Abstract
The archaeal Asgard superphylum currently stands as the most promising prokaryotic candidate, from which eukaryotic cells emerged. This unique superphylum encodes for eukaryotic signature proteins (ESP) that could shed light on the origin of eukaryotes, but the properties and function of these proteins is largely unresolved. Here, we set to understand the function of an Asgard archaeal protein family, namely the ESCRT machinery, that is conserved across all domains of life and executes basic cellular eukaryotic functions, including membrane constriction during cell division. We find that ESCRT proteins encoded in Loki archaea, express in mammalian and yeast cells, and that the Loki ESCRT-III protein, CHMP4-7, resides in the eukaryotic nucleus in both organisms. Moreover, Loki ESCRT-III proteins associated with chromatin, recruited their AAA-ATPase VPS4 counterpart to organize in discrete foci in the mammalian nucleus, and directly bind DNA. The human ESCRT-III protein, CHMP1B, exhibited similar nuclear properties and recruited both human and Asgard VPS4s to nuclear foci, indicating interspecies interactions. Mutation analysis revealed a role for the N terminal region of ESCRT-III in mediating these phenotypes in both human and Asgard ESCRTs. These findings suggest that ESCRT proteins hold chromatin binding properties that were highly preserved through the billion years of evolution separating Asgard archaea and humans. The conserved chromatin binding properties of the ESCRT membrane remodeling machinery, reported here, may have important implications for the origin of eukaryogenesis.
Similar content being viewed by others
Introduction
Encoded by all domains of life, the ESCRT machinery constitutes one of the most evolutionarily conserved cellular membrane remodeling machines [1]. ESCRT is a multiprotein complex that is composed of five subfamilies i.e., ESCRT- 0-III and the AAA-ATPase VPS4. Within this complex, ESCRT-III (named CHMPs in animal cells) and VPS4 manifest the minimal unit required for driving membrane fission [2, 3]. ESCRTs have been shown to execute membrane fission in numerous cellular processes in eukaryotes, including cell division, multivesicular bodies formation (MVBs), vesicle release, membrane repair, neural development, and nuclear envelop resealing [2, 4]. Additional noncanonical functions have also been proposed for this seminal complex [5,6,7,8,9]. Although the involvement of ESCRTs in canonical membrane-remodeling processes has been documented in primitive cellular life forms (i.e., cell division and vesicle release), its noncanonical functions have not been explored [1, 10].
Clues for the functional evolutionary origins of the ESCRT system could reside in the prokarya domain that houses the most ancient life forms of our planet. In bacteria, distant ESCRT homologs have recently been identified, but their cellular function is still largely unknown [11, 12], while in archaea, the complex was associated with cell division and vesicle release [1, 2, 5, 10, 13,14,15,16,17]. Genes encoding for ESCRT proteins have also been found in Asgard archaea, a newly discovered Asgard super-phyla, that encode for several eukaryotic signature proteins (ESPs) and is currently considered as the ancestor of the first eukaryotic cell [18,19,20]. To date, seven different ESCRT-related genes have been identified in Asgard archaea: homologs of two ESCRT-I proteins, two ESCRT-IIs, two ESCRT-IIIs (referred to here as CHMP1-3, CHMP4-7), and VPS4 [19, 20]. While homologs for ESCRT-III and VPS4 were previously identified in archaea of the TACK superphyla (named Cdv B and Cdv C, respectively) [1, 14, 16, 21], several lines of evidence suggest that Asgard ESCRTs are more closely related to eukaryotes. First, homologs for ESCRT-I and –II have only been found in Asgard archaea and were recently shown to carry similar function-related properties [22]. Second, domain analysis indicates that some domains of the Asgard ESCRTs are conserved in eukaryotes but are absent in other archaeal phyla [23, 24]. Finally, in recent experiments performed in yeast the Asgard VPS4 homolog was more efficient than the other archaeal homologs in reverting phenotypes associated with VPS4 absence [24]. Hence, elucidating the function of Asgard ESCRTs can shed light on the ancient cellular function of this conserved membrane remodeling machine in eukaryotes.
The Asgard super-phylum is highly diverse and encompasses a growing number of phyla, including Lokiarchaeota (Loki), Thorarchaeota (Thor), Odinarchaeota (Odi), Helarchaeota (Hela), Freyarcheota (Frey), and Heimdallarchaeota (Heimdall) [25]. Among these phyla, Loki was the first to be identified and is considered to be the most abundant, and Heimdall is thought to be the closest relative to eukaryotes [19, 20]. So far, most of the knowledge obtained on Asgard archaea was derived from metagenome sequence analysis. Moreover, there are currently no tools or appropriate model systems to study the biology of the Asgard archaea superphylum with the exception of one recent enrichment culture of Lokiarchaeota species population [26]. Hence, studying the functional role of ESCRT proteins encoded by Asgard archaea in their native environment is extremely challenging.
To overcome these challenges and to track back the functional origins of these cardinal proteins, we set out to characterize Asgard ESCRTs via their heterologous expression in mammalian cells. In this study, we find evidence to suggest that the functional origins of these proteins involve DNA binding. We achieved the expression of the genes encoding the two Asgard ESCRT-III subfamilies (CHMP1-3 and CHMP4-7) and VPS4 from both Heimdall and Loki origin in mammalian tissue culture cells. We further show that these proteins are working in concert; ESCRTIII/VPS4 homologs from Lokiarchaeota interacted with one another within mammalian cells, and changed their subcellular localization upon co-expression. Unexpectedly, Loki CHMP 4-7 was localized to the nucleus in both mammalian and yeast cells, when solely expressed, and reorganized into discrete foci in either the cytosol or the nucleus upon co-expression with VPS4 in mammalian cells. Consistently, CHMP4-7 was found in the chromatin fraction and recruited VPS4 to chromatin upon co-expression. Notably, mammalian ESCRT-III/VPS4 proteins (particularly CHMP1) were also found in the chromatin fraction, with human CHMP1B organizing in nuclear foci and recruiting both human and Asgard VPS4s to these foci when overexpressed in cells. Moreover, removing the homologous N terminal regions in both human and Asgard ESCRT-IIIs (CHMP1B and CHMP4-7, respectively) abolished their ability to organize in nuclear foci. Hence, specific human and Asgard ESCRT proteins associate with chromatin and functionally interact with one another inside mammalian cells, corroborating the conserved functionality of these proteins. Finally, recombinant Loki CHMP4-7 and human CHMP1B were able to directly bind DNA in vitro, indicating direct binding of ESCRT-III to DNA of both Asgard and human ESCRT-IIIs. Collectively, our findings highlight an ancient property of the ESCRT system to interact with chromatin that is conserved in humans.
Results
According to current models, the ESCRT-III and VPS4 complexes in eukaryotes constitute the core components of the ESCRT machine that execute membrane fission [27,28,29,30]. They do so by assembling ESCRT-III monomers into helical filaments and remodeling these filaments by VPS4. To track the functional eukaryotic origins of ESCRT protein family in the Asgard super-phyla, we first performed hierarchical clustering based on sequence homology and maximum likelihood phylogenetic analysis of all Asgard ESCRT-III/VPS4 genes deposited in the NCBI database and homologs encoded by representative unicellular eukaryotes and by humans. Our analysis showed that the Asgard genes share sequence homology and cluster together with their eukaryotic homologs into three subgroups according to the standard ESCRT protein classification (Fig. 1A, left panel). One group is composed of VPS4 homologs, and two groups are composed of ESCRT-III proteins, as previously shown using smaller cohorts [20, 23, 24]. The twelve human ESCRT-IIIs were divided between these two ESCRT-III subgroups so that CHMPs 1, 2, 3 (Vps2/24/46 in yeast) and IST1 resided in one cluster (named CHMP1-3) and CHMPs 4, 5, 6, 7 (Vps20/32/60 and Chm7 in yeast) in the second (named CHMP4-7). Sequence identity clustering analyses (Fig. 1A, right panel) show that the VPS4 cluster confers the highest sequence identity and evolutionary conservation between species while the sequences in the two ESCRT-III clusters are more diverged. Therefore, while VPS4 is relatively conserved between different archaea species, ESCRT-IIIs are highly diverse, querying a functional similarity for these homologs within the archaea phylum and between archaea and humans.
A Left panel: maximum likelihood phylogenetic tree of human and Asgard ESCRT III/VPS4. Colors of the branches denote the ESCRT classifications. Predicted domains for proteins in each cluster (InterPro) are denoted below cluster name. Right panel: a heatmap showing the sequence similarity of the three ESCRT groups (defined and colored according to their clustering on the phylogenetic tree), using Fitch–Margoliash distance. Proteins are colored in left and right panels according to their phylogenetic associations (as indicated in colored index on left). B Selected Robetta predicted structures of human ESCRT-III proteins examined (for confidence score, see Supplementary Fig. 1A). Full-length proteins are shown except for human IST1 in which amino acids 1–217 are shown. Colors encode to open (dark blue) and closed (light blue) ESCRT-III conformations. A Cryo-EM structure of the human IST1 and CHMP1B co-polymer (PDB ID, 3JC1) is shown below the human predictions. Single monomers of IST1, closed conformation (dark blue), and CHMP1B, open conformation (light blue) are highlighted in the filament structure. Overlay of structure predictions of all human ESCRT-III proteins from both Alphafold and Robetta as well as with the available structures in the protein database (PDB) is provided in Supplementary Fig. 1B. C Robetta predicted structures of the Asgard ESCRT-III proteins examined in this study (for confidence score, see Supplementary Fig. 1A). Overlay of structure predictions of the selected Asgard proteins obtained from both Alphafold and Robetta is provided in Supplementary Fig. 1C-D. D Density of averaged amino acids distances as measured based on Roebetta structure predictions of Loki and Hemidall (left panel) and of 83 Asgard CHMP1-3 and 77 CHMP4-7 sequences from our phylogenetic tree (right panel). The resulted averaged distances distributions strongly support two separate groups according to two-sample Kolmogorov-Smirnov test (Loki, D = 0.378, p value < 9.83e-14; Heimdall, D = 0.472, p value < 2.2e-16; All, D = 0.472, p value <2.2e-16).
Asgard ESCRT-III homologs from both clusters (CHMP1-3 and CHMP4-7) were previously reported to include the conserved ESCRT snf-7 polymerization domain and a C-terminal coil domain, in domain analysis (annotated in Fig. 1A), and to possess the conserved ESCRT protein-protein interaction motifs (MIM domains) [23, 24]. Asgard VPS4s had the conserved MIT domain, which governs the interaction between VPS4 and ESCRT-III in eukaryotes (annotated in Fig. 1A) [24, 31]. Therefore, Asgard ESCRT-III homologs encode for the polymerization and VPS4-interaction motifs identified in eukaryotes, suggesting conservation of function.
Following this notion, we asked whether the overall structure of ESCRT-III could be similar to Asgard and human homologs. For this purpose, we generated structural models for human ESCRT-III proteins and ESCRT-III homologs encoded by Loki and Heimdall species using the deep-learning-based structure prediction servers Robetta and AlphaFold (Fig. 1B and C, respectively and Supplementary Fig. 1, A–D) [32, 33]. The 12 human ESCRT-III proteins exhibited two main folds, previously termed the “open” and “closed” conformations [3]. Ten ESCRT-III proteins were predicted to reside in the open conformation and two (CHMP3 and IST1) in the closed conformation (Fig. 1B and Supplementary Fig. 1B). Interestingly, the structural models obtained for Asgard ESCRT-III homologs of both Loki and Heimdall resembled those obtained for human ESCRT-IIIs, with CHMP4-7 obtaining the so-called open conformation and CHMP1-3 the closed conformation (Fig. 1C and Supplementary Fig. 1, C, D). The number and length of helices in proteins that exhibited similar conformation (open or closed) were also similar between human and Asgard ESCRT-III homologs, except for Heimdall CHMP4-7, which had shorter helixes and an additional helix at the C terminal (Supplementary Fig. 1E). Plotting the averaged distances between amino acids in the Loki and Heimdall ESCRT-IIIs structural models supported a two-population distribution (CHMP1-3 and CHMP4-7) (Fig. 1E, left panels). Shorter averaged distances were obtained for the CHMP1-3 proteins, consistent with the more compact, closed conformation predicted for these proteins, hence, providing a good measure for separating the open and closed conformations. Expanding this analysis to 160 ESCRT-III Asgard genes from our phylogenetic tree, resulted in similar distributions, suggesting that the open and closed conformations are conserved in Asgard CHMP4-7 and CHMP1-3 proteins (Fig. 1E, right panel). Therefore, the human and Asgard ESCRT-III homologs are predicted to share similar 3D structural folds despite the low sequence homology and the evolutionary gap that separate them, supporting polymerization capabilities for Asgard ESCRT-IIIs. Moreover, the open and closed conformations of ESCRT-III proteins appear to be evolutionarily conserved and may therefore be crucial for the function of the machinery.
As our predictions suggest that the human and Asgard ESCRT-III homologs share structural similarities, we set forward to explore their function in mammalian and human tissue culture systems. To this end, we sub-cloned genes encoding Loki and Heimdall ESCRT-III proteins (CHMP1-3, CHMP4-7) and VPS4 into expression vectors (three proteins from each family). We specifically chose ESCRT-III and VPS4 genes that are coded from the same metagenomic assembled genomes (MAGs) to increase the likelihood of ESCRTs to interact with one another and carry out their function. All six Asgard ESCRT proteins were expressed in mammalian tissue culture cells (Fig. 2A). Slightly different subcellular distributions were obtained for Loki and Heimdall ESCRT proteins (Fig. 2B, C). Loki ESCRTs were found in both the soluble and insoluble fractions with CHMP1-3 and VPS4 enriched in the soluble fraction and CHMP4-7 equally distributed in both fractions (Fig. 2B, left panel). Heimdall ESCRTs CHMP4-7 and VPS4 were predominantly in the soluble fraction, while CHMP1-3 was found to be enriched in the insoluble fraction (Fig. 2B, right panel). In confocal images, fluorescently tagged versions of Heimdall ESCRTs were found to reside in both the nucleus and cytosol (Fig. 2C, right panel). Loki VPS4 was also distributed in both compartments, but Loki CHMP1-3 was mainly cytosolic and Loki CHMP4-7 was predominantly nuclear (Fig. 2C, middle panel). Similar subcellular distributions were observed for Loki ESCRTs in several cultured cell lines indicating that they reflect the general tendency of the proteins to distribute inside cells (Supplementary Fig. 2A). Therefore, genes encoding Asgard ESCRTs are expressed in human and mammalian cells and reside in both the nucleus and the cytoplasm. While these findings are consistent with the ability of human ESCRT-III/VPS4 to distribute between the cytosol and nucleus [34, 35], the realization that ESCRT homologs encoded by prokaryotic Asgard archaea enter the eukaryotic nucleus is noteworthy.
A–E MDCK cells were transfected with Emerald-CHMP1-3, Apple-CHMP4-7, or Emerald-VPS4 homologs from either Loki or Heimdall. Cells were lysed (A) or lysed and fractionated to supernatant (S) and pellet (P) (B) and subjected to western blot analysis using anti-mCh or anti-GFP antibodies. C Fluorescently tagged versions of ESCRTIII/VPS4 proteins from Human (left), Loki (middle), and Heimdall (right) were expressed in MDCK cells, as indicated. Following fixation and Hoechst staining (shown in merged images) cells were imaged using confocal microscopy. Note that while most of ESCRT proteins examined are distributed between the nucleus and cytosol when expressed alone, Loki CHMP4-7 is mainly nuclear. Representative single slice images are shown. Data obtained from at least three independent experiments, n ≥ 50 for each condition, scale = 10 µM. Similar results were obtained in HeLa and NIH3T3 cells (Supplementary Fig. 2A). D MDCK cells were co-transfected with fluorescently tagged Loki ESCRTIII/VPS4 combinations, as indicated, subjected to immunoprecipitation using anti-GFP antibodies, and immunoblotted (IB) using anti-mCherry antibodies. (I- cell extracts, Ub -unbound fraction, B- bound fraction). Data were reproduced in at least two independent experiments. E MDCK cells were co-transfected with fluorescently tagged Loki ESCRTIII/VPS4, as indicated. Followed fixation and Hoechst staining (shown in merged images), cells were imaged using confocal microscopy. Scale = 10 µM. Representative single slice images are shown. Note that while co-transfection of CHMP1-3 and VPS4 or CHMP1-3 and CHMP4-7 did not affect their distributions (first and second panels), co-expression of CHMP4-7 and VPS4 induced the accumulation of both proteins in discrete foci in the cytosol (~25% of the cells; third panel) or the nucleus (~30% of the cells; fourth panel). Images were obtained from at least three independent experiments, n ≥ 100 for each condition. Similar results were obtained in HeLa and NIH3T3 cells (Supplementary Fig. 3B). F SIM images of nuclear foci in MDCK cells co-transfected with Loki Apple-CHMP4-7 and Emerald-VPS4 and stained with DAPI. Representative single slice images are shown. White boxes indicated the regions shown in the enlarged images to the right. Scale = 2 µM (left panel), 1 µM (middle panel), 0.2 µM (right panel). n = 10. G Envy-VPS4 and yomKate2-CHMP4-7 were integrated in S. Cerevisiae genome individually, left panels (single slices, Scale = 5 µM) or together, right panels (maximum projections, Scale = 1 µM). Two representative examples are shown in right panel. Shown are individual fluorescence channels and an overlay of the fluorescence signal and DIC. Data was reproduced in three to four colonies obtained for each strain.
To find out whether Asgard ESCRT systems can be reconstituted in mammalian cells, we co-expressed pairs of ESCRT-III/VPS4 proteins in mammalian cells and examined their ability to interact with one another using Co-IP. While all three ESCRT Loki homologs were immunoprecipitated with one another, no interaction between Heimdall ESCRT homologs could be detected (Fig. 2D and Supplementary Fig. 2B, respectively). Therefore, while ESCRT homologs from both Loki and Heimdall are expressed in mammalian cells, we could only reconstitute interactions using the Loki homologs, suggesting that Loki ESCRT system is more functional than the Heimdall system, in the mammalian cell environment.
Given the ability of Loki ESCRT-III/VPS4 proteins to interact inside mammalian cells, we examined the subcellular localization of these proteins upon co-expression. Most co-expression conditions did not affect the subcellular distribution documented for each protein when expressed alone (compare Fig. 2C, middle panel with two upper panels in Fig. 2E). However, co-expression of Loki, CHMP4-7 and VPS4 homologs, shifted the localization pattern of both proteins (compare Fig. 2C, middle panel with two bottom panels in Fig. 2E). Upon co-expression, CHMP4-7 and VPS4 localized in discrete foci residing in either the cytosol (~25% of the cells, Fig. 2E, third panel) or in the nucleus (~30% of the cells, Fig. 2E bottom panel), exhibited a high degree of co-localization in both compartments (Averaged Manders’ coefficient in cytosol or nucleus >0.95, Supplementary Fig. 2C). Endogenous ESCRT-III proteins, CHMP4B and IST1, were found to reside in the cytosolic foci, but not in the nuclear foci, suggesting that the cytosolic foci represent the typical localization pattern of mammalian ESCRTs in MVBs, while the nuclear foci are unique (Supplementary Fig. 2D) [35].
Analysis of Loki CHMP4-7/VPS4 nuclear foci using structured illumination microscopy (SIM) showed that both proteins are in close proximity to one another, with VPS4 engulfing CHMP4-7, confirming co-localization between the proteins and suggesting an ordered organization of the complex inside the nucleus (Fig. 2F). Nuclear foci were not obtained upon co-expression of Heimdall CHMP4-7 and VPS4 (Supplementary Fig. 3A), but was reproduced for Loki ESCRTs in NIH3T3, and HeLa cells (Supplementary Fig. 3B), suggesting that this phenotype is specific to Loki ESCRTs. Moreover, nuclear foci formation relied on CHMP4-7 and VPS4 interactions in an intra-species manner, as no nuclear foci were observed upon co-expression of Loki CHMP4-7 with Heimdall VPS4 or vice versa (Supplementary Fig. 3C). Therefore, Heimdall and Loki ESCRTs exhibit distinct behaviors within cells and exclusive intra-species interaction in mammalian cells that results in a unique organization of Loki CHMP 4-7 and VPS4 in nuclear foci.
To examine whether the subcellular localization of Loki CHMP4-7 and VPS4, and their co-expression-induced redistribution is a general property of these proteins in eukaryotes, we incorporated the genes of Loki CHMP4-7 and VPS4 in the Saccharomyces Cerevisiae (S. Cerevisiae) yeast strain either individually or in concert. Consistent with the results obtained in mammalian cells, Loki CHMP4-7 was predominantly nuclear, while Loki VPS4 exhibited cytosolic localization (Fig. 2G, left panels). Upon co-expression, VPS4 was found in discrete foci in both the cytosol and the nucleus, but no accumulation of CHMP4-7 in these sites could be detected (Fig. 2G, right panels). These findings strongly indicate that Loki CHMP4-7 is targeted to the nucleus of eukaryotic cells, and that Loki VPS4 redistribute to discrete puncta in the cytosol and nucleus when co-expressed with Loki CHMP4-7 in eukaryotic cells.
To investigate the basis for Loki CHMP4-7 and VPS4 co-localization induced nuclear foci, observed in mammalian cells, we set out to examine the ability of Asgard ESCRT-III/VPS4 proteins to interact with one another and associate with chromatin using a fractionation assay. We find that both Loki CHMP4-7 and CHMP1-3 reside in the chromatin-bound fraction either when expressed alone or in the presence of an additional ESCRT partner. Loki VPS4, on the other hand, was found at low levels in the chromatin fraction, when expressed alone or together with CHMP1-3, but exhibited prominent chromatin association in the presence of CHMP4-7 (Fig. 3A). Heimdall CHMP4-7 and VPS4 were found in negligible amounts in the chromatin fraction, explaining the lack of ESCRT-III/VPS4 nuclear foci in cells (Supplementary Fig. 3D). Therefore, Loki CHMP4-7 resides in the chromatin fraction and recruit Loki VPS4 to chromatin upon their co-expression (a model for the interplay of Loki ESCRT-III/VPS4 in mammalian cells is presented in Fig. 3B).
A MDCK cells were transfected or co-transfected with fluorescently tagged versions of ESCRT-III/VPS4 Loki homologs (Emerald-CHMP1-3; CH1-3, Apple-CHMP4-7; CH4-7, Apple-CHMP1-3; CH1-3 or Emerald-VPS4). Cells were then fractionated using a protein fractionation kit (see Methods) and normalized portions of chromatin extracts (20 µg) were loaded and analyzed by western blot. H3 (histone 3) staining was used as chromatin-bound marker. Data were reproduced in at least three independent experiments. Top middle panel was subjected to longer exposure times relative to other panels due to extremely low levels of VPS4 in the chromatin fraction. B Schematic illustration of the interplay between Loki ESCRTIII/VPS4 proteins. All three proteins can interact with one another. CHMP4-7 and CHMP1-3 associate with chromatin, while VPS4 does not. Co-expression of CHMP4-7 with either CHMP1-3 or VPS4, increase their association with chromatin (indicated by the larger font size), but does not affect CHMP4-7 levels in chromatin. Co-expression of CHMP4-7 and VPS4 results accumulation in cytosolic and nuclear foci (indicated by blue dots). C Naïve MDCK cells were fractionated, and normalized amounts of cytoplasm (Cyto) and chromatin (Ch) extracts (20 µg) were loaded and subjected to western blot analysis using specific antibodies for the indicated proteins (see Supplementary Table 1). Data were reproduced in at least two independent experiments. D Expression of human HA-CHMP1B alone (upper panner) or together with Emerald-VPS4 human, Loki or Heimdall proteins (second to fourth panels) in MDCK cells. Cells were stained with anti-HA antibodies and imaged using confocal microscopy. Representative single slice images are shown. Scale = 10 µM. Images were obtained from at least two independent experiments, n ≥ 30 for each condition. E Schematic illustration summarizing the interactions between human CHMP1B and different human and Asgard VPS4 homologs (see Supplementary Fig. 4A).
Few endogenous ESCRT-III proteins, particularly CHMP1A and CHMP1B, were also found to reside predominantly in the chromatin fraction (Fig. 3C). Moreover, overexpression of CHMP1B gave rise to discrete foci in the nucleus with human VPS4 recruited to these Foci (Fig. 3D, first and second panels, respectively). Strikingly, Loki VPS4, but not Heimdall VPS4, was also recruited to human CHMP1B nuclear foci upon co-expression (Fig. 3D, third and fourth panels). Expression of VPS4 homologs from additional Asgard phyla revealed that the ability of Asgard VPS4 proteins to interact with human CHMP1B in nuclear foci is conserved in most Asgard phyla (i.e., Loki, Odin, Hela, and Frey), but is lost in few (i.e., Heimdall and Thor) (Supplementary Fig. 4A, and illustrated in Fig. 3E). We, therefore, concluded that: (a) the chromatin binding capabilities of ESCRT-III are conserved in humans and Asgard archaea and (b) human CHMP1B is capable of recruiting Loki and most Asgard VPS4 homologs to nuclear foci, suggesting that human ESCRT-IIIs can interact with Asgard VPS4 homologs and that Loki VPS4 better represent the Asgard ESCRT family than its Heimdall homolog.
To elucidate the basis for Loki ESCRT-III/VPS4 chromatin association and recruitment, we performed mutation analysis (Fig. 4A). The MIT domain in VPS4 is known to mediate interaction with ESCRT-III in eukaryotes, and was found to be conserved in Asgard VPS4 homologs [23, 24, 31]. We, therefore, asked how depleting the MIT domain in VPS4 would affect the ability of CHMP4-7 to recruit VPS4 to chromatin and organize in nuclear foci. A fluorescently tagged Loki VPS4 -ΔΜΙΤ mutant was expressed in cells and exhibited similar subcellular distribution as full-length Loki VPS4 (Supplementary Fig. 4B, top panel). In chromatin fractionation assay, Loki VPS4-ΔΜΙΤ was still recruited to chromatin in the presence of CHMP4-7, suggesting that the MIT domain is dispensable for VPS4 recruitment to chromatin (Fig. 4B, left panel). In confocal images, however, VPS4-ΔΜΙΤ co-localized with CHMP4-7 in discrete foci outside the nucleus, but the percentage of cells exhibiting VPS4/CHMP4-7 puncta in the nucleus was considerably reduced (~6%) (Fig. 4C, top panel and 4D). Loki VPS4-ΔΜΙΤ was not recruited to CHMP1B nuclear foci upon their co-expression, indicating that Loki VPS4 interacts with human CHMP1B via its MIT domain (Supplementary Fig. 4C). These data suggest that the MIT domain in Loki VPS4 is conserved enough to interact with human ESCRT-III proteins and is essential for the induction of nuclear foci by Loki ESCRT-III/VPS4 proteins. Yet, additional regions in Loki VPS4 contribute to its interaction with Loki CHMP4-7, allowing it to interact with- and be recruited by- Loki CHMP4-7 in the absence of the MIT domain.
A Schematic illustration of VPS4 and CHMP4-7 mutants used in B–D. The indicated regions were either deleted or replaced, as shown. B Chromatin fractionation. MDCK cells transfected with the indicated fluorescently tagged ESCRT-III/VPS4 Loki mutants were fractionated, and normalized amounts of chromatin extracts (20 µg) were loaded and analyzed by western blot. Data were obtained from two independent experiments. C Representative single-slice images of cells transfected with fluorescently tagged versions of the indicated ESCRT-III/VPS4 mutants, imaged using confocal microscopy. Scale = 10 µM. Data were obtained from at least three independent experiments, n ≥ 60 for each condition. D Percentages of cells that exhibited co-localization on nuclear foci, cytosolic foci or no co-localization, upon co-transfection with the indicated plasmids (Apple-CHMP4-7 and Emerald-VPS4, n = 73, Apple-CHMP4-7 and Emerald-VPS4ΔMIT, n = 60, Apple-CHMP4-7 C’ mutant and Emerald-VPS4, n = 75, Apple-CHMP4-7 Δ20–50 and Emerald-VPS4, n = 73, Apple-CHMP4-7 Δ13 and Emerald-VPS4 n = 75. Data were obtained from at least three independent experiments. Statistical analysis of the foci in the nucleus was calculated using Chi- square test two tail (**p value ≤ 0.01, ****p value ≤ 0.0001). E–F MDCK cells were transfected with human HA-CHMP1B or with CHMP1B N’ mutant in which the homolog residues of Loki CHMP4-7 were deleted (CHMP1B Δ7-32). Cells were subjected to chromatin fractionation assay (E) or imaged using confocal microscopy (F). E Cells were fractionated, and normalized portions of chromatin extracts (20 µg) were analyzed by western blotting using HA antibodies. Data were reproduced in at least two independent experiments. F Representative single slice images of cells imaged by confocal microscopy. Scale = 10 µM. Images obtained from at least two independent experiments, n ≥ 40 for each condition. G Multiple sequence alignment of the N-terminal secondary structures of CHMP4-7 proteins in Asgard archaea as predicted by AlphaFold2. The Lokiarchaeon homolog used in this study (red rectangle) is in accordance with this majority, whilst the Heimdallarchaeon homolog (blue rectangle) seems to be an outlier. X (peach) = no predicted secondary structure; H (gray) = helix. Surface potential field (calculated as described in material and methods) and the resulting consensus motifs are shown below the sequence alignment.
To identify putative chromatin binding regions in ESCRT-III, we performed multiple sequence alignment between Loki CHMP4-7 and human CHMP1 isoforms (Supplementary Fig. 4D). This comparison highlighted conserved regions in the N and C terminals of the proteins. Introducing point mutations to C-terminal in CHMP4-7 did not affect its nuclear localization, its ability to associate with chromatin or its ability to recruit VPS4 to chromatin (Fig. 4A and B, second panel and Supplementary Fig. 4B, second panel). Consistently, CHMP4-7/VPS4 nuclear foci could be readily detected in cells co-expressing CHMP4-7 C’ mutants together with VPS4 (Fig. 4C, second panel and 4D, ~20% in mutant compared to ~30% in WT). Therefore, the C-terminal of CHMP4-7 is not involved in nuclear localization, nor in chromatin binding and in recruiting VPS4 to chromatin. Depleting the conserved N-terminal region (AAs 20–50) in CHMP4-7, however, led to a reduced association of CHMP4-7 with chromatin and a complete loss of CHMP4-7/VPS4 nuclear foci (Fig. 4A, 4B third panel, 4C third panel, and 4D), but did not affect the nuclear localization of CHMP4-7 (Supplementary Fig. 4B, third panel). These results indicate that the conserved N-terminal region in CHMP4-7 is specifically involved in its ability to associate with chromatin in cells. Depleting the parallel region in human CHMP1B also abolished its ability to associate with chromatin and to organize in discrete nuclear foci (Fig. 4, E and F), indicating that the conserved N terminal region identified here is involved in chromatin association in both human and Loki ESCRT-III proteins.
One of the unique phenotypes observed for Loki CHMP4-7 was its prominent nuclear localization, observed in both mammalian and yeast cells. The extreme N terminal of Loki CHMP4-7 is highly enriched in positively charged AAs (7 out of 13 AAs are Lysine or Arginine), suggesting its role as a nuclear targeting signal (NLS). While this region is not predicted as a classical NLS, structure-based bioinformatics analysis indicates a conserved unstructured positively charged region at the extreme N terminal of Asgard CHMP4-7 proteins, inferring that this region is functionally important (Fig. 4G). A CHMP4-7 mutant truncated of the first 13 AAs was distributed to both the nucleus and the cytosol in mammalian cells, suggesting that this region is involved in nuclear localization (Supplementary Fig. 4B, bottom panel). Despite the mild nuclear localization phenotype (averaged nuclear/cytosol ratio of 5:1 CHMP4-7 vs. 2.5:1 in CHMP4-7 Δ13), the CHMP4-7 Δ13 mutant exhibited the lowest chromatin association observed in our mutant analysis and failed to induce nuclear foci formation when co-expressed with VPS4 (Figs. 4A, 4B right panel, 4C bottom panel, 4D, and Supplementary Fig. 4E). Co-localization in cytosolic foci was rare but could be observed (detected in ~7% of the cells), suggesting that CHMP4-7/VPS4 interactions were not completely abolished. Hence, the first 13 AAs in Loki CHMP4-7 are involved in nuclear localization but, more significantly, are crucial for chromatin association and nuclear foci formation.
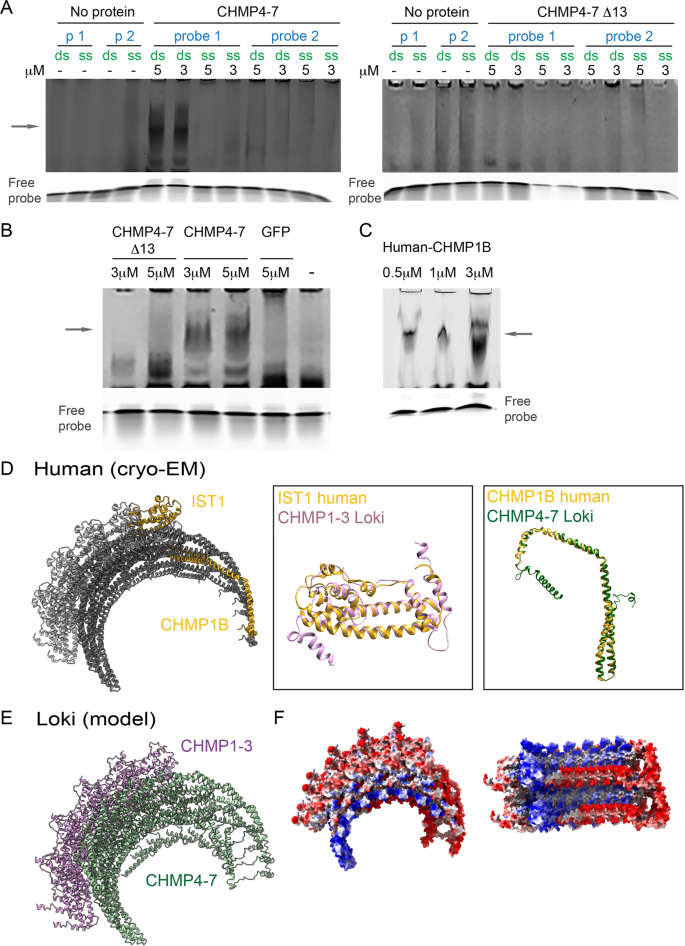
As our data indicated a specific association of Asgard and human ESCRT-IIIs with chromatin, we set to examine their ability to bind DNA directly. To this end, we expressed and purified full-length and Δ13 Loki CHMP4-7 proteins from E. coli and performed a gel shift assay using 5′ Cy5-tagged double-strand or single strand nucleotide DNA probes at different lengths (40 and 22 bases long. Using this assay, Loki CHMP4-7 was found to bind only the 40 bases long probe and only in the double strand form, as indicated by the presence of fluorescently labeled higher MW bands (Fig. 5A, left panel and 5B). No gel shift was observed, upon repeating the experiment with the CHMP4-7 Δ13 mutant, confirming a role for the N terminal region in DNA binding (Fig. 5A, right panel and 5B). Human CHMP1B was recently suggested to bind nucleic acids, in vitro [36]. Consistently, using our assay conditions, CHMP1B was found to bind DNA, albeit at lower protein concentrations than those calibrated for Loki CHMP4-7 (Fig. 5C). Therefore, Asgard and human ESCRT-IIIs directly bind DNA, in vitro, suggesting that the chromatin association observed for ESCRT-III in cells is mediated by their direct binding to DNA.
A–C Electrophoretic mobility shift assay (EMSA) performed in the presence of Cy5-5’ end labeled ssDNA or dsDNA probes (2.5 µM, probe 1, 40 bases; probe 2, 22 bases, see sequences in method section) and the purified proteins: Loki CHMP4-7, Loki CHMP4-7 Δ13, GFP (negative control) and, human CHMP1B at the indicated concentrations. Arrows in upper panel indicate complex formation. Lower panel, free DNA probe as seen by shorter exposure time. Data were reproduced in at least three independent experiments. D–F Predicted structural organization of Loki ESCRT-III filament. D Left: Sectioned representation of the cryo-EM human IST1 and CHMP1B co-polymer structure (PDB ID 3JC1). Middle to right panels: overlay of the predicted structure of Loki CHMP1-3 (Robbeta) on the human IST1 cryo-EM monomer structure, and the predicted Loki CHMP4-7 structure (Robbeta) on human CHMP1B cryo-EM monomer structure, respectively. E The predicted Loki CHMP1-3 and CHMP4-7 structures (Robetta) were overlaid on the co-polymer cryo-EM structure of human IST1 and CHMP1B, respectively, to generate a theoretical 3D structural organization model of Loki ESCRT-III proteins. Shown are Loki CHMP1-3 (purple) and Loki CHMP4-7 (green). F Electrostatic map of the theoretical Loki ESCRT-III co-polymer. Positive charge, blue; Negative charge, red; Neutral, white. Left panel, side view; Right panel axial view, showing the tube from the inside. Note, the accumulation of positively charged amino acids in the inner side of the tube which may infer affinity to negatively charged DNA.
Discussion
We show that ESCRT-III proteins of Asgard and human lineages exhibit chromatin/DNA binding properties and provide evidence for the involvement of conserved protein–protein interactions in this process. Based on these findings we propose that, in addition to their canonical role in membrane remodeling, a conserved role for ESCRT proteins involving DNA binding should also be considered.
Our study was based on the heterogeneous expression of predicted coding sequences from Asgard archaea in mammalian cells. We found that genes encoding Asgard ESCRT-III and VPS4 homologs are expressed in several cell lines and exhibit similar subcellular distributions, which also resemble their localization when co-expressed in the unicellular eukaryote S. cerevisiae. Moreover, Loki ESCRT-III/VPS4 proteins were found to interact with one another inside mammalian cells. These results indicate that Asgard ESCRT proteins can be studied inside mammalian cells and suggest that heterologous expression of Asgard genes in mammalian cells can be used as a tool to study the biochemical and cellular properties of ESPs encoded by Asgard archaea.
The low sequence identity values observed between predicted ESCRT-III proteins from different Asgard species (Fig. 1A) motivated us to sample ESCRT-III/VPS4 modules taken from two different species. As eukaryotic ESCRTs rely on an intimate protein-protein interactions network to mediate their function, we specifically chose coding sequences obtained from the same MAG. Indeed, we found that ESCRT-III/VPS4 Loki and Heimdall homologs exhibited different cellular properties and did not interact with one another inside cells (Fig. 2C and Supplementary Fig. 3C). While based on sequence homology, Heimdall archaea were considered closer to humans [19, 20], our data suggest that Loki ESCRTs are functionally closer to their human homologs and better represent the Asgard superphyla. First, Loki ESCRT-III/VPS4 complexes were found to interact with one another inside mammalian cells while no interactions were detected for Heimdall ESCRT-III/VPS4 complexes. Second, Loki ESCRT-III and VPS4 organized in nuclear foci, similar to their human homologs (CHMP1B and VPS4), while Heimdall ESCRT-III/VPS4 did not. Third, Loki VPS4, and most Asgard VPS4 homologs examined, were recruited to CHMP1B induced nuclear foci, while the Heimdall VPS4 homolog failed to do so. Interestingly, while the predicted structure of Loki CHMP4-7 highly resembled the overall fold of the human ESCRT-III open conformation, Heimdall CHMP4-7 had a slightly different fold, exhibiting shorter helixes and an additional C-terminal helix (Fig. 1C and Supplementary Fig. 1E). Moreover, the unstructured positively-charged N terminal region that was identified in Asgard CHMP4-7 was not conserved in the Heimdall homolog (Fig. 4G). It is, therefore, possible that the basis for the conservation of function in the ESCRT kingdom lies in their structural similarity more than in their sequence identity.
The predictions obtained by the deep-learning-based servers AlphaFold and Robetta highlighted conserved structural features of ESCRT-III proteins. Traditionally, ESCRT-III proteins were shown to obtain an autoinhibitory loop that opens upon activation and allows polymerization of the ESCRT-III filament [3, 35]. The inactive and active conformations were termed “closed” and “open”, respectively. Among the twelve human ESCRT-III proteins, ten were predicted to have an open conformation and two a closed conformation. Interestingly, in both Asgard species used in this study, one ESCRT-III protein was predicted to be in an open conformation and one in a closed conformation, suggesting that both conformations are conserved in Asgard ESCRT-IIIs (Fig. 1C and Supplementary Fig. 1). Expanding this analysis to 160 Asgard ESCRT-III sequences from our phylogenetic tree further supported this notion (Fig. 1D). Notably, the structure of eukaryotic CHMP1B and IST1 co-polymer is composed of an ESCRT-III pair exhibiting open and closed conformations [37, 38], raising the possibility that both conformations are needed for building a functional ESCRT filament. To test this notion, we overlaid the deep-learning-based structures we generated for Loki CHMP1-3 and CHMP4-7 on the solved cryo-EM structure of the CHMP1B-IST1 co-polymer (PDB ID 3JC1, Fig. 5D) [38]. Specifically, we overlaid Loki CHMP1-3 on human IST1, both in the closed conformation, and Loki CHMP4-7 with human CHMP1B, both in the open conformation. Despite extremely low sequence identity values (20.74% between Loki CHMP1-3 and IST1 and 17.52% between Loki CHMP4-7 and CHMP1B), a very good fit was observed between the Loki and human proteins, which enabled us to generate a putative structural model for Loki ESCRT-III filaments (Fig. 5E). Mapping the electrostatic surface of the modeled Loki ESCRT-III filament revealed a patch of positively charged amino acids that faces its inner surface (Fig. 5F). This positive patch of amino acids is part of the N-terminal helix of CHMP4-7, which we found to govern CHMP4-7-chromatin association. Moreover, this region is equivalent to the N-terminal helix in CHMP1B, which includes a patch of positively charged amino acids that were suggested to drive its interactions with nucleic acids, in vitro [36]. It may be that, like their human homologs, Loki ESCRT-III organize into helical filament and are able to bind DNA through electrostatic interactions with positive amino acids that reside in the inner side of the filament. However, this notion currently relies on theoretical models, and further experimental work is required to examine whether the chromatin binding properties of ESCRT-IIIs rely on their organization into high-ordered filaments.
Our findings that ESCRT-III proteins associate with chromatin are in line with previous findings in mammalian cells. In 2001 Stauffer and colleagues reported that CHMP1 concentrates at chromatin-condensed regions and protects them from nuclease degradation [8]. More recently, a truncated version of CHMP7 was shown to accumulate in discrete foci in the nucleus and to recruit CHMP4B to these foci [39], and a cryo-EM structure of IST1 and CHMP1B filament was solved in the presence of nucleic acids, showing direct interactions between the ESCRT-III filament and nucleic acids [38]. Our findings that these properties are shared in both Asgard and mammalian ESCRTs and are mediated by the same domain, conserved in both human and Loki homologs, strongly suggest that the ability of ESCRT-III proteins to bind DNA is physiologically relevant.
Our data point out the involvement of chromatin binding in the function of ancient ESCRTs. ESCRTs have been shown to be involved in cell division in both TACK archaea and in eukaryotes [1, 10]. In TACK archaea, CdvA and the ESCRT-III-VPS4 homologs CdvB and CdvC have been designated as the cell division machinery [14, 16, 21], and CdvA was found to assemble into filaments in the presence of DNA [40]. Given the established role of ESCRT in membrane remodeling [4, 41], it is possible that ESCRTs originally evolved as basic machinery for driving cell division by coupling DNA segregation and membrane constriction.
An alternative explanation to the observed data is that ESCRTs may have a conserved role in genome stability and activity. Eukaryotic ESCRT-IIIs have been documented to be involved in genome retrotransposition via the LINE complex, and to localize to micronuclei [7, 39, 42]. In addition, CHMP1B was shown to reside in chromatin-condensed regions and protect them from nuclease degradation [8], and CHMP2A (also called BC-2) was shown to accumulate in discrete regions in the nucleus where it co-localized with the histone3 phosphorylation marker PH3 and with the transcriptional silencing protein BM1 [6]. Lastly, ESCRTs have been identified as nuclear matrix and chromatin binding proteins in several large-scale proteomic screens [42,43,44]. The archaeal DNA is wrapped in a hypernuclesome structure that is stabilized by archaeal histones. Interestingly, based on sequence similarity, some eukaryotic histones (H3–H4) have been suggested to emerge from archaeal histones [45]. Histone genes were also found to be encoded in Asgard archaea, at different abundancies, with some phyla encoding no/few histone homologs and others, i.e., Heimdall, encoding ten different histones [45]. The reported findings in mammalian cells, the similarity of archaeal and eukaryotic histones, and the data reported here raise the possibility that ESCRTs may have a role in genome organization and regulation in both Asgard archaea and eukaryotes.
Our findings that Loki CHMP4-7 reside in the nucleus when expressed in mammalian and yeast cells strongly suggest that Asgard ESCRT-IIIs are actively targeted to the nuclear membrane. Although no NLS was previously described for ESCRT-III proteins, most mammalian ESCRT-IIIs were found to distribute between the cytosol and nucleus [34, 35] and a nuclear export signal (NES) was recently identified in the C’ of the mammalian ESCRT-III protein CHMP7 [39]. Mutating the homologous region in Loki CHMP4-7 (Fig. 4, CHMP4-7 C’ mut) did not affect its nuclear localization, suggesting that this region does not function as an NES in Loki ESCRT-III. Nonetheless, we identified a conserved region at the extreme N terminal of Asgard CHMP4-7, which partially mediates nuclear localization in Loki CHMP4-7. Therefore, both mammalian and Asgard ESCRT-IIIs encode for specific signals that direct their localization to the nucleus, suggesting an active interplay between the ESCRT complex and the nucleus. In this respect, Loki and Heimdall archaea enriched from sediments in Arhus Bay were recently reported to have a condensed DNA organization [46]. Although a membrane engulfing these condensed DNA structures has not been shown yet, it is possible that the ability of Asgard ESCRTs to enter the nuclear membrane of eukaryotes, reported here, represents their physiological properties in Asgard archaea and that the evolutionarily conserved role of ESCRTs involves genome stability and protection.
Material and methods
Phylogenic analysis
Multiple sequence alignments of ESCRT-III and VPS4 protein sequences were produced using MAFFT (version v7.464) [47]. An approximately maximum likelihood tree was created using the JTT (Jones–Taylor–Thornton) + CAT model in FastTree (version 2.1.10) and visualized using iTOL [48, 49]. Separate heatmaps were then created for these ESCRT-III and VPS4 protein sequences according to their clustering in the tree. The matrix of pairwise distances from these aligned sequences was calculated by using dissimilarity (Fitch matrix), seqinr package (in “R”) and visualized with the “heatmap.2” package in “R” ([50,51,52]).
3D structure prediction
Amino acid sequences of all human ESCRT-III proteins the Asgard ESCRT-III proteins from Loki GC14_75 (KKK42122.1, KKK44605.1, KKK42121.1) and Heimdall AB_125 (OLS31933.1, OLS31932.1, OLS31934.1) were subjected to structure prediction using the Robetta server using the RoseTTAFold [33] and using AlphaFold [32] via ChimeraX software. Overlay of predicted structures from both servers and previously solved structures of human ESCRT-III proteins available in the PDB (Supplementary Fig. 1, A–D) show a relatively high agreement increasing the confidence of the predicted structures. A relatively high confidence score was given to the first 100 AAs in the sequence representing the two core helixes, H1 and H2. Structure predictions were additionally generated for 77 Asgard CHMP4-7 and 83 Asgard CHMP1-3 homologs using the Robetta server. Then, the averaged distance of each amino acid against all amino acids in the predicted structure was calculated by measuring the distances between the beta carbons in each AAs (or alpha carbons in Glycine) (proDy python package) [53] and was used as an indication for the compactness of the predicted structure. Smaller averaged distances indicate a more compact structure and were therefore referred to as the closed conformation (Fig. 1D).
Sequence variety among the N-terminal regions in Asgard ESCRT-III Chmp 4–7 proteins, was examined for 77 sequences from our CHMP4-7 cluster (Fig. 4G). Sequencing bias, was avoided by clustering sequences with 70% identity using cd-hit [54], and selecting one random sequence per cluster or the Loki and Heimdall sequences used in this work (for clusters containing these sequences). This resulted in a reduced set of 33 sequences. Then multiple sequence alignment was calculated in mafft (BLOSUM45) [47] and the predicted structures of these sequences was generated in AlphaFold2 version 2.2.0. Finally, secondary structure was extracted from the resulting PDB files, using STRIDE [55], and was aligned with the MSA using python script, (Fig. 4G, top panel). The sequence logo below the MSA was calculated using Jalview [56]. Surface potential field of the predicted protein structures was calculated in protein-sol patches [57]. A python script was used to average the potential over all atoms per amino acid for each protein, providing one potential value per amino acid for each protein.
A model for Loki ESCRT-III filament was generated in Coot [58] by fitting the structure predictions obtained in Robetta for Loki CHMP1-3 and CHMP4-7 on the solved cryo-EM structure of the human IST1 and CHMP1B co-polymer (PDB ID: 3JC1) (Fig. 5D–F). The helices from the modeled structures were manually overlapped on the relevant 3JC1 helices, followed by geometry regularization of the junctions between the helices.
Cell culture and transfection
MDCK cells were grown in MEM supplemented with 5% fetal bovine serum (FBS), 2 mM glutamine, 10,000 U/ml penicillin, and 10 mg/ml streptomycin. NIH3T3 and HeLa cells were grown in DMEM supplemented with 10% FBS supplemented with 2 mM glutamine, 10,000 U/ml penicillin, and 10 mg/ml streptomycin. Transfection was carried out using Lipofectamine 2000 (Life Technologies, Carlbad, CA) according to the manufacturer’s guidelines.
Plasmid constructs
The MAG of Loki sp. GC14_75 and Heimdall archaeon AB_125, each contained open reading frames (ORFs) for CHMP1-3, CHMP4-7, and VPS4 genes; were used for our study. ORFs obtained by the NCBI accession numbers KKK42122.1, KKK44605.1, KKK42121.1, OLS31932.1, OLS31933.1, OLS31934.1, respectively were amplified by PCR and cloned to either Emerald-C1 or Apple-C1 vector (Clontech). Loki CHMP4-7 Δ20-50: AAs 20-50 were removed from Apple-CHMP4-7 by overlapping PCR. Loki CHMP4-7 Δ13: AAs 1-13 were removed from Apple-CHMP4-7 by overlapping PCR. Loki CHMP4-7 C’ mut: Apple-CHMP4-7 was mutated using overlapping PCR as follows: V205 codon GTT was replaced by GCT; I209 codon ATC was replaced by GCT; and L212 codon CTT was replaced by GCT. Loki VPS4 ΔMIT: The predicted MIT domain (AAs 1-82) were deleted from Emerald -VPS4 by overlapping PCR. ORFs of VPS4 proteins from Thorarchaeota (Thor), Odinarchaeota (Odin), Helarchaeota (Hela), and Freyarcheota (Frey) obtained by the NCBI accession numbers TFG35021.1, OLS18192.1, NVM30765.1, MBS7247407.1 respectively, were amplified by PCR and cloned to Emerald-C1. HA- CHMP1B plasmid was a kind gift from Craig Blackstone (NIDDK, NIH). Human CHMP1B Δ7-32 - AAs 7-32 were removed from HA-CHMP1B plasmid by overlapping PCR.
All plasmids were verified by sequencing.
Integration of VPS4 and CHMP4-7 in yeast
Yeast strains were generated in the background of W1588 MATa S. cerevisiae. Envy-VPS4 and yomKate2-CHMP4-7 fusion genes were cloned via Gibson assembly and expressed under a constitutive promoter in URA3 and ADE1 loci respectively. For microscope experiments, yeast strains were incubated overnight in synthetic complete media with 4% glucose (SC Glc 4%). Next day, the cell cultures were diluted at O.D. 0.1–0.2 in warm SC Glc 4% and were incubated for 2–3 h. 400 μl of cell solutions were transferred into eight-well Ibidi slides (cat #80826) and yeast cells were imaged using a confocal spinning-disk microscope.
Cellular imaging
Immunostaining
Cells were permeabilized with 0.5% Triton X-100 for 10 min and blocked with 10% FBS for 15 min. HA-CHMP1B transfected cells were stained with rabbit polyclonal anti-HA antibodies and subjected to a secondary antibody staining using Alexa Fluor 594 anti-rabbit (1:1000, Life Technologies). All cells were stained with Hoechst 33342. Finally, cells were mounted with Fluoromount-G (SouthernBiotech, Birmingham, AL).
Microscopy
Cells were plated at low density on #1.0 (Menzel, Braunschweig, Germany) or high-resolution #1.5 coverslips (Marienfeld, Lauda-Konigshfen, Germany). In the following day, cells were transfected with the indicated plasmids, fixed 24 h later using 4% paraformaldehyde and stained with Hoechst. Cells transfected with HA-CHMP1B were further subjected to immunostaining as described above. Low expressing cells were then imaged in 3D using confocal spinning-disk microscope (Marianas; Intelligent Imaging, Denver, CO) with a 63× oil objective (numerical aperture, 1.4) and an electron-multiplying charge-coupled device camera (pixel size, 0.079 μM; Evolve; Photometrics, Tucson, AZ). Images were processed using SlideBook version 6 (Intelligent Imaging). 3D SIM imaging was performed using the ELYRA PS.1 microscope (Carl Zeiss MicroImaging). Thin z-sections (0.11–0.15 μM) of high-resolution images were collected in three rotations and five phases for each channel. Image reconstruction and processing were performed in ZEN (Carl Zeiss MicroImaging).
Statistical analysis
Statistical analysis was performed using Graph Pad Prism version 9.00 for Windows (La Jolla, CA, USA). Comparison between two groups was assessed by chi-square test. For our analysis, p value < 0.05 was considered statistically significant.
Biochemical analysis
Sedimentation assay
Assay was performed according to Soomin Shim et al. [35]. Briefly, transfected MDCK cells, were harvested 24 h post transfection, washed with PBS, and solubilized in lysis buffer on ice for 40 minutes. Supernatant and pellet fractions were separated by centrifuging samples at 10,000 g for 15 min at 4 °C. Pellets were resuspended in lysis buffer to the same volume as supernatant, and equal volumes of fractions were analyzed by western blot.
Chromatin fractionation
Chromatin was extracted using a chromatin extraction kit (Thermo Scientific, cat# 78840), according to manufacturer’s instructions. Chromatin-bound nuclear extract fractions were transferred to a new vial and total protein concentrations were measured using BCA Protein Assay Kit (Pierce Biotechnology, Rockford, IL). Equal total protein amounts (20 µg) were loaded in each lane. H3 antibodies (histone 3) were used as chromatin-bound marker.
Immunoprecipitation
Immunoprecipitation was perform using GFP-Trap Agarose kit (ChromoTek) according to the manufacturer’s instructions with the following modifications: transfected MDCKS cells were harvested 24 h post transfection, washed with PBS and lysed in lysis buffer [10 mM Tris/Cl pH 7.5, 100 mM NaCl, 0.5 mM EDTA, 0.5% NP40, 1 mM PMSF, a complete protease inhibitor (Roche Diagnostics, Alameda, CA, USA)] on ice for 30 min. Cell lysates were then centrifuged (20,000 g for 10 min, 4 °C) and supernatants were diluted in dilution buffer [10 mM Tris/Cl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 1 mM PMSF, complete protease] and incubated for 1 h with GFP- Trap-A beads. Following incubation, lysates were centrifuged at 2500 g for 2 min at 4 °C to separate between the unbound (Ub) and the bound (B) fractions. Samples were loaded on SDS-PAGE gel and analyzed by Western blot.
Western blot
MDCK Cells were lysed as described above or using RIPA lysis buffer [150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris (pH 8.0)] supplemented with complete protease inhibitor (Roche Diagnostics, Mannheim, Germany) for 30 min at 4 °C. Total protein concentrations were measured using the BCA Protein Assay Kit (Pierce Biotechnology, Rockford, IL), and equal total protein amounts were loaded in each lane. Membranes were stained with primary antibodies for endogenous proteins or with mouse anti-GFP antibodies (for Emerald-tagged proteins detection) or rabbit anti-mCherry antibodies (for Apple-tagged proteins detection) for 16 h at 4 °C followed by anti-rabbit or anti mouse-peroxidase secondary antibodies for 1 h (1:10,000; Jackson ImmunoResearch, West Grove, PA). For complete antibodies list, see Supplementary Material.
Electrophoretic mobility shift assay (EMSA)
EMSA assay was performed according to Adrian Zander et al. with few modifications [59]. Briefly 5′ Cys- labeled oligonucleotides (dsDNA or ssDNA) ATCCACCTGTACATCAACTCGCCCGGCGGCTCGATCAGCG (40 bases probe) or ATTCGATCG GGGCGGGGCGAGC (22 bases probe), 2.5 µM each, were incubated with purified GFP, CHMP1B, CHMP4-7 or CHMP4-7 Δ13 at different molar ratios (see the corresponding figure legends) for 20 min at 65 °C in 1x NaGT buffer [50 mM Tris/HCl, pH7.4, 100 mM NaCl, 5 mM MgCl2, 2% glycerol, 10 mM Dithiothreitol, 67 µg/ml BSA]. Reactions (final volume 20 μL) were resolved by electrophoresis at 4 °C through native gel containing 5% polyacrylamide (37.5:1acrylamide: bisacrylamide) in 0.5x TBE [45 mM Tris-Borate, 1 mM EDTA] and 2.5% glycerol, as described in Yi-Wen Hsieh et al. [60]. Fluorescently labeled oligonucleotides and protein–nucleic acid complexes were imaged at the appropriate wavelength using a Typhoon FLA 7000 biomolecular imager (GE Healthcare, PA, USA) to reveal specific Fl-dye labeling.
Protein expression and purification
Escherichia coli BL21, transformed with plasmids expressing either His-MBP-TEV CHMP4-7, His-MBP-TEV CHMP4-7 Δ13, or His-MBP-TEV CHMP1B, were grown LB medium. Bacteria were harvested by centrifugation after IPTG induction and lysed in high salt buffer by sonication. Proteins were purified using Ni–NTA beads (Thermofisher) followed by dialysis in the presence of TEV protease. Protein solutions were then subjected to a second Ni–NTA purification round and flow-through was collected. CHMP4-7 and CHMP4-7 Δ13 proteins were kept in 50 mM Tris pH = 7.4, 10% glycerol, 500 mM NaCl, 15 mM 2-mercaptoethanol. For CHMP1B, flowthrough was loaded on SCE column using ÄKTA gel filtration system (20 mM Tris pH = 7.4 150 mM NaCl). GFP was expressed and purified from E. coli, using standard protocols by the protein purification unit at the Weizmann Institute (Rehovot, Israel). Fresh protein preparations were used in gel shift assays.
Illustrations were generated in Biorender.
Data availability
All raw data that support the findings of this study are available upon request from the corresponding author NE.
References
Caspi Y, Dekker C. Dividing the archaeal way: the ancient cdv cell-division machinery. Front Microbiol. 2018;9:174.
Alonso YAM, Migliano SM, Teis D. ESCRT-III and Vps4: a dynamic multipurpose tool for membrane budding and scission. FEBS J. 2016;283:3288–302.
McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem. 2013;82:663–92.
Hurley JH. ESCRTs are everywhere. EMBO J. 2015;34:2398–407.
Gatta AT, Carlton JG. The ESCRT-machinery: closing holes and expanding roles. Curr Opin Cell Biol. 2019;59:121–32.
Hodges E, Redelius JS, Wu W, Hoog C. Accelerated discovery of novel protein function in cultured human cells. Mol Cell Proteom. 2005;4:1319–27.
Sagona AP, Nezis IP, Stenmark H. Association of CHMP4B and autophagy with micronuclei: implications for cataract formation. Biomed Res Int. 2014;2014:974393.
Stauffer DR, Howard TL, Nyun T, Hollenberg SM. CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. J Cell Sci. 2001;114:2383–93.
Stuffers S, Brech A, Stenmark H. ESCRT proteins in physiology and disease. Exp Cell Res. 2009;315:1619–26.
Tarrason Risa G, Hurtig F, Bray S, Hafner AE, Harker-Kirschneck L, Faull P, et al. The proteasome controls ESCRT-III-mediated cell division in an archaeon. Science. 2020;369:eaaz2532.
Junglas B, Huber ST, Heidler T, Schlosser L, Mann D, Hennig R, et al. PspA adopts an ESCRT-III-like fold and remodels bacterial membranes. Cell. 2021;184:3674–88. e3618
Liu J, Tassinari M, Souza DP, Naskar S, Noel JK, Bohuszewicz O, et al. Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. Cell. 2021a;184:3660–73. e3618
Deatherage BL, Cookson BT. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect Immun. 2012;80:1948–57.
Lindas AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci USA. 2008;105:18942–6.
Mantel PY, Marti M. The role of extracellular vesicles in Plasmodium and other protozoan parasites. Cell Microbiol. 2014;16:344–54.
Samson RY, Obita T, Hodgson B, Shaw MK, Chong PL, Williams RL, et al. Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol Cell. 2011;41:186–96.
Yagisawa F, Fujiwara T, Takemura T, Kobayashi Y, Sumiya N, Miyagishima SY, et al. ESCRT machinery mediates cytokinetic abscission in the unicellular red alga Cyanidioschyzon merolae. Front Cell Dev Biol. 2020;8:169.
MacLeod F, Kindler GS, Wong HL, Chen R, Burns BP. Asgard archaea: diversity, function, and evolutionary implications in a range of microbiomes. AIMS Microbiol. 2019;5:48–61.
Spang A, Saw JH, Jorgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521:173–9.
Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Backstrom D, Juzokaite L, Vancaester E, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–8.
Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science 2008;322:1710–3.
Hatano T, Palani S, Papatziamou D, Salzer R, Souza DP, Tamarit D, et al. Asgard archaea shed light on the evolutionary origins of the eukaryotic ubiquitin-ESCRT machinery. Nat Commun. 2022;13:3398.
Frohn BP, Hartel T, Cox J, Schwille P. Tracing back variations in archaeal ESCRT-based cell division to protein domain architectures. PLoS One. 2022;17:e0266395.
Lu Z, Fu T, Li T, Liu Y, Zhang S, Li J, et al. Coevolution of eukaryote-like Vps4 and ESCRT-III subunits in the Asgard Archaea. mBio. 2020;11:e00417–20.
Liu Y, Makarova KS, Huang WC, Wolf YI, Nikolskaya AN, Zhang X, et al. Expanded diversity of Asgard archaea and their relationships with eukaryotes. Nature 2021b;593:553–7.
Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, et al. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 2020;577:519–25.
Henne WM, Buchkovich NJ, Zhao Y, Emr SD. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell. 2012;151:356–71.
Schoneberg J, Pavlin MR, Yan S, Righini M, Lee IH, Carlson LA, et al. ATP-dependent force generation and membrane scission by ESCRT-III and Vps4. Science 2018;362:1423–8.
Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature 2009;458:172–7.
de Franceschi N, Alqabandi M, Weissenhorn W, Bassereau P. Dynamic and sequential protein reconstitution on negatively curved membranes by giant vesicles fusion. Bio Protoc. 2019;9:e3294.
Caillat C, Maity S, Miguet N, Roos WH, Weissenhorn W. The role of VPS4 in ESCRT-III polymer remodeling. Biochem Soc Trans. 2019;47:441–8.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021;596:583–9.
Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021;373:871–6.
Adar-Levor S, Goliand I, Elbaum M, Elia N. Studying the spatial organization of ESCRTs in cytokinetic abscission using the high-resolution imaging techniques SIM and Cryo-SXT. Methods Mol Biol. 2019;1998:129–48.
Shim S, Kimpler LA, Hanson PI. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–79.
Talledge NMJ, Wenzel D, Nguyen H, Lalonde MBM, Skalicky J, Frost A, et al. The ESCRT-III proteins IST1 and CHMP1B assemble around nucleic acids. bioRxiv, 2018.
Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, et al. Helical structures of ESCRT-III are disassembled by VPS4. Science 2008;321:1354–7.
McCullough J, Clippinger AK, Talledge N, Skowyra ML, Saunders MG, Naismith TV, et al. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science 2015;350:1548–51.
Vietri M, Schultz SW, Bellanger A, Jones CM, Petersen LI, Raiborg C, et al. Unrestrained ESCRT-III drives micronuclear catastrophe and chromosome fragmentation. Nat Cell Biol. 2020;22:856–67.
Moriscot C, Gribaldo S, Jault JM, Krupovic M, Arnaud J, Jamin M, et al. Crenarchaeal CdvA forms double-helical filaments containing DNA and interacts with ESCRT-III-like CdvB. PLoS One. 2011;6:e21921.
Elia N, Ott C, Lippincott-Schwartz J. Incisive imaging and computation for cellular mysteries: lessons from abscission. Cell. 2013;155:1220–31.
Horn AV, Celic I, Dong C, Martirosyan I, Han JS. A conserved role for the ESCRT membrane budding complex in LINE retrotransposition. PLoS Genet. 2017;13:e1006837.
Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–9.
Schweppe DK, Huttlin EL, Harper JW, Gygi SP. BioPlex display: an interactive suite for large-scale AP-MS protein-protein interaction data. J Proteome Res. 2018;17:722–6.
Henneman B, van Emmerik C, van Ingen H, Dame RT. Structure and function of archaeal histones. PLoS Genet. 2018;14:e1007582.
Avci B, Brandt J, Nachmias D, Elia N, Albertsen M, Ettema TJG, et al. Spatial separation of ribosomes and DNA in Asgard archaeal cells. ISME J. 2022;16:606–10.
Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66.
Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296.
Price MN, Dehal PS, Arkin AP. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490.
Fitch WM. An improved method of testing for evolutionary homology. J Mol Biol. 1966;16:9–16.
Charif D, Lobry JR. SeqinR 1.0-2: a contributed package to the r project for statistical computing devoted to biological sequences retrieval and analysis. Biological and Medical Physics, Biomedical Engineering Springer, Berlin, Heidelberg, 2007.
G. Warnes BB, L Bonebakker, R Gentleman, W Huber, A Liaw, T Lumley, et al. g plots: various R programming tools for plotting data. R Package Vers. 2015.
Bakan A, Meireles LM, Bahar I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics 2011;27:1575–7.
Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006;22:1658–9.
Heinig M, Frishman D. STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004;32:W500–502.
Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009;25:1189–91.
Hebditch M, Warwicker J. Web-based display of protein surface and pH-dependent properties for assessing the developability of biotherapeutics. Sci Rep. 2019;9:1969.
Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501.
Zander A, Holzmeister P, Klose D, Tinnefeld P, Grohmann D. Single-molecule FRET supports the two-state model of Argonaute action. RNA Biol. 2014;11:45–56.
Hsieh YW, Alqadah A, Chuang CF. An optimized protocol for electrophoretic mobility shift assay using infrared fluorescent dye-labeled oligonucleotides. J Vis Exp. 2016;29:54863.
Acknowledgements
We thank Dan Levi (BGU) and Eran Meshorer (HUJI) for fruitful discussions on chromatin-binding proteins and constructive advice. We thank Chen Keasar (BGU) for assistance with structure prediction measurements. We also thank, Dr. Ran Zalk (BGU) for helpful feedback and technical help with structural analysis. We thank all members of the Elia lab for critical feedback throughout the project. The Elia laboratory is funded by the Israeli Science Foundation (ISF) Grant no. 1323/18. I. M. acknowledges support from the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant no. 640384) and the Israel Science Foundation (grant no. 1947/19).
Author information
Authors and Affiliations
Contributions
DN, IM, and NE conceptualized the project. DN designed, performed, and analyzed all cell biology and biochemical experiments. NM purified recombinant proteins. AZ performed phylogenetic analysis and generated density plot. MS performed experiments. MS ad RY performed and analyzed structure predictions. YD assisted in experiments. IT and AA assisted with experiments in yeast. BF and PS performed bioinformatics analysis of Asgard CHMP4-7 N-terminal regions. RZ generated structural model and helped with structural data interpretation. NE wrote the manuscript with the help of DN and IM. All authors read and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nachmias, D., Melnikov, N., Zorea, A. et al. Asgard ESCRT-III and VPS4 reveal conserved chromatin binding properties of the ESCRT machinery. ISME J 17, 117–129 (2023). https://doi.org/10.1038/s41396-022-01328-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-022-01328-2