Abstract
Coastal upwelling zones are hotspots of oceanic productivity, driven by phytoplankton photosynthesis. Bacteria, in turn, grow on and are the principal remineralizers of dissolved organic matter (DOM) produced in aquatic ecosystems. However, the molecular processes that key bacterial taxa employ to regulate the turnover of phytoplankton-derived DOM are not well understood. We therefore carried out comparative time-series metatranscriptome analyses of bacterioplankton in the Northwest Iberian upwelling system, using parallel sampling of seawater and mesocosms with in situ-like conditions. The mesocosm experiment uncovered a taxon-specific progression of transcriptional responses from bloom development (characterized by a diverse set of taxa in the orders Cellvibrionales, Rhodobacterales, and Pelagibacterales), over early decay (mainly taxa in the Alteromonadales and Flavobacteriales), to senescence phases (Flavobacteriales and Saprospirales taxa). Pronounced order-specific differences in the transcription of glycoside hydrolases, peptidases, and transporters were found, supporting that functional resource partitioning is dynamically structured by temporal changes in available DOM. In addition, comparative analysis of mesocosm and field samples revealed a high degree of metabolic plasticity in the degradation and uptake of carbohydrates and nitrogen-rich compounds, suggesting these gene systems critically contribute to modulating the stoichiometry of the labile DOM pool. Our findings suggest that cascades of transcriptional responses in gene systems for the utilization of organic matter and nutrients largely shape the fate of organic matter on the time scales typical of upwelling-driven phytoplankton blooms.
Similar content being viewed by others
Introduction
Wind-induced upwelling of nutrient-rich subsurface water triggers pronounced phytoplankton blooms along eastern boundary coastal zones. Despite their small surface area (~7% of global ocean), coastal seas contribute 14-30% of the global oceanic primary production [1]. Phytoplankton produce a diverse mixture of organic compounds by photosynthesis [2,3,4], which is rapidly processed by prokaryotes into biomass and CO2 [5]. Typically, substrate availability and interactions among microorganisms determine spatiotemporal differences in microbial community composition and function [6,7,8,9,10,11]. However, compared to the temporal dynamics in bacterial community composition [12, 13], little is known about the microbially mediated processes involved in the degradation and uptake of phytoplankton-derived organic matter on the time scales of a few days to weeks that characterize upwelling blooms.
Although bacterioplankton communities are composed of thousands of bacterial populations, typically only a few become dominant during phytoplankton blooms [6]. Hence, it has been postulated that the utilization of phytoplankton-derived dissolved organic matter (DOM) is partitioned among specialized bacteria that thrive under rapidly changing environmental conditions [6]. This is underscored by both field studies of naturally occurring phytoplankton blooms [11, 14,15,16,17] and enrichment experiments [18,19,20,21]. For example, well-known phytoplankton-associated bacterial taxa such as the Flavobacteriaceae and Alteromonadaceae are efficient degraders of algal polysaccharides, proteins, and glycoproteins [6, 11, 13], and the Roseobacter clade (Rhodobacteraceae) are exceptionally competitive utilizers of low-molecular-weight DOM compounds like dimethylsulfoniopropionate (DMSP), polyamines, and taurine [6, 22]. Collectively, these studies show that DOM compositional characteristics are important for structuring bacterioplankton community composition, potentially driving bacterial succession over longer time scales.
Several studies that investigated bacterial responses to phytoplankton-derived organic matter [18, 23,24,25,26], concentrated seawater DOM [27, 28] and DOM model compounds [4, 29, 30], found pronounced functional resource partitioning among distinct bacterial clades. Metabolic functions that are typically detected in bloom-associated bacteria involve the degradation of polymers (e.g., carbohydrate-active enzymes that hydrolyze glycosidic bonds found in polysaccharides or proteolytic enzymes that hydrolyze peptide bonds), and the transport of hydrolysis products, besides features like surface adhesion, bacterial signaling, biofilm formation, and motility [11, 24]. While there is ample knowledge of the molecular mechanisms driving DOM utilization by bacteria in the open ocean, corresponding knowledge for coastal seas is limited.
Given the rapidly shifting phytoplankton bloom dynamics in upwelling areas, we carried out a shipboard mesocosm experiment over seven days to investigate bacterial growth and transcription responses associated with different bloom phases (i.e., bloom development, through early decay to senescence phases). In parallel with the experiment, we sampled the field station from where the mesocosm water was collected, to contrast the responses measured in the experiment with dynamics taking place in the natural environment. We hereby hypothesize that the detection of transcriptional differences between key taxa across bloom phases in a replicated experimental setting would have the potential to inform on the ecology of these taxa in their natural environment.
Materials and methods
Study site, sampling, and experimental setup
Seawater was collected during the ENVISION-III cruise [31], at shelf station 3 (Stn 3; 42° 7’ 42.3984” N, 8° 55’ 44.9724” W) (Fig. S1). Detailed methods on the experimental setup are given in Supplementary Material. Briefly, for initiating the mesocosm experiment on 5 August 2016 (day 0), each of three mesocosms received a mix of nutrient-rich water from 20 m depth (152 L; derived from a recent upwelling) and water from 5 m (38 L; with a phytoplankton bloom under development) (Fig. S2). All water was filtered through a 200 µm mesh. Based on previous experience [32], we expected this would induce pronounced and replicated phytoplankton blooms in the mesocosms, of comparable magnitude to the blooms occurring in natural surface waters. This experimental design allowed us to determine the transcriptional responses of bacteria to different bloom phases, by avoiding advective processes that would disrupt the temporal connectivity between samples sequentially collected at sea. Mesocosms were incubated onboard and to maintain in situ temperature (~15 °C) the mesocosms were placed in a tank with flow-through seawater from ~2 m depth. Subsamples of ~5 L were collected daily for total prokaryotic cell counts and 3H-leucine incorporation rates, chlorophyll a, and NH4+, NO2−, NO3−, PO43−, and SiO44− concentrations, and on days 0, 1, 3, 5, and 7 for DNA and RNA extractions. In parallel, corresponding samples were collected from 5 m depth at Stn 3.
Chlorophyll a and nutrients
Size-fractionated chlorophyll a (Chl a) concentrations were determined by sequentially filtering 300 mL seawater samples through 3.0- and 0.2-μm-pore-size polycarbonate filters [31]. Chl a was extracted with 90% acetone at 4 °C overnight in the dark. Fluorescence was determined using the non-acidification technique [33], with a TD-700 fluorometer (Turner Designs) calibrated with a pure Chl a solution [34]. Samples for inorganic nutrient analysis (NH4+, NO2−, NO3−, PO43−, and SiO44−) were collected in 50 mL polyethylene bottles and stored at –20 °C until analysis by standard colorimetric methods [35]. Dissolved organic carbon (DOC) concentrations were measured with a Shimadzu TOC-V analyzer according to [36] (Supplementary Material).
Prokaryote abundance and heterotrophic production
Samples (1.8 mL) for prokaryote abundance were preserved with a mix of 1% and 0.05% (final concentrations) of paraformaldehyde and glutaraldehyde, respectively, and frozen at −80 °C until analysis. Samples were stained with 2.5 mM SybrGreen DNA fluorochrome and enumerated with a FACSCalibur flow cytometer (Becton Dickinson) [37]. Bacterial heterotrophic production was estimated through the 3H-leucine incorporation method according to [38]. Samples (1 mL in triplicates) were amended with 40 nM radioactive leucine (final concentration) and dark-incubated at in situ temperature for 1 h. Production was calculated using a conversion factor of 3.1 kg C mol Leu−1 [38] (Supplementary Material).
Microbial community composition analysis
Prokaryotic and eukaryotic community composition were determined from ~2 L water samples, which were sequentially filtered through 3-μm-pore-size Nuclepore polycarbonate filters (Whatman) and 0.22-μm-pore-size Sterivex filters (EMD Millipore) and analyzed as described in [31] and Supplementary Material. In brief, for prokaryotes, the 16S rRNA gene V4-V5 region was amplified using the universal primers “515 F” and “926 R” [39] from the <3.0-µm and >0.2-µm size fraction. Eukaryotic 18S rRNA genes were amplified using the primers TAReuk454FWD1 and TAReukREV3 [40] from both size fractions. Amplicons were sequenced on a MiSeq platform (Illumina, Inc.) to obtain 2 × 300 bp paired-end reads. Raw reads were processed using the Ampliseq (v2.2.0) pipeline [41] and SILVA reference database (v138.1) [42] for taxonomic assignments of 16S rRNA gene amplicon sequence variants (ASVs). The databases PR2 together with the marine protist database from the BioMArKs project were used to infer taxonomy of 18S rRNA gene ASVs.
Metatranscriptomics analysis
For metatranscriptomics, ~3.5 L water samples from each of the triplicate mesocosms and the field samples were sequentially filtered through 3-μm-pore-size polycarbonate filters (Whatman) and collected on Sterivex filters (GP 0.22-μm-pore-size), preserved in 2 mL RNAlater (Qiagen, Hilden, Germany), and immediately flash-frozen in liquid nitrogen. RNA was extracted using RNeasy (Qiagen), treated to remove DNA and rRNA, and linearly amplified with minor modifications [29, 43]. Sequencing was done at the Swedish National Genome Infrastructure, on a HiSeq 2500 platform (Illumina, Inc.) in rapid mode to obtain 2 × 125 bp paired-end reads. Sequencing summary statistics are in Table S1.
Details on metatranscriptomics analyses are in Supplementary Material. In brief, Illumina adapter sequences were removed with Cutadapt [44] and reads were trimmed with Sickle using default settings [45]. Reads aligning to an in-house database of stable RNA sequences were removed with ERNE [46] and quality reads de-novo assembled with MEGAHIT [47] separately for mesocosm and field samples. Open reading frames (ORFs) were determined with Prodigal [48] in single mode. The ORFs were clustered at a 99% level with VSEARCH [49] and aligned to the NCBI Refseq protein database (release date: December 20, 2018) with DIAMOND [50]. Taxonomic annotations were assigned using MEGAN [51]. For details on annotations and analyses of transcribed DOM active genes and phylogenetic marker genes see Supplementary Material. In brief, glycoside hydrolases (GHs), peptidases (PEPs), transporters (TPs), and sulfatases (STs) were detected and classified with HMMER3 using HMM profiles specific for each in the PFAM, MEROPS, Transporter Classification, and SulfAtlas databases, respectively. GHs were additionally classified with run-dbcan (v2.0.11) against the dbCAN2 database (release date: July 31, 2018) [52]). To further identify transcriptionally active taxa, we carried out phylogenetic analyses on two expressed marker genes broadly distributed in bacteria: the genes coding for ribosomal protein L12 (the most highly expressed ribosomal protein gene in our data set) and for RecA (necessary for maintaining DNA integrity).
Statistics, normalizations and visualization
Detailed description of statistics and normalizations are provided in the Supplementary Material. In brief, for principal component analysis (PCA), raw counts were transformed into centered log ratios (clr) using CoDaSeq [53] (v0.99.6) and Euclidean distances computed with the function dist (vegan v2.5–7). PCAs were performed using the function prcomp (stats v4.1.0) in R v4.1.0 [54]. For order-specific PCA analyses, clrs were calculated for each order separately. Redundancy analysis (RDA) was performed on the same input data as described above (clr). Environmental variables used in the RDA were selected based on pairwise Pearson correlations coefficients <0.9 and variance inflation factors <10. To detect differences in gene transcription between bacterial orders, we normalized individual gene transcript counts to the total transcription within each order, attempting to favor changes in bacterial transcription over changes in abundance. The same principle was applied to analyses at the genus level, whereby genus level information on transcription of the studied gene systems was obtained by grouping order-normalized transcript counts at the respective taxonomic genus levels and functional GH family or PFAM levels (Supplementary Material).
For visualization in Ternary plots, we grouped order-normalized transcript counts at the genus and GH family or PFAM level into development phase (DP - mean of day 0 and day 1; n = 4), early decay (ED - mean of day 3; n = 3), and senescence phase (SP - mean of day 5 and 7; n = 6), and standardized the counts to equal row sums (Supplementary Material). To obtain additional insight into the transcriptional dynamics of the coastal upwelling system, we compared the expression of these genes at GH family and PFAM levels between the parallel mesocosm and field samples through linear regressions based on log2-transformed order-normalized transcripts (Supplementary Material).
Results
Microbial dynamics during an upwelling-driven phytoplankton bloom
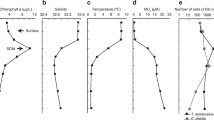
Sampling in the NW Iberian Peninsula coastal upwelling system captured a phytoplankton bloom during its development and subsequent decaying phases (Fig. S1). Chl a increased from 3.8 mg m−3 to a peak of 14 mg m−3 on day 2, progressively decreasing to 2.5 mg m−3 on day 7 (Fig. 1A). The 18S rRNA gene analysis showed that dinoflagellates (mostly Dinophyceae) were dominant components of the eukaryotic community along with ciliates (Ciliophora) during bloom development. From day 5 onwards, the relative abundance of diatoms (e.g., Chaetoceros and Thalassiosira) increased (Fig. S3A, [31]). DOC concentrations ranged from 82 µM C (day 2) to 53 µM C (day 7) (Fig. 1A). The continuous increase in inorganic nutrient concentrations measured from day 4 onwards at station 3 indicated an upwelling pulse (Fig. 1B, Fig. S2). Bacterial production was highest on day 2, coinciding with the phytoplankton peak, and bacterial abundance increased until day 5 (Fig. 1C).
A, E Chlorophyll a (Chl a) and dissolved organic carbon (DOC) concentrations, (B, F) Nitrate (NO3−) and phosphate (PO43−) concentrations, (C, G) Bacterial abundance (BA) and bacterial production (BP), and (D, H) Transcription of the 15 most active prokaryotic taxa. Arrows indicate the time points when metatranscriptome samples were taken. Error bars denote standard deviation of triplicate mesocosms, except for day 0, which were taken from pooled water used for filling the tanks (n = 1).
The 16S rRNA gene analysis showed that the bacterial community was largely dominated by e.g., the Flavobacteriaceae genera Aurantivirga and Polaribacter making up ~11% of the community along with Flavicella and Formosa (up to 6%) and the Rhodobacteraceae genera Planktomarina and Yoonia-Loktanella accounting for 8.6% and 6.1%, respectively. Gammaproteobacteria such as the genus Glaciecola and the clades SAR92 and OM60/NOR5 showed relative abundances ≤2% (Fig. S3B).
The prokaryotic community transcription was dominated by Cellvibrionales, which nearly doubled to ~30% of total transcripts on day 3 (Fig. 1D), primarily due to the families Porticoccaceae and Halieaceae (identified through phylogenetic analyses of the genes encoding ribosomal protein L12 and RecA; Figs. S4, S5). The other taxa remained fairly stable, with Flavobacteriales (mainly the Flavobacteriaceae genera Polaribacter and Tenacibaculum; Figs. S6, S7) and Rhodobacterales (genus Planktomarina; Figs. S8, S9) each contributing ~10% of total transcription. Pelagibacterales (genus Pelagibacter; Figs. S8, S9) and Alteromonadales (genera Alteromonas and Glaciecola; Figs. S4, S5) accounted for ~4% each (Fig. 1D).
Microbial dynamics during an experimental mesocosm bloom
As in the field, Chl a peaked on day 2 (~18.6 mg m−3) and decreased to 1.5 mg m−3 on day 7 (Fig. 1E). 18S rRNA gene analysis showed a mixed phytoplankton community, primarily composed of dinoflagellates (mostly Dinophyceae) and diatoms (e.g., Chaetoceros and Thalassiosira) (Fig. S3A). Chlorophyta peaked on day 3 and marine alveolates (MALV), and marine stramenopiles (MAST) increased on day 7 (Fig. S3A). The transition from bloom development to early decay was associated with pronounced changes in nutrient concentrations (Figs. 1E, F, S2). Upon the Chl a decrease, dissolved organic carbon (DOC) doubled from day 2 to 3, reaching ~145 µM C (Fig. 1E), whereas dissolved inorganic nutrients sharply decreased (Figs. 1F, S2). Still, bacterial production reached maximum rates later than in the field, peaking on day 5 - one day after the peak in bacterial abundance (Fig. 1G).
The 16S rRNA gene analysis showed that the genera Alteromonas (Alteromonadaceae) and Pseudoalteromonas (Pseudoalteromonadaceae) increased substantially on days 3 and 5 (up to ~10% of community) along with Rhodobacteraceae genera like Planktomarina and Yoonia-Loktanella (up to ~14%) (Fig. S3B). During bloom senescence, the Flavobacteriales genus Polaribacter (Flavobacteriaceae) became dominant (up to ~40% on day 7).
The prokaryotic transcriptional responses in the mesocosms were comparable to those in the field during the phytoplankton bloom development phase (days 0 and 1; Fig. 1D, H, S10); dominated by Cellvibrionales (families Porticoccaceae and Halieaceae; Figs. S4, S5), Rhodobacterales (family Rhodobacteraceae and genus Planktomarina; Figs. S8, S9), and Flavobacteriales (Flavobacteriaceae genera Polaribacter and Tenacibaculum; Figs. S6, S7). The transition from bloom development to early decay, characterized by the rapid decrease in Chl a along with inorganic nutrients and a concomitant increase in DOC until day 3 (Fig. 1E), substantially induced Alteromonadales transcription (from 3% to 38% of total transcripts; Fig. 1H), mainly the genera Alteromonas and Glaciecola (Alteromonadaceae) and Pseudoalteromonas (Pseudoalteromonadaceae) (Figs. S4, S5). At this time, the transcription of both Flavobacteriales and Rhodobacterales remained fairly stable. Thereafter, Alteromonadales transcription decreased to 20% on day 7, whereas Flavobacteriales transcription increased to 22% of transcripts. Simultaneously, a few orders with initially low transcription (e.g., Saprospirales and Vibrionales) increased toward day 7 (Fig. 1H). Bacterial richness and Shannon diversity based on the transcriptional data on the phylogenetic marker genes for L12 and RecA in the mesocosms were highest during bloom development and decreased during bloom senescence. In the field, dynamics in diversity were less pronounced (Fig. S11).
A principal component analysis (PCA) performed on the metatranscriptomic data further emphasized shifts from the bloom development phase (day 0 and 1) to the early decay phase (day 3) and to the senescence phase (day 5 and 7) (Fig. 2A; PC1 explained 68% of the temporal transcriptional variation). The temporal shift in prokaryotic transcription was significant (PERMANOVA, R2 = 0.79, p < 0.001). A redundancy analysis (RDA) showed that Chl a (0.2 and 3.0 µm size fractions), DOC, and NH4+, explained ~55% of the variation on RDA1 and ~19% on RDA2, and collectively accounted for ~68% of the variation in community transcription (Monte Carlo permutation test, p = 0.001). Variance partitioning analysis further showed that Chl a (3.0 µm fraction) explained ~27% and DOC ~11% of variation in transcription (Chl a 3.0–0.2 µm and NH4+ were not significant) (Figs. 2B, S12), emphasizing the coupling between phytoplankton bloom development and bacterial gene expression.
A Principal component analysis (PCA) of clr-transformed transcript counts and pairwise Euclidean distances of open reading frames (ORFs) with at least 5 counts per million (CPM) in at least 3 samples (50533 ORFs accounting for ~68% of total transcripts - TPM). Values were scaled to unit variance. B Redundancy analysis (RDA) based on the same data and preprocessing steps. Environmental variables were log-transformed, centered, and scaled to unit variance prior to RDA analysis.
Dynamics in DOM utilization gene transcription in the mesocosm bloom phases
We performed PCAs on order-normalized expression that distinguishes the contribution of transcriptional regulation from changes due to growth (Fig. S13). These analyses showed that Alteromonadales expression shifted strongly between day 1 and 3 (Fig. S13A), as did Flavobacteriales and Saprospirales to a somewhat lesser degree (Fig. S13B, C), largely mirroring the changes in community transcription (Fig. 1H). In contrast, the expression of Rhodobacterales, Cellvibrionales, and Pelagibacterales remained relatively stable until day 5 (Fig. S13D–F), indicating little immediate responsiveness to the DOC increase on day 3. This indicated a divergence between orders in sensing and utilizing changes in the organic matter pool during phytoplankton bloom progression.
Analysis of the transcribed genes involved in the utilization of phytoplankton-derived labile organic matter or nutrients showed taxon-specific transcription patterns throughout the phytoplankton bloom (Fig. 3). Cellvibrionales had the highest order-normalized levels of GH expression until day 3 (Fig. 3A, y-axis); and the highest relative expression of GHs, as normalized to the entire metatranscriptome, was recorded for Alteromonadales on day 3 (Fig. 3A, size of filled circles). These taxa were primarily represented by the Porticoccaceae family and the genera Alteromonas plus Glaciecola, respectively (Figs. 3D, S14). In contrast, transcription of flavobacterial GHs increased three-fold during bloom progression to ~1% of order-normalized transcription on day 7 (Fig. 3A), mainly due to the genus Polaribacter (Flavobacteriaceae) (Figs. 3D, S14). Also PEPs showed pronounced differences between bacterial taxa in both temporal expression dynamics and in relative transcription. For all orders, PEP transcription remained fairly constant during the bloom development phase (days 0 and 1), after which the transcriptional investment steadily increased in Saprospirales and Flavobacteriales (Fig. 3B), especially in Polaribacter (Figs. 3D, S14). Alteromonadales, in turn, showed a pronounced peak in PEP expression on day 3 (Fig. 3B), due mainly to the genera Alteromonas and Pseudoalteromonas (Figs. 3D, S14). Rhodobacterales generally had high relative PEP expression (reaching ~2.4%, Fig. 3B) compared to GHs (~0.3% of order-normalized transcription, Fig. 3A), and were primarily represented by unclassified Rhodobacteraceae (Figs. 3D, S14). Membrane transporters (TPs) accounted for ~10–20% of order-normalized transcription in the studied bacteria (Fig. 3C). While Cellvibrionales, Alteromonadales, and Flavobacteriales primarily invested in the transcription of TonB-dependent transporters (TBDTs; on average accounting for around 40%, 30%, and 20% of their total TP transcription, respectively), Rhodobacterales and Pelagibacterales favored ABC-type transporter transcription (45–40% of TP transcription) (Fig. S15). The Flavobacteriales (e.g., Polaribacter) and Saprospirales (dominated by the genus Phaeodactylibacter and an unclassified taxon) depicted a relatively constant investment in transporters throughout the phytoplankton bloom (Figs. 3C, D, S14); note though that for Flavobacteriales this stability resulted from shifts between taxa within the Flavobacteriaceae, from”Unclassified” to Polaribacter (Fig. 3D). In the other orders, expression generally decreased one-fourth over time, although Pelagibacteraceae transporter expression increased ~3-fold during the bloom development phase (day 0 to 1), to a peak at 37% of order-normalized transcripts (dominated by Candidatus Pelagibacter, Figs. 3C, D, S14). Altogether, these results emphasize the broad range in temporal adjustments of transcriptional investment in DOM and nutrient scavenging processes that marine bacterioplankton perform during phytoplankton bloom progression.
A Glycoside hydrolases (GHs), (B) peptidases (PEPs), and (C) transporters (TPs). Data for the six transcriptionally most active bacterial orders are shown as transcripts per million (TPMs; denoted by size of circles) and as order-normalized proportions of transcription (as percent of TPMs for each order). Error bars denote standard deviations (n = 3 mesocosms, except for day 0 where n = 1). D Relative order-normalized transcript abundances at the genus level for each of the studied target gene systems (i.e., GH, PEP, and TP).
Transcription of DOM-utilization genes as putative drivers of functional succession during the mesocosm bloom
To further investigate the dynamics in bacterial substrate usage across bloom phases, we visualized the transcription of GHs, PEPs, and TPs in ternary plots (Fig. 4). We found pronounced differences between orders in grouping patterns resulting from: (i) differences in temporal expression of individual genes that were shared between orders (Fig. 4 and S15), and (ii) the expression of genes restricted to particular orders (or shared in different combinations; Fig. S16). Moreover, changes in GH, PEP, and TP transcription over time within orders, especially in the Alteromonadales and Flavobacteriales, resulted from changes in transcription ascribed to successional dynamics of different genera, as seen by comparing individual data points in the function plots to the corresponding taxon plots (Fig. 4). In the three transcribed gene systems, this was most evident as shifts in dominance in transcription from Glaciecola to e.g., Alteromonas and Pseudoalteromonas (in Alteromonadales) and from unclassified Flavobacteriaceae to Polaribacter (in Flavobacteriales).
A Glycoside hydrolases (GHs), (B) peptidases (PEPs), and (C) transporters (TPs). Bloom phases are the development phase (DP, n = 4; days 0 and 1), early decay (ED, n = 3; day 3), and senescence phase (SP, n = 6; days 5 and 7). Bubble sizes denote normalized transcript abundances of individual GH families or PFAMs per genus averaged over all days in percent. The top 12 most abundant GH families are shown in color, all others in grey. All PEPs are color-coded according to their proteolytic families in the MEROPS database. The top 12 most abundant TP families in addition to TC TTT, MR, and PRC are shown in color. Abbreviation of TC families: Mot/Exb - The H+- or Na+-translocating Bacterial Flagellar Motor/ExbBD Outer Membrane Transport Energizer; OMR - Outer Membrane Receptor (here TonB-dependent transporters; TBDTs); OOP - OmpA-OmpF Porin; TRAP-T - Tripartite ATP-independent Periplasmic Transporter; TTT - Tricarboxylate Transporter; ABC - ATP-binding Cassette; F-ATPase - H+- or Na+-translocating F-type, V-type and A-type ATPase; Sec - General Secretory Pathway; NaT-DC - Na+-transporting Carboxylic Acid Decarboxylase; QCR - Proton-translocating Quinol:Cytochrome c Reductase; COX - Proton-translocating Cytochrome Oxidase; Na-NDH - Na+-translocating NADH:Quinone Dehydrogenase; FeoB - Ferrous Iron Uptake; MR - Ion-translocating Microbial Rhodopsin, PRC - Photosynthetic Reaction Center.
Divergent grouping of transcribed GH genes affiliated with Alteromonadales and/or Flavobacteriales was observed along the early decay axis or toward the senescence phase axis, respectively (Fig. 4A), due to e.g., genes that likely code mostly for endo-acting laminarinases (e.g., GH17, GH16), exo-acting laminarinases, and α-amylases (GH3 and GH13, respectively) (Fig. 4 and S15). The other orders transcribed only few GHs (note Saprospirales GH3 and GH13 in the bloom development and early decay phases), or one as in Pelagibacterales. Sulfatase transcription was generally very low, with potential utilization of sulfated carbon compounds among Planctomycetes, Rhizobiales, and Rhodobacteraceae (Fig. S17).
Transcribed peptidase genes of Alteromonadales grouped primarily along the early decay axis (Fig. 4B), although not directly on the axis - indicating their transcription was maintained into the senescence phase to a higher degree than GHs. Differences over time were observed for diverse sets of metallo- (e.g., M23, M20, M41), serine- (e.g., S8, S9, and S24), and cysteine peptidases (e.g., C26). In contrast, expressed Flavobacteriales PEP genes, similar to their GHs, grouped away from the central part of the ternary plot toward the senescence axis (Fig. 4B). The relative transcriptional investment, in particular of intracellular cysteine peptidase C56, metallo- (e.g., M50, M20, and M1), and serine peptidases (e.g., S24 and S41) differed between Flavobacteriales and Alteromonadales (Fig. 4B and S15). The transcribed Rhodobacterales and Cellvibrionales PEP genes formed fairly tight clusters toward the center of the ternary plot, indicating that transcription of these genes remained largely stable during the bloom. Also Saprospirales showed a relatively stable investment, although several peptidases deviated (e.g., C14, M24, and M41) or were not expressed during the development phase (Fig. 4B and S15B).
Clustering of the many transcribed transporter genes affiliated with Alteromonadales (Fig. 4C) was in line with their overall strong response primarily during early decay (i.e., day 3; Figs. 1H, 2A). Alteromonadales were particularly active in transcribing TBDTs and general secretory pathway (Sec) family genes (Fig. S15). For Flavobacteriales transporters, there was a tight cluster centered in the ternary and a second cluster tightly aligning with the bloom senescence axis. The first cluster included secretory pathway and cation transport systems (e.g., 3.B.1 NaT-DC and 3.D.5 Na-NDH), whereas the second cluster was enriched in ABC-type transporters and the outer membrane factor (1.B.17 OMF). In contrast to the tight clustering of Rhodobacterales transporters (3.A.1 ABC for sugars, branched-chain amino acids, and DMSP), the spread of Cellvibrionales indicated a pronounced divergence in temporal expression of particular transporters. Cellvibrionales showed a high transcriptional investment in TBDTs (up to ~60% of their total TP transcription on day 7) and had the highest proteorhodopsin transcription during bloom development together with Pelagibacterales (Fig. S15C). In Pelagibacterales, the most abundant transporters were the Na+-transporting carboxylic acid decarboxylase (NaT-DC) that peaked on day 2 and the ABC Superfamily (Fig. 4C).
Responsiveness of functional gene expression (GHs, PEPs, and TPs) in the field compared to mesocosms
Regression analyses showed that transcription of GHs, PEPs, and TPs were strongly and positively correlated between mesocosms and field samples, except for GHs in Saprospirales (Fig. S18, Supplemental Material Table S1). Overall, GHs and PEPs (Fig. S18A, B) showed some variability, with slopes between 0.5 and 1.2 and R2adj values between 0.32 and 0.97, whereas TPs (Figs. S18C, S19) were similarly expressed (slopes: 0.9–1.1 and R2adj: 0.7–0.96). The early decay phase (day 3), during which major nutrient transitions occurred in the mesocosms, showed that GHs for the orders Alteromonadales, Rhodobacterales, and Cellvibrionales were highly correlated between mesocosms and the field (slopes > 0.8). Nevertheless, putative cellulases, α-amylase, exo- and endo-acting glucanases/laminarinases and chitinase (e.g., GH3, 13, 16, and 19) deviated from this pattern and were more responsive to nutritional conditions in the field. In contrast, Flavobacteriales showed a higher transcriptional investment in putative GHs in the mesocosms compared to the field (slopes < 0.8, Supplementary Material Table S2). These included e.g., GH3 and GH23, and exo-acting β-N-acetylglucosaminidases (GH20) targeting amino-sugars, and endo-β−1,4-mannanases (e.g., GH26) for hydrolyzing plant polysaccharides (Fig. 5A). In the field, there was a tendency to higher transcription of GHs involved in the degradation of structural polymers such as chitin and peptidoglycan (e.g., GH15, 73, 81, 144, 158) (Fig. 5A, S18). The negative correlation of GH transcription between mesocosm and field for Saprospirales, together with a tendency of higher transcription of PEPs and TPs in mesocosms, indicated a minor role of these Bacteroidetes in the upwelling (Figs. S18, S19).
A Glycoside hydrolases (GHs), (B) peptidases (PEPs), and (C) transporters (TPs). The 1:1 line is shown in gray, whereas the red line shows a linear fit considering all target PFAMs. Color-legend same as Fig. 4. A Top 12 most abundant GH families, (B) all PEP classes, and in (C) top 12 most abundant TC families and TTT, MR, and PRC. Detailed information on gene annotations, transcripts per million (TPM) and percentage of order-specific transcription are given in Table S1 and linear regression statistics in Supplementary Material Tables S1 and S2.
For PEPs, most orders showed a fairly similar transcriptional investment in both systems (Fig. 5B; PEPs aligning with the 1:1 line). Still, Alteromonadales and Pelagibacterales tended to have higher expression in the mesocosms (Fig. 5B, slope < 1); note though those relationships changed on day 7 (Figs. S18B, S19). Compared to GHs and PEPs, transporter expression was highly consistent between the mesocosms and field (slopes: 0.9–1) (Fig. 5C)—especially for Flavobacteriales and Rhodobacterales (slopes > 0.96). Inorganic phosphate transporters (PiTs) showed higher proportions in Flavobacteriales in the field, whereas ABC transporters were more important in mesocosms. Pelagibacterales showed the lowest number of regulated genes (slope ~1) (Fig. 5C, Supplementary Material Table S2), but an exceptional transcriptional investment in transporters during bloom development (Figs. 3C, S19).
We generated a conceptual model of the transcriptional cascades of major bacterial taxa across an upwelling bloom (Fig. 6) that builds on the genetic analyses in general and on the comparative analysis of order-specific gene transcription patterns of GHs, PEPs, and TPs between the mesocosms and the field in particular (Figs. 5 and S18). The model outlines inferred changes in DOM and the identity of bacterial orders and families (and genera where possible) that dominate the transcription in different bloom phases.
In the graph circles, cell sizes of the six studied bacterial orders denote their relative contribution to overall community transcription. Gray shapes outside the graph circles indicate tentative compositional changes in the dissolved organic matter pool. In the lower circles, the transcriptional response of the bacterial orders characteristic to each of the bloom phases is indicated. Doughnuts denote the transcription of gene systems for dissolved organic matter utilization and nutrient uptake: glycoside hydrolases (GHs), peptidases (PEPs), and transporters (TPs). The size of doughnut sections reflects differences in the relative allocation of transcriptional effort to the different gene systems.
Discussion
Transcriptional cascades across phytoplankton bloom phases
Our findings portray rapid adjustments in molecular mechanisms underlying functional traits of marine bacteria that can influence carbon cycling in upwelling waters. Since transcriptional responses in the mesocosms distinctly changed as a function of the phytoplankton bloom development phase, to the sharp peak in Chl a into the early decay and senescence phases, we here discuss responses of the studied bacteria according to bloom phases. Still, the taxonomic detail obtained from different genes or by different molecular approaches differed depending on the representation of taxa in current databases. Nevertheless, our analyses of 16S rRNA genes in community DNA and of total metatranscriptomes, including expressed targeted gene systems (i.e., GHs, PEPs, TPs, and STs) along with phylogenetic analyses of transcribed genes conserved across broad groups of bacteria (i.e., genes for RecA and ribosomal protein L12), provided a coherent view on the taxonomic identity of the key bacterial players in the studied upwelling bloom system.
The transcriptional dominance during bloom development of such phylogenetically different bacterial orders as Pelagibacterales, Rhodobacterales, and Cellvibrionales was noteworthy. The Pelagibacterales (SAR11 clade; here Pelagibacteraceae) are adapted to oligotrophic conditions, with streamlined genomes and fairly low transcriptional plasticity in combination with highly expressed high-affinity ABC transporter systems [55, 56]. In contrast, the Rhodobacterales (here mostly Rhodobacteraceae represented by the Roseobacter clade) life strategies range from streamlined oligotrophs to metabolically versatile opportunists [6, 57]. Yet, both Pelagibacteraceae and Rhodobacteraceae typically engage in the assimilation of phytoplankton-derived metabolites, in particular low-molecular-weight dissolved organic matter (LMW-DOM) [56, 58,59,60,61]. Knowledge of the ecophysiology of Cellvibrionales is limited, although they are common players in the global ocean [31, 62,63,64]. The order, in our study mainly represented by Halieaceae and Porticoccaceae, includes bacteria isolated from macroalgae or seaweed that possess agarolytic capabilities, such as Agarilytica rhodophyticola [65] and Gilvimarinus polysaccharolyticus [66]. Our results indicated that the Cellvibrionales families were similar to Rhodobacteraceae with respect to the number of expressed GHs and PEPs, but with substantially higher expression levels for the former. Moreover, Cellvibrionales dominated transcription of TBDTs, which together with their high transcription of GHs suggests a crucial role in the turnover of labile carbohydrates. As such, polysaccharide hydrolysis by different Cellvibrionales families potentially complements the Rhodobacteraceae and Pelagibacteraceae toward the turnover of LMW-DOM compounds.
A remarkable feature shared by the Pelagibacteraceae, Rhodobacteraceae, and Cellvibrionales families was their very limited transcriptional response to the strong DOC pulse on day 3. This could have been expected for pelagibacters, given their oligotrophic life strategy and their recognized specialization on LMW-DOM like carboxylic acids and DMSP [56, 67]. Nevertheless, Pelagibacteraceae were highly dynamic in their membrane transporter expression before the DOC pulse, focused on organic acids and DMSP (e.g., TTT, NaT-DC, TRAP-T, and ABC). However, the limited response by Rhodobacteraceae was surprising given that these bacteria (i.e., roseobacters such as Planktomarina) typically are referred to as dominant components of the bacterial community during phytoplankton blooms [6]. Incidentally, a lack of pronounced responses to phytoplankton decay DOM by both Pelagibacteraceae and Rhodobacteraceae was also observed across a spring bloom in the North Sea [11]. These findings and other studies suggest a limited involvement by Rhodobacteraceae in the degradation of high-molecular-weight DOM (HMW-DOM) compounds from massive phytoplankton decay [6]; in part due to being outcompeted by HMW-DOM specialists among e.g., Gammaproteobacteria and Flavobacteriia [68]. Our results provide the environmental context to model organism work showing Rhodobacteraceae as sensitive interaction partners that benefit from their proximity to active phytoplankton with which they can exchange metabolites [69,70,71].
An important feature of the early decay phase was the burst in Alteromonadales transcription, especially transcripts affiliated with the genera Alteromonas (Alteromonadaceae) and Pseudoalteromonas (Pseudoalteromonadaceae). These genera are widespread opportunists [27, 68, 72] and efficient scavengers of (algal) polysaccharides (e.g., laminarin, alginate, and pectin) [29, 30, 73, 74]. Beyond being opportunists in experiments, Alteromonas relatives are increasingly observed in various natural waters [74, 75]. Indeed, the pronounced transcriptional response in several GH genes showed that Alteromonadales rapidly exploited diverse polysaccharides or possibly glycoproteins. Notably though, this was combined with a broad set of PEPs, potentially related to the use of released proteins [76], and ABC transporters (e.g., for cobalamin) and outer membrane porins (potentially allowing surface attachment for utilization of polymers in aggregates or decaying phytoplankton). Altogether, our transcriptional analyses suggest that the success of Alteromonadales, and especially Alteromonas and Pseudoalteromonas, under bloom decay conditions fundamentally relies on the ability to exploit a palette of labile biopolymers, particularly polysaccharides and proteins [72, 76].
The early decay phase triggered a pronounced transcriptional response also by Bacteroidetes, which in the senescence phase further increased in relative transcription levels compared to Alteromonadales. Early bacterioplankton community composition studies showed that, in particular, members of the Flavobacteriaceae thrive upon phytoplankton bloom demise, suggesting a preference for HMW-DOM [77, 78]. Indeed, Bacteroidetes are efficient degraders of algal polysaccharides (e.g., laminarin, alpha-glucans, and sulfated xylans) [11, 79, 80] and proteins [29, 81,82,83,84]. These bacteria engaged little in transcription of sulfatases as compared to Planctomycetes and Rhizobiales. This is in line with previous reports showing that members of the Plactomycetes and Verrucomicrobia are specialized in the degradation of complex polysaccharides such as fucoidan [85,86,87]. Here, Flavobacteriales and Alteromonadales overall expressed a similar number of GH and PEP. In fact, they shared several GHs (e.g., GH3, 17, and 16) that accounted for similar proportions of their transcription, suggesting an important role of laminarin (a beta-1,3-glucan used for carbon storage in phytoplankton, particularly in diatoms), thus indicating an overlapping bacterial substrate range (i.e., niche space). However, Flavobacteriales showed a broader suite of expressed enzymes for hydrolyzing plant polysaccharides (endo-β-1,4-mannanases—e.g., GH26). While the source of mannans during algae blooms remains unclear, the cell walls of some diatoms contain mannans [88]. Thus, our findings suggest that certain Flavobacteriales mediate alpha- and beta-mannan degradation. Alteromonadales, in turn, transcribed higher levels of TBDTs and one-third more ABC-transporters (26 PFAMs) compared to Flavobacteriales and the temporal development of e.g., TBDT and porin genes differed substantially. These findings confirm previous observations on taxon-specific differences in transporter expression [11] and the importance of TBDTs for organic matter acquisition during phytoplankton blooms [15, 89], and provide novel mechanistic understanding on the divergent temporal evolution of hydrolytic enzyme and transporter transcription between the two taxa. In particular, the proportionately increased Flavobacteriaceae transcription of diverse enzymes toward bloom senescence suggested these bacteria are capable of exploiting a broader variety of phytoplankton-derived biopolymers.
Interpreting bacterioplankton transcriptional responses to upwelling-driven blooms
The annual cycle of phytoplankton biomass in the NW Iberian upwelling system is characterized by a spring and summer bloom season representative of temperate shelf seas [90,91,92]. Rather than continuous blooms in each season, intermittent upwelling events induce variability in the duration of sequential bloom cycles from two to 20 days from initiation to complete dissipation [93,94,95,96,97,98]. The duration of the field and mesocosm phytoplankton blooms in our study (eight days) falls within this range, although the amplitude of changes in Chl a and DOC concentrations was more pronounced in the mesocosms. These differences are consistent with the higher initial nutrient concentrations in the mesocosms (intentionally obtained by water mass mixing) compared to the field, and the lack of loss factors such as advective or diffusive processes. It should also be noted that the mesocosm senescence phase was not observed in the field, likely due to the upwelling-driven injection of nutrients to the surface waters from day 4 onward. Nevertheless, the cascade of order-level transcriptional responses in gene systems for utilization of organic matter and nutrients that developed in parallel in the field and in the mesocosms was remarkably similar (except Saprospirales responses). Thus, it is pertinent to interpret the experimental results on resource utilization in the context of the natural settings.
What initially appeared a puzzling result was the substantial increase in Cellvibrionales transcription with bloom progression in the field but decrease in the mesocosm. Based on the generally high expression levels of GHs during mesocosm bloom development, along with the limited transcriptional response to the DOC pulse in the mesocosms on day 3, we propose that the Cellvibrionales families Haliaceae and Porticoccaceae largely rely on the utilization of polysaccharides released from physiologically deteriorating phytoplankton (but not from mass lysis). In contrast, the strong increase in transcription of recognized opportunist Alteromonadales genera like Alteromonas and Pseudoalteromonas [72, 99] in the mesocosms was less surprising. However, Alteromonadales accounted for up to 4% of the relative transcription in the field samples, as represented primarily by the genus Glaciecola, and for example Alteromonas is not uncommon in field studies of natural phytoplankton blooms [14], placing it as a potentially important player in upwelling systems. Our mesocosm findings suggest that in ecosystems with recurrent phytoplankton blooms, a diverse set of Alteromonadales genera are not merely opportunists, but rather, fine-tuned scavengers able to take advantage of labile biopolymers from lysing phytoplankton cells (selected polysaccharides and proteins; preferably supplied at a reasonably stable rate). These gammaproteobacterial taxa were accompanied by pelagibacters and roseobacters, which appear to be competitive in the quest for various labile LMW-DOM compounds under the transient high-nutrient conditions associated with phytoplankton blooms in upwelling systems (see details on these taxa above). Particularly, the pelagibacters are highly oligotrophic bacteria, yet some lineages of the SAR11 clade prefer coastal zones [56]. Lastly, we emphasize the recognized importance of Flavobacteriaceae in organic matter degradation during bloom senescence [11, 77], probably due to their combined ability to engage unusually diverse sets of enzyme systems for utilization for both polysaccharides and proteins [82]. For Saprospirales, we foresee that the high investment in peptidase transcription could provide an advantage over Flavobacteriaceae upon massive release of protein from phytoplankton lysis, resulting in resource partitioning of proteins between distinct Bacteroidetes taxa.
Our transcriptomics results on resource partitioning suggest that the chemical characteristics of important components of the DOM pool rapidly change during phytoplankton bloom succession. Bacteria can contribute to such DOM remodeling through differences between bacterial groups in the demands for, or utilization efficiency of, key elements like carbon and nitrogen, as observed for roseobacters compared to Bacteroidetes [100]. Accordingly, adjustment of glycoside hydrolase and peptidase expression, and corresponding membrane transporters, to selectively target carbon from carbohydrates or nitrogen-rich compounds like proteins could influence DOM pool stoichiometry. Our findings also suggest that transcriptional analysis of metabolic plasticity in nutrient acquisition can provide novel knowledge of mechanisms that underlie bacterioplankton succession under upwelling conditions leading to phytoplankton blooms, and how this relates to the labile DOM pool that accounts for a large fraction of surface ocean carbon fluxes [3, 23]. Given coastal zones contribute disproportionately to ocean productivity [1], rapid dynamics in bloom progression and bacterial responses would ultimately shape the biogeochemistry of the contemporary ocean. These lines of reasoning indicate that uncovering the linkages between bacterial activity and spatiotemporal variability in DOM chemical composition represents a tangible pursuit for microbial oceanography.
Data availability
Ribosomal RNA gene sequence data have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI (https://www.ebi.ac.uk/ena), under project accession numbers PRJEB36188 (16S rRNA gene) and PRJEB36099 (18S rRNA gene). Metatranscriptome sequences are available at the EMBL-EBI European Nucleotide Archive repository (https://www.ebi.ac.uk/ena), under the project accession PRJEB36727 (mesocosms; ERS5512667-ERS5512693) and PRJEB36728 (field samples; ERS5513557-ERS5513582).
References
Gattuso JP, Frankignoulle M, Wollast R. Carbon and carbonate metabolism in coastal aquatic ecosystems. Annu Rev Ecol Syst. 1998;29:405–34.
Arnosti C, Wietz M, Brinkhoff T, Hehemann JH, Probandt D, Zeugner L, et al. The biogeochemistry of marine polysaccharides: sources, inventories, and bacterial drivers of the carbohydrate cycle. Ann Rev Mar Sci. 2021;13:81–108.
Moran MA, Kujawinski EB, Stubbins A, Fatland R, Aluwihare LI, Buchan A, et al. Deciphering ocean carbon in a changing world. Proc Natl Acad Sci USA. 2016;113:3143–51.
Poretsky RS, Sun S, Mou X, Moran MA. Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ Microbiol. 2010;12:616–27.
Azam F. Microbial control of oceanic carbon flux: The plot thickens. Science.1998;280:694–96.
Buchan A, LeCleir GR, Gulvik CA, González JM. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol. 2014;12:686–98.
Bunse C, Bertos-Fortis M, Sassenhagen I, Sildever S, Sjoqvist C, Godhe A, et al. Spatio-temporal interdependence of bacteria and phytoplankton during a baltic sea spring bloom. Front Microbiol. 2016;7:517.
Cram JA, Chow CE, Sachdeva R, Needham DM, Parada AE, Steele JA, et al. Seasonal and interannual variability of the marine bacterioplankton community throughout the water column over ten years. ISME J. 2015;9:563–80.
Seymour JR, Amin SA, Raina JB, Stocker R. Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat Microbiol. 2017;2:17065.
Taylor JD, Cottingham SD, Billinge J, Cunliffe M. Seasonal microbial community dynamics correlate with phytoplankton-derived polysaccharides in surface coastal waters. ISME J. 2014;8:245–8.
Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science.2012;336:608–11.
Hernando-Morales V, Varela MM, Needham DM, Cram J, Fuhrman JA, Teira E. Vertical and seasonal patterns control bacterioplankton communities at two horizontally coherent coastal upwelling sites off galicia (NW Spain). Microb Ecol. 2018;76:866–84.
Needham DM, Fuhrman JA. Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat Microbiol. 2016;1:16005.
Tada Y, Taniguchi A, Nagao I, Miki T, Uematsu M, Tsuda A, et al. Differing growth responses of major phylogenetic groups of marine bacteria to natural phytoplankton blooms in the western North Pacific Ocean. Appl Environ Microbiol. 2011;77:4055–65.
Williams TJ, Wilkins D, Long E, Evans F, DeMaere MZ, Raftery MJ, et al. The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ Microbiol. 2013;15:1302–17.
Ottesen EA, Young CR, Eppley JM, Ryan JP, Chavez FP, Scholin CA, et al. Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc Natl Acad Sci USA. 2013;110:E488–97.
Ottesen EA, Young CR, Gifford SM, Eppley JM, Marin R 3rd, Schuster SC, et al. Ocean microbes. Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages. Science. 2014;345:207–12.
Beier S, Rivers AR, Moran MA, Obernosterer I. The transcriptional response of prokaryotes to phytoplankton-derived dissolved organic matter in seawater. Environ Microbiol. 2015;17:3466–80.
Sarmento H, Gasol JM. Use of phytoplankton-derived dissolved organic carbon by different types of bacterioplankton. Environ Microbiol. 2012;14:2348–60.
Shi Y, McCarren J, DeLong EF. Transcriptional responses of surface water marine microbial assemblages to deep-sea water amendment. Environ Microbiol. 2012;14:191–206.
Vorobev A, Sharma S, Yu M, Lee J, Washington BJ, Whitman WB, et al. Identifying labile DOM components in a coastal ocean through depleted bacterial transcripts and chemical signals. Environ Microbiol. 2018;20:3012–30.
Moran MA, Belas R, Schell MA, González JM, Sun F, Sun S, et al. Ecological genomics of marine Roseobacters. Appl Environ Microbiol. 2007;73:4559–69.
Nowinski B, Moran MA. Niche dimensions of a marine bacterium are identified using invasion studies in coastal seawater. Nat Microbiol. 2021;6:524–32.
Rinta-Kanto JM, Sun S, Sharma S, Kiene RP, Moran MA. Bacterial community transcription patterns during a marine phytoplankton bloom. Environ Microbiol. 2012;14:228–39.
Sharma AK, Becker JW, Ottesen EA, Bryant JA, Duhamel S, Karl DM, et al. Distinct dissolved organic matter sources induce rapid transcriptional responses in coexisting populations of Prochlorococcus, Pelagibacter and the OM60 clade. Environ Microbiol. 2013;16:2815–30.
Kieft B, Li Z, Bryson S, Hettich RL, Pan C, Mayali X, et al. Phytoplankton exudates and lysates support distinct microbial consortia with specialized metabolic and ecophysiological traits. Proc Natl Acad Sci USA. 2021;118:e2101178118.
McCarren J, Becker JW, Repeta DJ, Shi Y, Young CR, Malmstrom RR, et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci USA. 2010;107:16420–7.
Sosa OA, Gifford SM, Repeta DJ, DeLong EF. High molecular weight dissolved organic matter enrichment selects for methylotrophs in dilution to extinction cultures. ISME J. 2015;9:2725–39.
Pontiller B, Martínez-García S, Lundin D, Pinhassi J. Labile dissolved organic matter compound characteristics select for divergence in marine bacterial activity and transcription. Front Microbiol. 2020;11:588778.
Bryson S, Li Z, Chavez F, Weber PK, Pett-Ridge J, Hettich RL, et al. Phylogenetically conserved resource partitioning in the coastal microbial loop. ISME J. 2017;11:2781–92.
Joglar V, Prieto A, Barber-Lluch E, Hernández-Ruiz M, Fernández E, Teira E. Spatial and temporal variability in the response of phytoplankton and prokaryotes to B-vitamin amendments in an upwelling system. Biogeosciences.2020;17:2807–23.
Martínez-García S, Fernández E, Álvarez-Salgado XA, González J, Lønborg C, Marañón E, et al. Differential responses of phytoplankton and heterotrophic bacteria to organic and inorganic nutrient additions in coastal waters off the NW Iberian Peninsula. Mar Ecol Prog Ser. 2010;416:17–33.
Welschmeyer NA. Fluorometric analysis of chlorophyll a in the presence of chlorophyll b and pheopigments. Limnol Oceanogr. 1994;39:1985–92.
Parsons TR, Maita Y, Lalli CM. Fluorometric determination of chlorophylls. A manual of chemical & biological methods for seawater analysis: Oxford, UK: Pergamon Press; 1984. p. 107–09.
Hansen H, Grasshoff K. Automated chemical analysis. Methods of seawater analysis. 2nd ed: Verlag Chemie, Weinheim; 1983. p. 347–95.
Álvarez-Salgado XA, Miller AEJ. Simultaneous determination of dissolved organic carbon and total dissolved nitrogen in seawater by high temperature catalytic oxidation: conditions for precise shipboard measurements. Mar Chem. 1998;62:325–33.
Calvo-Díaz A, Morán XAG. Seasonal dynamics of picoplankton in shelf waters of the southern Bay of Biscay. Aquat Micro Ecol. 2006;42:159–74.
Smith DC, Farooq A. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar Micro Food Webs. 1992;6:107–14.
Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–14.
Logares R, Sunagawa S, Salazar G, Cornejo-Castillo FM, Ferrera I, Sarmento H, et al. Metagenomic 16S rDNA Illumina tags are a powerful alternative to amplicon sequencing to explore diversity and structure of microbial communities. Environ Microbiol. 2014;16:2659–71.
Straub D, Blackwell N, Langarica-Fuentes A, Peltzer A, Nahnsen S, Kleindienst S. Interpretations of environmental microbial community studies are biased by the selected 16S rRNA (Gene) amplicon sequencing pipeline. Front Microbiol. 2020;11:1–18.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6.
Poretsky RS, Gifford S, Rinta-Kanto J, Vila-Costa M, Moran MA. Analyzing gene expression from marine microbial communities using environmental transcriptomics. J Vis Exp. 2009;24:1–6.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 2011;17:10–12.
Joshi NA, Fass JN. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files. 2011. https://github.com/najoshi/sickle.
Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS One. 2013;8:1–13.
Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics.2015;31:1674–6.
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119.
Rognes T, Flouri T, Nichols B, Quince C, Mahe F. VSEARCH: a versatile open source tool for metagenomics. PeerJ.2016;4:e2584.
Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60.
Huson DH, Beier S, Flade I, Górska A, El-Hadidi M, Mitra S, et al. MEGAN community edition - interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol. 2016;12:1–12.
Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, et al. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–101.
Gloor GB, Wu JR, Pawlowsky-Glahn V, Egozcue JJ. It’s all relative: analyzing microbiome data as compositions. Ann Epidemiol. 2016;26:322–9.
R Core Team. R: A language and environment for statistical computing. 4.1.0 ed. Vienna, Austria: R Foundation for Statistical Computing; 2021.
Cottrell MT, Kirchman DL. Transcriptional control in marine copiotrophic and oligotrophic bacteria with streamlined genomes. Appl Environ Microbiol. 2016;82:6010–18.
Giovannoni SJ. SAR11 bacteria: The most abundant plankton in the oceans. Ann Rev Mar Sci. 2017;9:231–55.
Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE, et al. Genome characteristics of a generalist marine bacterial lineage. ISME J. 2010;4:784–98.
Allers E, Gomez-Consarnau L, Pinhassi J, Gasol JM, Simek K, Pernthaler J. Response of Alteromonadaceae and Rhodobacteriaceae to glucose and phosphorus manipulation in marine mesocosms. Environ Microbiol. 2007;9:2417–29.
Alonso C, Pernthaler J. Roseobacter and SAR11 dominate microbial glucose uptake in coastal North Sea waters. Environ Microbiol. 2006;8:2022–30.
Noell SE, Giovannoni SJ. SAR11 bacteria have a high affinity and multifunctional glycine betaine transporter. Environ Microbiol. 2019;21:2559–75.
Sowell SM, Wilhelm LJ, Norbeck AD, Lipton MS, Nicora CD, Barofsky DF, et al. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 2009;3:93–105.
Chenard C, Wijaya W, Vaulot D, Lopes Dos Santos A, Martin P, Kaur A, et al. Temporal and spatial dynamics of bacteria, archaea and protists in equatorial coastal waters. Sci Rep. 2019;9:16390.
Pajares S, Varona-Cordero F, Hernández-Becerril DU. Spatial distribution patterns of bacterioplankton in the oxygen minimum zone of the tropical mexican pacific. Micro Ecol. 2020;80:519–36.
Signori CN, Pellizari VH, Enrich-Prast A, Sievert SM. Spatiotemporal dynamics of marine bacterial and archaeal communities in surface waters off the northern Antarctic Peninsula. Deep-Sea Res Pt Ii. 2018;149:150–60.
Ling SK, Xia J, Liu Y, Chen GJ, Du ZJ. Agarilytica rhodophyticola gen. nov., sp. nov., isolated from Gracilaria blodgettii. Int J Syst Evol Microbiol. 2017;67:3778–83.
Cheng H, Zhang S, Huo YY, Jiang XW, Zhang XQ, Pan J, et al. Gilvimarinus polysaccharolyticus sp. nov., an agar-digesting bacterium isolated from seaweed, and emended description of the genus Gilvimarinus. Int J Syst Evol Microbiol. 2015;65:562–69.
Malmstrom RR, Kiene RP, Cottrell MT, Kirchman DL. Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic ocean. Appl Environ Microbiol. 2004;70:4129–35.
Kirchman DL. Growth rates of microbes in the oceans. Ann Rev Mar Sci. 2016;8:285–309.
Durham BP, Dearth SP, Sharma S, Amin SA, Smith CB, Campagna SR, et al. Recognition cascade and metabolite transfer in a marine bacteria-phytoplankton model system. Environ Microbiol. 2017;19:3500–13.
Ferrer-González FX, Widner B, Holderman NR, Glushka J, Edison AS, Kujawinski EB, et al. Resource partitioning of phytoplankton metabolites that support bacterial heterotrophy. ISME J. 2021;15:762–73.
Landa M, Burns AS, Roth SJ, Moran MA. Bacterial transcriptome remodeling during sequential co-culture with a marine dinoflagellate and diatom. ISME J. 2017;11:2677–90.
Pedler BE, Aluwihare LI, Azam F. Single bacterial strain capable of significant contribution to carbon cycling in the surface ocean. Proc Natl Acad Sci USA. 2014;111:7202–7.
Koch H, Dürwald A, Schweder T, Noriega-Ortega B, Vidal-Melgosa S, Hehemann JH, et al. Biphasic cellular adaptations and ecological implications of Alteromonas macleodii degrading a mixture of algal polysaccharides. ISME J. 2019;13:92–103.
López-Pérez M, Gonzaga A, Martin-Cuadrado AB, Onyshchenko O, Ghavidel A, Ghai R, et al. Genomes of surface isolates of Alteromonas macleodii: the life of a widespread marine opportunistic copiotroph. Sci Rep. 2012;2:696.
Bergauer K, Fernandez-Guerra A, Garcia JAL, Sprenger RR, Stepanauskas R, Pachiadaki MG, et al. Organic matter processing by microbial communities throughout the Atlantic water column as revealed by metaproteomics. Proc Natl Acad Sci USA. 2018;115:E400–8.
Hou S, López-Pérez M, Pfreundt U, Belkin N, Stüber K, Huettel B, et al. Benefit from decline: the primary transcriptome of Alteromonas macleodii str. Te101 during Trichodesmium demise. ISME J. 2018;12:981–96.
Pinhassi J, Sala MM, Havskum H, Peters F, Guadayol O, Malits A, et al. Changes in bacterioplankton composition under different phytoplankton regimens. Appl Environ Microbiol. 2004;70:6753–66.
Riemann L, Steward GF, Azam F. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl Environ Microbiol. 2000;66:578–87.
Cottrell MT, Kirchman DL. Natural assemblages of marine Proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol. 2000;66:1692–7.
Krüger K, Chafee M, Ben Francis T, Glavina Del Rio T, Becher D, Schweder T, et al. In marine Bacteroidetes the bulk of glycan degradation during algae blooms is mediated by few clades using a restricted set of genes. ISME J. 2019;13:2800–16.
Ben Hania W, Joseph M, Bunk B, Spröer C, Klenk HP, Fardeau ML, et al. Characterization of the first cultured representative of a Bacteroidetes clade specialized on the scavenging of cyanobacteria. Environ Microbiol. 2017;19:1134–48.
Fernández-Gómez B, Richter M, Schüler M, Pinhassi J, Acinas SG, González JM, et al. Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J. 2013;7:1026–37.
Orsi WD, Smith JM, Liu S, Liu Z, Sakamoto CM, Wilken S, et al. Diverse, uncultivated bacteria and archaea underlying the cycling of dissolved protein in the ocean. ISME J. 2016;10:2158–73.
Xing P, Hahnke RL, Unfried F, Markert S, Huang S, Barbeyron T, et al. Niches of two polysaccharide-degrading Polaribacter isolates from the North Sea during a spring diatom bloom. ISME J. 2015;9:1410–22.
Sichert A, Corzett CH, Schechter MS, Unfried F, Markert S, Becher D, et al. Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat Microbiol. 2020;5:1026–39.
Ivanova AA, Naumoff DG, Miroshnikov KK, Liesack W, Dedysh SN. Comparative genomics of four Isosphaeraceae Planctomycetes: a common pool of plasmids and glycoside hydrolase genes shared by Paludisphaera borealis PX4(T), Isosphaera pallida IS1B(T), Singulisphaera acidiphila DSM 18658(T), and Strain SH-PL62. Front Microbiol. 2017;8:412.
Vidal-Melgosa S, Sichert A, Francis TB, Bartosik D, Niggemann J, Wichels A, et al. Diatom fucan polysaccharide precipitates carbon during algal blooms. Nat Commun. 2021;12:1150.
Le Costaouëc T, Unamunzaga C, Mantecon L, Helbert W. New structural insights into the cell-wall polysaccharide of the diatom Phaeodactylum tricornutum. Algal Res. 2017;26:172–9.
Francis TB, Bartosik D, Sura T, Sichert A, Hehemann JH, Markert S, et al. Changing expression patterns of TonB-dependent transporters suggest shifts in polysaccharide consumption over the course of a spring phytoplankton bloom. ISME J. 2021;15:2336–50.
Bode A, Estévez MG, Varela M, Vilar JA. Annual trend patterns of phytoplankton species abundance belie homogeneous taxonomical group responses to climate in the NE Atlantic upwelling. Mar Environ Res. 2015;110:81–91.
Cermeño P, Marañón E, Pérez V, Serret P, Fernández E, Castro CG. Phytoplankton size structure and primary production in a highly dynamic coastal ecosystem (Ría de Vigo, NW-Spain): Seasonal and short-time scale variability. Estuar Coast Shelf Sci. 2006;67:251–66.
Nogueira E, Pérez FF, Rı́os AF. Modelling thermohaline properties in an estuarine upwelling ecosystem (Rı́a de Vigo: NW Spain) using box-jenkins transfer function models. Estuar Coast Shelf Sci. 1997;44:685–702.
Broullón E, López-Mozos M, Reguera B, Chouciño P, Doval MD, Fernández-Castro B, et al. Thin layers of phytoplankton and harmful algae events in a coastal upwelling system. Prog Oceanogr. 2020;189:102449.
Fraga F. Upwelling off the Galician Coast, Northwest Spain. In: Richards FA, editor. Coastal Upwelling. Washington: American Geophysical Union; 1981. p. 176–82.
Nogueira E, Figueiras FG. The microplankton succession in the Ría de Vigo revisited: species assemblages and the role of weather-induced, hydrodynamic variability. J Mar Syst. 2005;54:139–55.
Pitcher GC, Walker DR, Mitchellinnes BA, Moloney CL. Short-term variability during an anchor station study in the southern Benguela Upwelling System - phytoplankton dynamics. Prog Oceanogr. 1991;28:39–64.
Smayda TJ, Trainer VL. Dinoflagellate blooms in upwelling systems: Seeding, variability, and contrasts with diatom bloom behaviour. Prog Oceanogr. 2010;85:92–107.
Wilkerson FP, Lassiter AM, Dugdale RC, Marchi A, Hogue VE. The phytoplankton bloom response to wind events and upwelled nutrients during the CoOP WEST study. Deep-Sea Res Pt Ii. 2006;53:3023–48.
Reintjes G, Arnosti C, Fuchs B, Amann R. Selfish, sharing and scavenging bacteria in the Atlantic Ocean: a biogeographical study of bacterial substrate utilisation. ISME J. 2019;13:1119–32.
Mayali X, Weber PK, Pett-Ridge J. Taxon-specific C/N relative use efficiency for amino acids in an estuarine community. FEMS Microbiol Ecol. 2013;83:402–12.
Acknowledgements
We thank all the people involved in the project ENVISION for helping with sampling and analytical work. We thank the crew on the R/V Ramon Margalef for their hospitality and professionalism during the cruise. From IIM-CSIC, V. Vieitez and M.J. Pazó performed the inorganic and organic nutrient analysis. We are grateful to Celia Marrasé and Carlos Pedrós-Alió for insightful and constructive comments on the manuscript, and Martin Brusin for artistic contribution to the conceptual model. We thankfully acknowledge the insightful and constructive criticism by three reviewers on our manuscript. This research was supported by a grant from the Swedish Research Council to JP and the Spanish Ministry of Economy and Competitiveness through the ENVISION (CTM2014–59031-P) and INTERES (CTM2017-83362-R) projects to EF and ET, and TRAITS PID2019-110011RB-C33 to SM-G and PID2019-110011RB-C32 to JMG (Spanish Ministry of Science and Innovation, Spanish State Research Agency, doi: 10.13039/501100011033). BP was supported by the Faculty of Health and Life Sciences at Linnaeus University and VJ was supported by an FPI fellowship from the Spanish Ministry of Economy and Competitiveness. The scintillation counter was supported by the 2006-06 ICTS project of the Ministry of Science and Innovation at the Spanish Institute of Oceanography. The authors would like to acknowledge support from Science for Life Laboratory, the National Genomics Infrastructure, NGI, and Uppmax (compute project SNIC 2017/7-419 and storage project SNIC 2020/16-76), Sweden, for providing assistance in massive parallel sequencing and computational infrastructure.
Funding
Open access funding provided by Linnaeus University.
Author information
Authors and Affiliations
Contributions
ET, EF, SMG, and JP conceptualized and designed the study. BP, VJ, and ET collected the samples. BP, DL, and JMG processed metatranscriptomics data. BP analyzed the data with contributions from DL and JMG. BP and JP wrote the paper with input from all authors.
Corresponding author
Ethics declarations
COMPETING interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pontiller, B., Martínez-García, S., Joglar, V. et al. Rapid bacterioplankton transcription cascades regulate organic matter utilization during phytoplankton bloom progression in a coastal upwelling system. ISME J 16, 2360–2372 (2022). https://doi.org/10.1038/s41396-022-01273-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-022-01273-0
This article is cited by
-
Coral thermal stress and bleaching enrich and restructure reef microbial communities via altered organic matter exudation
Communications Biology (2024)
-
Jellyfish detritus supports niche partitioning and metabolic interactions among pelagic marine bacteria
Microbiome (2023)
-
Carbohydrates and carbohydrate degradation gene abundance and transcription in Atlantic waters of the Arctic
ISME Communications (2023)