune checkpoint inhibitors. J Thorac Dis. 2019;11:S71–S80.
Anagnostou V, Bardelli A, Chan TA, Turajlic S. The status of tumor mutational burden and immunotherapy. Nat Cancer. 2022;3:652–6.
Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30:44–56.
Marabelle A, Fakih M, Lopez J, Shah M, Shapira-Frommer R, Nakagawa K, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21:1353–65.
McGrail DJ, Pilie PG, Rashid NU, Voorwerk L, Slagter M, Kok M, et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol. 2021;32:661–72.
Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202–6.
Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8.
Heeke S, Benzaquen J, Hofman V, Long-Mira E, Lespinet V, Bordone O, et al. Comparison of three sequencing panels used for the assessment of tumor mutational burden in NSCLC reveals low comparability. J Thorac Oncol. 2020;15:1535–40.
Ricciuti B, Wang X, Alessi JV, Rizvi H, Mahadevan NR, Li YY, et al. Association of high tumor mutation burden in non-small cell lung cancers with increased immune infiltration and improved clinical outcomes of PD-L1 blockade across PD-L1 expression levels. JAMA Oncol. 2022;8:1160–8.
Angelova M, Mlecnik B, Vasaturo A, Bindea G, Fredriksen T, Lafontaine L, et al. Evolution of metastases in space and time under immune selection. Cell. 2018;175:751–65 e16.
Galon J, Bruni D. Tumor immunology and tumor evolution: intertwined histories. Immunity. 2020;52:55–81.
Banik G, Betts CB, Liudahl SM, Sivagnanam S, Kawashima R, Cotechini T, et al. High-dimensional multiplexed immunohistochemical characterization of immune contexture in human cancers. Methods Enzymol. 2020;635:1–20.
De Guillebon E, Dardenne A, Saldmann A, Seguier S, Tran T, Paolini L, et al. Beyond the concept of cold and hot tumors for the development of novel predictive biomarkers and the rational design of immunotherapy combination. Int J Cancer. 2020;147:1509–18.
Schoffski P, Tan DSW, Martin M, Ochoa-de-Olza M, Sarantopoulos J, Carvajal RD, et al. Phase I/II study of the LAG-3 inhibitor ieramilimab (LAG525) +/- anti-PD-1 spartalizumab (PDR001) in patients with advanced malignancies. J Immunother Cancer. 2022;10:e003776.
Thakkar D, Paliwal S, Dharmadhikari B, Guan S, Liu L, Kar S, et al. Rationally targeted anti-VISTA antibody that blockades the C-C’ loop region can reverse VISTA immune suppression and remodel the immune microenvironment to potently inhibit tumor growth in an Fc independent manner. J Immunother Cancer. 2022;10:e003382.
Curigliano G, Gelderblom H, Mach N, Doi T, Tai D, Forde PM, et al. Phase I/Ib clinical trial of sabatolimab, an anti-TIM-3 antibody, alone and in combination with spartalizumab, an anti-PD-1 antibody, in advanced solid tumors. Clin Cancer Res. 2021;27:3620–9.
Aroldi F, Saleh R, Jafferji I, Barreto C, Saberian C, Middleton MR. Lag3: from bench to bedside. Cancer Treat Res. 2022;183:185–99.
Niu J, Maurice-Dror C, Lee DH, Kim DW, Nagrial A, Voskoboynik M, et al. First-in-human phase 1 study of the anti-TIGIT antibody vibostolimab as monotherapy or with pembrolizumab for advanced solid tumors, including non-small-cell lung cancer(☆). Ann Oncol. 2022;33:169–80.
De Mello RA, Zhu JH, Iavelberg J, Potim AH, Simonetti D, Silva JA Jr., et al. Current and future aspects of TIM-3 as biomarker or as potential targeted in non-small cell lung cancer scope: is there a role in clinical practice? Transl Lung Cancer Res. 2020;9:2311–4.
Bai R, Lv Z, Xu D, Cui J. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomark Res. 2020;8:34.
Sholl LM. Biomarkers of response to checkpoint inhibitors beyond PD-L1 in lung cancer. Mod Pathol. 2022;35:66–74.
Shirasawa M, Yoshida T, Imabayashi T, Okuma K, Matsumoto Y, Masuda K, et al. Baseline PD-L1 expression and tumour-infiltrated lymphocyte status predict the efficacy of durvalumab consolidation therapy after chemoradiotherapy in unresectable locally advanced patients with non-small-cell lung cancer. Eur J Cancer. 2022;162:1–10.
Gettinger SN, Choi J, Mani N, Sanmamed MF, Datar I, Sowell R, et al. A dormant TIL phenotype defines non-small cell lung carcinomas sensitive to immune checkpoint blockers. Nat Commun. 2018;9:3196.
Anagnostou V, Luke JJ. Quantitative spatial profiling of TILs as the next step beyond PD-L1 testing for immune checkpoint blockade. Clin Cancer Res. 2022;28:4835–7.
Kim HR, Park SM, Seo SU, Jung I, Yoon HI, Gabrilovich DI, et al. The ratio of peripheral regulatory T cells to Lox-1(+) polymorphonuclear myeloid-derived suppressor cells predicts the early response to anti-PD-1 therapy in patients with non-small cell lung cancer. Am J Respir Crit Care Med. 2019;199:243–6.
Khatir W, Humbert O, Benzaquen J, Bontoux C, Neels J, Berland L, et al. Identification of a circulating immunological signature predictive of response to immune checkpoint inhibitors in patients with advanced non-small cell lung cancer. Clin Transl Med. 2022;12:e1018.
Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577:549–55.
Patil NS, Nabet BY, Muller S, Koeppen H, Zou W, Giltnane J, et al. Intratumoral plasma cells predict outcomes to PD-L1 blockade in non-small cell lung cancer. Cancer Cell. 2022;40:289–300 e4.
Philip M, Schietinger A. CD8(+) T cell differentiation and dysfunction in cancer. Nat Rev Immunol. 2022;22:209–23.
Datar I, Sanmamed MF, Wang J, Henick BS, Choi J, Badri T, et al. Expression analysis and significance of PD-1, LAG-3, and TIM-3 in human non-small cell lung cancer using spatially resolved and multiparametric single-cell analysis. Clin Cancer Res. 2019;25:4663–73.
Eiva MA, Omran DK, Chacon JA, Powell DJ Jr. Systematic analysis of CD39, CD103, CD137, and PD-1 as biomarkers for naturally occurring tumor antigen-specific TILs. Eur J Immunol. 2022;52:96–108.
Fridman WH, Zitvogel L, Sautes-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14:717–34.
Remark R, Merghoub T, Grabe N, Litjens G, Damotte D, Wolchok JD, et al. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci Immunol. 2016;1:aaf6925.
Glass G, Papin JA, Mandell JW. SIMPLE: a sequential immunoperoxidase labeling and erasing method. J Histochem Cytochem. 2009;57:899–905.
Zhang W, Hubbard A, Jones T, Racolta A, Bhaumik S, Cummins N, et al. Fully automated 5-plex fluorescent immunohistochemistry with tyramide signal amplification and same species antibodies. Lab Invest. 2017;97:873–85.
Morrison LE, Lefever MR, Behman LJ, Leibold T, Roberts EA, Horchner UB, et al. Brightfield multiplex immunohistochemistry with multispectral imaging. Lab Invest. 2020;100:1124–36.
Akturk G, Sweeney R, Remark R, Merad M, Gnjatic S. Multiplexed immunohistochemical consecutive staining on single slide (MICSSS): multiplexed chromogenic IHC assay for high-dimensional tissue analysis. Methods Mol Biol. 2020;2055:497–519.
Tsujikawa T, Kumar S, Borkar RN, Azimi V, Thibault G, Chang YH, et al. Quantitative multiplex immunohistochemistry reveals myeloid-inflamed tumor-immune complexity associated with poor prognosis. Cell Rep. 2017;19:203–17.
Moles Lopez X, Barbot P, Van Eycke YR, Verset L, Trepant AL, Larbanoix L, et al. Registration of whole immunohistochemical slide images: an efficient way to characterize biomarker colocalization. J Am Med Inf Assoc. 2015;22:86–99.
Day WA, Lefever MR, Ochs RL, Pedata A, Behman LJ, Ashworth-Sharpe J, et al. Covalently deposited dyes: a new chromogen paradigm that facilitates analysis of multiple biomarkers in situ. Lab Invest. 2017;97:104–13.
Polaske NW, Kelly BD, Ashworth-Sharpe J, Bieniarz C. Quinone methide signal amplification: covalent reporter labeling of cancer epitopes using alkaline phosphatase substrates. Bioconjug Chem. 2016;27:660–6.
Ilie M, Beaulande M, Ben Hadj S, Chamorey E, Schiappa R, Long-Mira E, et al. Chromogenic multiplex immunohistochemistry reveals modulation of the immune microenvironment associated with survival in elderly patients with lung adenocarcinoma. Cancers. 2018;10:326.
Ilie M, Beaulande M, Hamila M, Erb G, Hofman V, Hofman P. Automated chromogenic multiplexed immunohistochemistry assay for diagnosis and predictive biomarker testing in non-small cell lung cancer. Lung Cancer. 2018;124:90–4.
Ilie M, Beaulande M, Long-Mira E, Bontoux C, Zahaf K, Lalvee S, et al. Analytical validation of automated multiplex chromogenic immunohistochemistry for diagnostic and predictive purpose in non-small cell lung cancer. Lung Cancer. 2022;166:1–8.
Schubert W, Bonnekoh B, Pommer AJ, Philipsen L, Bockelmann R, Malykh Y, et al. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat Biotechnol. 2006;24:1270–8.
Wang N, Li X, Wang R, Ding Z. Spatial transcriptomics and proteomics technologies for deconvoluting the tumor microenvironment. Biotechnol J. 2021;16:e2100041.
Lin JR, Izar B, Wang S, Yapp C, Mei S, Shah PM, et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife. 2018;7:e31657.
Cheng Y, Burrack RK, Li Q. Spatially resolved and highly multiplexed protein and rna in situ detection by combining CODEX with RNAscope in situ hybridization. J Histochem Cytochem. 2022;70:571–81.
Black S, Phillips D, Hickey JW, Kennedy-Darling J, Venkataraaman VG, Samusik N, et al. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat Protoc. 2021;16:3802–35.
Herbel C, Reiß S, Jungblut M, Eckardt D, Bosio A. MACSima™ Imaging Platform provides new insights into cancer biology and target discovery by cyclic immunofluorescence-based imaging. MACS More. 2019;18:16–20.
Berghmans E, Van Raemdonck G, Schildermans K, Willems H, Boonen K, Maes E, et al. MALDI mass spectrometry imaging linked with top-down proteomics as a tool to study the non-small-cell lung cancer tumor microenvironment. Methods Protoc. 2019;2:44.
Francisco-Cruz A, Parra ER, Tetzlaff MT, Wistuba II. Multiplex immunofluorescence assays. Methods Mol Biol. 2020;2055:467–95.
Kriegsmann M, Zgorzelski C, Casadonte R, Schwamborn K, Muley T, Winter H, et al. Mass spectrometry imaging for reliable and fast classification of non-small cell lung cancer subtypes. Cancers. 2020;12:2704.
Ptacek J, Locke D, Finck R, Cvijic ME, Li Z, Tarolli JG, et al. Multiplexed ion beam imaging (MIBI) for characterization of the tumor microenvironment across tumor types. Lab Invest. 2020;100:1111–23.
Tan WCC, Nerurkar SN, Cai HY, Ng HHM, Wu D, Wee YTF, et al. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun. 2020;40:135–53.
Cooper WA, Lantuejoul S, Mino-Kenudson M. Predicting response to programmed cell death protein-1 or programmed death-ligand 1 blockade in NSCLC—is multiplex immunohistochemistry or immunofluorescence the answer? J Thorac Oncol. 2021;16:1247–9.
Shakya R, Nguyen TH, Waterhouse N, Khanna R. Immune contexture analysis in immuno-oncology: applications and challenges of multiplex fluorescent immunohistochemistry. Clin Transl Immunol. 2020;9:e1183.
Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70:46–58.
Choi YJ, Nakano K, Ide T, Sakae T, Ichikawa R, Hizawa T, et al. Demonstrating a Filter-free wavelength sensor with double-well structure and its application. Biosens. 2022;12:1033.
Hofman P, Badoual C, Henderson F, Berland L, Hamila M, Long-Mira E, et al. Multiplexed immunohistochemistry for molecular and immune profiling in lung cancer-just about ready for prime-time? Cancers. 2019;11:283.
Laberiano-Fernandez C, Hernandez-Ruiz S, Rojas F, Parra ER. Best practices for technical reproducibility assessment of multiplex immunofluorescence. Front Mol Biosci. 2021;8:660202.
Parra ER, Francisco-Cruz A, Wistuba II. State-of-the-art of profiling immune contexture in the era of multiplexed staining and digital analysis to study paraffin tumor tissues. Cancers. 2019;11:247.
Mungenast F, Fernando A, Nica R, Boghiu B, Lungu B, Batra J, et al. Next-generation digital histopathology of the tumor microenvironment. Genes. 2021;12:538.
Surace M, Rognoni L, Rodriguez-Canales J, Steele KE. Characterization of the immune microenvironment of NSCLC by multispectral analysis of multiplex immunofluorescence images. Methods Enzymol. 2020;635:33–50.
Bankhead P. Developing image analysis methods for digital pathology. J Pathol. 2022;257:391–402.
Evans AJ, Salama ME, Henricks WH, Pantanowitz L. Implementation of whole slide imaging for clinical purposes: issues to consider from the perspective of early adopters. Arch Pathol Lab Med. 2017;141:944–59.
Akturk G, Parra ER, Gjini E, Lako A, Lee JJ, Neuberg D, et al. Multiplex tissue imaging harmonization: a multicenter experience from CIMAC-CIDC immuno-oncology biomarkers network. Clin Cancer Res. 2021;27:5072–83.
Taube JM, Roman K, Engle EL, Wang C, Ballesteros-Merino C, Jensen SM, et al. Multi-institutional TSA-amplified multiplexed immunofluorescence reproducibility evaluation (MITRE) study. J Immunother Cancer. 2021;9:e002197.
Taube JM, Akturk G, Angelo M, Engle EL, Gnjatic S, Greenbaum S, et al. The Society for Immunotherapy of Cancer statement on best practices for multiplex immunohistochemistry (IHC) and immunofluorescence (IF) staining and validation. J Immunother Cancer. 2020;8:e000155.
Fraggetta F, L’Imperio V, Ameisen D, Carvalho R, Leh S, Kiehl TR, et al. Best practice recommendations for the implementation of a digital pathology workflow in the anatomic pathology laboratory by the European Society of Digital and Integrative Pathology (ESDIP). Diagnostics. 2021;11:2167.
Rojas F, Hernandez S, Lazcano R, Laberiano-Fernandez C, Parra ER. Multiplex immunofluorescence and the digital image analysis workflow for evaluation of the tumor immune environment in translational research. Front Oncol. 2022;12:889886.
Coulter C, McKay F, Hallowell N, Browning L, Colling R, Macklin P, et al. Understanding the ethical and legal considerations of digital pathology. J Pathol Clin Res. 2022;8:101–15.
Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16:703–15.
Berry S, Giraldo NA, Green BF, Cottrell TR, Stein JE, Engle EL, et al. Analysis of multispectral imaging with the AstroPath platform informs efficacy of PD-1 blockade. Science. 2021;372:eaba2609.
Bosisio FM, Van Herck Y, Messiaen J, Bolognesi MM, Marcelis L, Van Haele M, et al. Next-generation pathology using multiplexed immunohistochemistry: mapping tissue architecture at single-cell level. Front Oncol. 2022;12:918900.
Ghaffari Laleh N, Ligero M, Perez-Lopez R, Kather JN. Facts and hopes on the use of artificial intelligence for predictive immunotherapy biomarkers in cancer. Clin Cancer Res. 2022;29:316–23.
Cheng JY, Abel JT, Balis UGJ, McClintock DS, Pantanowitz L. Challenges in the development, deployment, and regulation of artificial intelligence in anatomic pathology. Am J Pathol. 2021;191:1684–92.
Hoyt CC. Multiplex immunofluorescence and multispectral imaging: forming the basis of a clinical test platform for immuno-oncology. Front Mol Biosci. 2021;8:674747.
Locke D, Hoyt CC. Companion diagnostic requirements for spatial biology using multiplex immunofluorescence and multispectral imaging. Front Mol Biosci. 2023;10:1051491.
Parra ER, Jiang M, Solis L, Mino B, Laberiano C, Hernandez S, et al. Procedural requirements and recommendations for multiplex immunofluorescence tyramide signal amplification assays to support translational oncology studies. Cancers. 2020;12:255.
Lewis SM, Asselin-Labat ML, Nguyen Q, Berthelet J, Tan X, Wimmer VC, et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat Methods. 2021;18:997–1012.
Hegde PS, Chen DS. Top 10 challenges in cancer immunotherapy. Immunity. 2020;52:17–35.
Hofman P, Heeke S, Alix-Panabieres C, Pantel K. Liquid biopsy in the era of immuno-oncology: is it ready for prime-time use for cancer patients? Ann Oncol. 2019;30:1448–59.
Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyo D, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24:1559–67.
Courtiol P, Maussion C, Moarii M, Pronier E, Pilcer S, Sefta M, et al. Deep learning-based classification of mesothelioma improves prediction of patient outcome. Nat Med. 2019;25:1519–25.
Wang X, Barrera C, Bera K, Viswanathan VS, Azarianpour-Esfahani S, Koyuncu C, et al. Spatial interplay patterns of cancer nuclei and tumor-infiltrating lymphocytes (TILs) predict clinical benefit for immune checkpoint inhibitors. Sci Adv. 2022;8:eabn3966.
Jiang Y, Yang M, Wang S, Li X, Sun Y. Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun. 2020;40:154–66.
Baxi V, Edwards R, Montalto M, Saha S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod Pathol. 2022;35:23–32.
Park S, Ock CY, Kim H, Pereira S, Ma M, Choi S, et al. Artificial intelligence-powered spatial analysis of tumor-infiltrating lymphocytes as complementary biomarker for immune checkpoint inhibition in non-small-cell lung cancer. J Clin Oncol. 2022;40:1916–28.
Hanna MG, Ardon O, Reuter VE, Sirintrapun SJ, England C, Klimstra DS, et al. Integrating digital pathology into clinical practice. Mod Pathol. 2022;35:152–64.
Vanguri RS, Luo J, Aukerman AT, Egger JV, Fong CJ, Horvat N, et al. Multimodal integration of radiology, pathology and genomics for prediction of response to PD-(L)1 blockade in patients with non-small cell lung cancer. Nat Cancer. 2022;3:1151–64.
Chen RJ, Lu MY, Williamson DFK, Chen TY, Lipkova J, Noor Z, et al. Pan-cancer integrative histology-genomic analysis via multimodal deep learning. Cancer Cell. 2022;40:865–78 e6.
Acknowledgements
The authors received no additional support for the research or authorship of this article.
Funding
There was no financial support for the research or authorship of this article.
Author information
Authors and Affiliations
Contributions
EP, MI, IW and PH and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This article does not contain studies involving human participants or animals that were performed by the authors.
Consent for publication
This article does not contain any individual person’s data in any form.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Parra, E.R., Ilié, M., Wistuba, I.I. et al. Quantitative multiplexed imaging technologies for single-cell analysis to assess predictive markers for immunotherapy in thoracic immuno-oncology: promises and challenges. Br J Cancer 129, 1417–1431 (2023). https://doi.org/10.1038/s41416-023-02318-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-023-02318-7