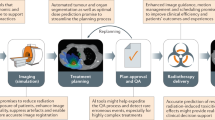
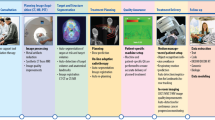
Abstract
Artificial intelligence (AI) applications have enabled remarkable advancements in healthcare delivery. These AI tools are often aimed to improve accuracy and efficiency of histopathology assessment and diagnostic imaging interpretation, risk stratification (i.e., prognostication), and prediction of therapeutic benefit for personalized treatment recommendations. To date, multiple AI algorithms have been explored for prostate cancer to address automation of clinical workflow, integration of data from multiple domains in the decision-making process, and the generation of diagnostic, prognostic, and predictive biomarkers. While many studies remain within the pre-clinical space or lack validation, the last few years have witnessed the emergence of robust AI-based biomarkers validated on thousands of patients, and the prospective deployment of clinically-integrated workflows for automated radiation therapy design. To advance the field forward, multi-institutional and multi-disciplinary collaborations are needed in order to prospectively implement interoperable and accountable AI technology routinely in clinic.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 4 print issues and online access
$259.00 per year
only $64.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel L, Torre A, Ahmedin D. GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Glob Cancer Stat. 2018;68:394–424.
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33.
Wang L, Lu B, He M, Wang Y, Wang Z, Du L. Prostate cancer incidence and mortality: global status and temporal trends in 89 countries from 2000 to 2019. Front Public Health. 2022;10:811044.
Walz J, Graefen M, Chun FK, Erbersdobler A, Haese A, Steuber T, et al. High incidence of prostate cancer detected by saturation biopsy after previous negative biopsy series. Eur Urol. 2006;50:498–505.
Fuchsjäger M, Shukla-Dave A, Akin O, Barentsz J, Hricak H. Prostate cancer imaging. Acta Radiol. 2008;49:107–20.
Farolfi A, Calderoni L, Mattana F, Mei R, Telo S, Fanti S, et al. Current and emerging clinical applications of PSMA PET diagnostic imaging for prostate cancer. J Nucl Med. 2021;62:596–604.
Stabile A, Dell’Oglio P, Gandaglia G, Fossati N, Brembilla G, Cristel G, et al. Not all multiparametric magnetic resonance imaging–targeted biopsies are equal: the impact of the type of approach and operator expertise on the detection of clinically significant prostate cancer. European Urology. Oncology. 2018;1:120–8.
Cackowski FC, Mahal B, Heath EI, Carthon B. Evolution of disparities in prostate cancer treatment: is this a new normal? Am Soc Clin Oncol Educ Book. 2021;41:e203–e14.
Spratt DE, Zhang J, Santiago-Jiménez M, Dess RT, Davis JW, Den RB, et al. Development and validation of a novel integrated clinical-genomic risk group classification for localized prostate cancer. J Clin Oncol. 2018;36:581.
Shariat SF, Karakiewicz PI, Suardi N, Kattan MW. Comparison of nomograms with other methods for predicting outcomes in prostate cancer: a critical analysis of the literature. Clin Cancer Res. 2008;14:4400–7.
Kim HL, Li P, Huang H-C, Deheshi S, Marti T, Knudsen B, et al. Validation of the Decipher Test for predicting adverse pathology in candidates for prostate cancer active surveillance. Prostate Cancer Prostatic Dis. 2019;22:399–405.
Reese AC, Pierorazio PM, Han M, Partin AW. Contemporary evaluation of the National Comprehensive Cancer Network prostate cancer risk classification system. Urology. 2012;80:1075–9.
Terada N, Akamatsu S, Kobayashi T, Inoue T, Ogawa O, Antonarakis ES. Prognostic and predictive biomarkers in prostate cancer: latest evidence and clinical implications. Ther Adv Med Oncol. 2017;9:565–73.
Spratt DE, Sun Y, Van der Wal D, Huang S-C, Mohamad O, Armstrong AJ, et al. An AI-derived digital pathology-based biomarker to predict the benefit of androgen deprivation therapy in localized prostate cancer with validation in NRG/RTOG 9408. Am Soc Clin Oncol. 2022. https://ascopubs.org/doi/abs/10.1200/JCO.2022.40.6_suppl.223.
Litwin MS, Tan HJ. The diagnosis and treatment of prostate cancer: a review. JAMA 2017;317:2532–42.
Schaeffer EM, Srinivas S, Adra N, An Y, Barocas D, Bitting R, et al. NCCN Guidelines® insights: prostate cancer, version 1.2023: featured updates to the NCCN guidelines. J Natl Compr Cancer Netw. 2022;20:1288–98.
Veloso SG, Lima MF, Salles PG, Berenstein CK, Scalon JD, Bambirra EA. Interobserver agreement of Gleason score and modified Gleason score in needle biopsy and in surgical specimen of prostate cancer. Int Braz J Urol. 2007;33:639–51.
Brimo F, Epstein JI. Immunohistochemical pitfalls in prostate pathology. Hum Pathol. 2012;43:313–24.
Kweldam CF, van der Kwast T, van Leenders GJ. On cribriform prostate cancer. Transl Androl Urol. 2018;7:145–54.
Ross AE, D’Amico AV, Freedland SJ. Which, when and why? Rational use of tissue-based molecular testing in localized prostate cancer. Prostate Cancer Prostatic Dis. 2016;19:1–6.
Ma MW, Gao XS, Lyu F, Gu XB, Yin H, Li HZ, et al. Development of a nomogram predicting metastatic disease and the assessment of NCCN, AUA and EAU guideline recommendations for bone imaging in prostate cancer patients. World J Urol. 2021;39:1815–23.
Dall’Era MA, Maddala T, Polychronopoulos L, Gallagher JR, Febbo PG, Denes BS. Utility of the Oncotype DX® prostate cancer assay in clinical practice for treatment selection in men newly diagnosed with prostate cancer: a retrospective chart review analysis. Urol Pract. 2015;2:343–8.
Bostrom PJ, Bjartell AS, Catto JW, Eggener SE, Lilja H, Loeb S, et al. Genomic predictors of outcome in prostate cancer. Eur Urol. 2015;68:1033–44.
Kornberg Z, Cooperberg MR, Spratt DE, Feng FY. Genomic biomarkers in prostate cancer. Transl Androl Urol. 2018;7:459–71.
Koenig IR, Fuchs O, Hansen G, von Mutius E, Kopp MV. What is precision medicine? Eur Res J. 2017;50:1700391.
Roychowdhury S, Chinnaiyan AM. Advancing precision medicine for prostate cancer through genomics. J Clin Oncol. 2013;31:1866.
Copeland J (ed). The Essential Turing: the ideas that gave birth to the computer age. Oxford: Clarendon Press; 2004. p. viii+613. Br J Hist Sci 2006;39:470–1.
Fetzer JH. What is Artificial Intelligence? Artificial Intelligence: Its Scope and Limits: Springer, 1990. p. 3–27.
Ongsulee P, editor Artificial intelligence, machine learning and deep learning. 2017 15th international conference on ICT and knowledge engineering (ICT&KE); 2017: IEEE.
Singh A, Thakur N, Sharma A, editors. A review of supervised machine learning algorithms. 2016 3rd International Conference on Computing for Sustainable Global Development (INDIACom); 2016: IEEE.
Avanzo M, Wei L, Stancanello J, Vallieres M, Rao A, Morin O, et al. Machine and deep learning methods for radiomics. Med Phys. 2020;47:e185–e202.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44.
He K, Zhang X, Ren S, Sun J, editors. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition; 2016.
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial networks. Commun Acm 2020;63:139–44.
Baydoun A, Xu K, Heo JU, Yang H, Zhou F, Bethell LA, et al. Synthetic CT generation of the pelvis in patients with cervical cancer: a single input approach using generative adversarial network. IEEE Access. 2021;9:17208–21.
Ronneberger O, Fischer P, Brox T, editors. Dental X-ray image segmentation using a U-shaped Deep Convolutional network. International Symposium on Biomedical Imaging; 2015.
Carbonneau M-A, Cheplygina V, Granger E, Gagnon G. Multiple instance learning: a survey of problem characteristics and applications. Pattern Recognit. 2018;77:329–53.
Salcedo-Sanz S, Rojo-Álvarez JL, Martínez-Ramón M, Camps-Valls G. Support vector machines in engineering: an overview. Wiley Interdiscip Rev Data Min Knowl Discov. 2014;4:234–67.
Liang F, Qian P, Su KH, Baydoun A, Leisser A, Van Hedent S, et al. Abdominal, multi-organ, auto-contouring method for online adaptive magnetic resonance guided radiotherapy: An intelligent, multi-level fusion approach. Artif Intell Med. 2018;90:34–41.
Huang F, Xie G, Xiao R. Research on Ensemble Learning. In 2009 International Conference on Artificial Intelligence and Computational Intelligence. IEEE; 2009. vol. 3, pp. 249–52.
Rigatti SJ. Random forest. J Insur Med. 2017;47:31–9.
Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck Krauss Silva V, Busam KJ, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25:1301–9.
Raciti P, Sue J, Ceballos R, Godrich R, Kunz JD, Kapur S, et al. Novel artificial intelligence system increases the detection of prostate cancer in whole slide images of core needle biopsies. Mod Pathol. 2020;33:2058–66.
Perincheri S, Levi AW, Celli R, Gershkovich P, Rimm D, Morrow JS, et al. An independent assessment of an artificial intelligence system for prostate cancer detection shows strong diagnostic accuracy. Mod Pathol. 2021;34:1588–95.
Ström P, Kartasalo K, Olsson H, Solorzano L, Delahunt B, Berney DM, et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. Lancet Oncol. 2020;21:222–32.
Mun Y, Paik I, Shin S-J, Kwak T-Y, Chang H. Yet another automated Gleason grading system (YAAGGS) by weakly supervised deep learning. npj Digital Med. 2021;4:1–9.
Bulten W, Pinckaers H, van Boven H, Vink R, de Bel T, van Ginneken B, et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol. 2020;21:233–41.
Pantanowitz L, Quiroga-Garza GM, Bien L, Heled R, Laifenfeld D, Linhart C, et al. An artificial intelligence algorithm for prostate cancer diagnosis in whole slide images of core needle biopsies: a blinded clinical validation and deployment study. Lancet Digital Health. 2020;2:e407–e16.
Nagpal K, Foote D, Tan F, Liu Y, Chen PC, Steiner DF, et al. Development and validation of a deep learning algorithm for gleason grading of prostate cancer from biopsy specimens. JAMA Oncol. 2020;6:1372–80.
Bulten W, Kartasalo K, Chen PC, Strom P, Pinckaers H, Nagpal K, et al. Artificial intelligence for diagnosis and Gleason grading of prostate cancer: the PANDA challenge. Nat Med. 2022;28:154–63.
Cifci D, Foersch S, Kather JN. Artificial intelligence to identify genetic alterations in conventional histopathology. J Pathol. 2022;257:430–44.
Schaumberg AJ, Rubin MA, Fuchs TJ. H&E-stained whole slide image deep learning predicts SPOP mutation state in prostate cancer. BioRxiv. 2018:064279.
Wang Z, Wang Y, Zhang J, Hu Q, Zhi F, Zhang S, et al. Significance of the TMPRSS2: ERG gene fusion in prostate cancer. Mol Med Rep. 2017;16:5450–8.
Dadhania V, Gonzalez D, Yousif M, Cheng J, Morgan TM, Spratt DE, et al. Leveraging artificial intelligence to predict ERG gene fusion status in prostate cancer. BMC Cancer. 2022;22:494.
Leo P, Chandramouli S, Farre X, Elliott R, Janowczyk A, Bera K, et al. Computationally derived cribriform area index from prostate cancer hematoxylin and eosin images is associated with biochemical recurrence following radical prostatectomy and is most prognostic in gleason grade group 2. Eur Urol Focus. 2021;7:722–32.
Esteva A, Feng J, van der Wal D, Huang SC, Simko JP, DeVries S, et al. Prostate cancer therapy personalization via multi-modal deep learning on randomized phase III clinical trials. NPJ Digit Med. 2022;5:71.
Hinata N, Fujisawa M. Racial differences in prostate cancer characteristics and cancer-specific mortality: an overview. World J Mens Health. 2022;40:217–27.
Bhargava HK, Leo P, Elliott R, Janowczyk A, Whitney J, Gupta S, et al. Computationally derived image signature of stromal morphology is prognostic of prostate cancer recurrence following prostatectomy in African American PatientsStroma Predicts Prostate Cancer Outcome in African Americans. Clin Cancer Res. 2020;26:1915–23.
Roach M, Zhang J, Esteva A, Mohamad O, Van der Wal D, Simko J, et al. Prostate cancer risk in African American men evaluated via digital histopathology multi-modal deep learning models developed on NRG Oncology phase III clinical trials. Am Soc Clin Oncol. 2022. https://ascopubs.org/doi/pdf/10.1200/JCO.2022.40.16_suppl.108?role=tab.
Suarez-Ibarrola R, Sigle A, Eklund M, Eberli D, Miernik A, Benndorf M, et al. Artificial intelligence in magnetic resonance imaging-based prostate cancer diagnosis: where do we stand in 2021? Eur Urol Focus. 2022;8:409–17.
Turkbey B, Haider MA. Deep learning-based artificial intelligence applications in prostate MRI: brief summary. Br J Radio. 2022;95:20210563.
Jiang KW, Song Y, Hou Y, Zhi R, Zhang J, Bao ML, et al. Performance of artificial intelligence-aided diagnosis system for clinically significant prostate cancer with MRI: a diagnostic comparison study. J Magn Reson Imaging. 2023;57:1352–64.
Baydoun A, Xu K, Bethell LA, Zhou F, Heo JU, Zhao K, et al. Auto-contouring FDG-PET/MR images for cervical cancer radiation therapy: An intelligent sequential approach using focally trained, shallow U-Nets. Intell Based Medicine 2021;5:100026.
Hiremath A, Shiradkar R, Fu P, Mahran A, Rastinehad AR, Tewari A, et al. An integrated nomogram combining deep learning, Prostate Imaging-Reporting and Data System (PI-RADS) scoring, and clinical variables for identification of clinically significant prostate cancer on biparametric MRI: a retrospective multicentre study. Lancet Digit Health. 2021;3:e445–e54.
Algohary A, Shiradkar R, Pahwa S, Purysko A, Verma S, Moses D, et al. Combination of peri-tumoral and intra-tumoral radiomic features on Bi-Parametric MRI accurately stratifies prostate cancer risk: a multi-site study. Cancers (Basel). 2020;12:2200.
Li L, Shiradkar R, Leo P, Algohary A, Fu P, Tirumani SH, et al. A novel imaging based Nomogram for predicting post-surgical biochemical recurrence and adverse pathology of prostate cancer from pre-operative bi-parametric MRI. EBioMedicine. 2021;63:103163.
Shiradkar R, Ghose S, Jambor I, Taimen P, Ettala O, Purysko AS, et al. Radiomic features from pretreatment biparametric MRI predict prostate cancer biochemical recurrence: preliminary findings. J Magn Reson Imaging. 2018;48:1626–36.
Mitterberger M, Horninger W, Aigner F, Pinggera GM, Steppan I, Rehder P, et al. Ultrasound of the prostate. Cancer Imaging. 2010;10:40.
Lei Y, Tian S, He X, Wang T, Wang B, Patel P, et al. Ultrasound prostate segmentation based on multidirectional deeply supervised V-Net. Med Phys. 2019;46:3194–206.
Zhu N, Najafi M, Han B, Hancock S, Hristov D. Feasibility of image registration for ultrasound-guided prostate radiotherapy based on similarity measurement by a convolutional neural network. Technol cancer Res Treat. 2019;18:1533033818821964.
Guo H, Kruger M, Xu S, Wood BJ, Yan P. Deep adaptive registration of multi-modal prostate images. Comput Med Imaging Graph. 2020;84:101769.
Chiu PK-F, Shen X, Wang G, Ho C-L, Leung C-H, Ng C-F, et al. Enhancement of prostate cancer diagnosis by machine learning techniques: an algorithm development and validation study. Prostate Cancer Prostatic Dis. 2022;25:672–6.
Wang K, Chen P, Feng B, Tu J, Hu Z, Zhang M, et al. Machine learning prediction of prostate cancer from transrectal ultrasound video clips. Front Oncol. 2022;12:948662.
El Naqa I. Prospective clinical deployment of machine learning in radiation oncology. Nat Rev Clin Oncol. 2021;18:605–6.
Laskar SG, Sinha S, Krishnatry R, Grau C, Mehta M, Agarwal JP. Access to radiation therapy: from local to global and equality to equity. JCO Glob Oncol. 2022;8:e2100358.
Agazaryan N, Chow P, Lamb J, Cao M, Raldow A, Beron P, et al. The timeliness initiative: continuous process improvement for prompt initiation of radiation therapy treatment. Adv Radiat Oncol. 2020;5:1014–21.
McIntosh C, Conroy L, Tjong MC, Craig T, Bayley A, Catton C, et al. Clinical integration of machine learning for curative-intent radiation treatment of patients with prostate cancer. Nat Med. 2021;27:999–1005.
Field M, Hardcastle N, Jameson M, Aherne N, Holloway L. Machine learning applications in radiation oncology. Phys Imaging Radiat Oncol. 2021;19:13–24.
Largent A, Barateau A, Nunes JC, Mylona E, Castelli J, Lafond C, et al. Comparison of deep learning-based and patch-based methods for Pseudo-CT generation in MRI-based prostate dose planning. Int J Radiat Oncol Biol Phys. 2019;105:1137–50.
Isola P, Zhu J-Y, Zhou T, Efros AA, editors. Image-to-image translation with conditional adversarial networks. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017.
Cusumano D, Lenkowicz J, Votta C, Boldrini L, Placidi L, Catucci F, et al. A deep learning approach to generate synthetic CT in low field MR-guided adaptive radiotherapy for abdominal and pelvic cases. Radiother Oncol. 2020;153:205–12.
Fu J, Yang Y, Singhrao K, Ruan D, Chu FI, Low DA, et al. Deep learning approaches using 2D and 3D convolutional neural networks for generating male pelvic synthetic computed tomography from magnetic resonance imaging. Med Phys. 2019;46:3788–98.
Maspero M, Savenije MHF, Dinkla AM, Seevinck PR, Intven MPW, Jurgenliemk-Schulz IM, et al. Dose evaluation of fast synthetic-CT generation using a generative adversarial network for general pelvis MR-only radiotherapy. Phys Med Biol. 2018;63:185001.
Hsu SH, Han Z, Leeman JE, Hu YH, Mak RH, Sudhyadhom A. Synthetic CT generation for MRI-guided adaptive radiotherapy in prostate cancer. Front Oncol. 2022;12:969463.
Brouwer CL, Dinkla AM, Vandewinckele L, Crijns W, Claessens M, Verellen D, et al. Machine learning applications in radiation oncology: current use and needs to support clinical implementation. Phys Imaging Radiat Oncol. 2020;16:144–8.
Duan J, Bernard M, Downes L, Willows B, Feng X, Mourad WF, et al. Evaluating the clinical acceptability of deep learning contours of prostate and organs-at-risk in an automated prostate treatment planning process. Med Phys. 2022;49:2570–81.
Liu C, Gardner SJ, Wen N, Elshaikh MA, Siddiqui F, Movsas B, et al. Automatic segmentation of the prostate on CT images using deep neural networks (DNN). Int J Radiat Oncol Biol Phys. 2019;104:924–32.
Ma L, Guo R, Zhang G, Tade F, Schuster DM, Nieh P, et al. Automatic segmentation of the prostate on CT images using deep learning and multi-atlas fusion. Proc SPIE Int Soc Opt Eng. 2017;10133:1013320.
Jimenez-Pastor A, Lopez-Gonzalez R, Fos-Guarinos B, Garcia-Castro F, Wittenberg M, Torregrosa-Andrés A, et al. Automated prostate multi-regional segmentation in magnetic resonance using fully convolutional neural networks. Eur Radiol. 2023:1–10. https://doi.org/10.1007/s00330-023-09410-9.
Wortel G, Eekhout D, Lamers E, van der Bel R, Kiers K, Wiersma T, et al. Characterization of automatic treatment planning approaches in radiotherapy. Phys Imaging Radiat Oncol. 2021;19:60–5.
Nawa K, Haga A, Nomoto A, Sarmiento RA, Shiraishi K, Yamashita H, et al. Evaluation of a commercial automatic treatment planning system for prostate cancers. Med Dosim. 2017;42:203–9.
Cilla S, Romano C, Morabito VE, Macchia G, Buwenge M, Dinapoli N, et al. Personalized treatment planning automation in prostate cancer radiation oncology: a comprehensive dosimetric study. Front Oncol. 2021;11:636529.
Cilla S, Ianiro A, Romano C, Deodato F, Macchia G, Buwenge M, et al. Template-based automation of treatment planning in advanced radiotherapy: a comprehensive dosimetric and clinical evaluation. Sci Rep. 2020;10:423.
Tillman-Schwartz E, Tillman G, Tansky J, Paly J, Efstathiou J, Wang Y. Prospective analysis of a deep learning auto-contouring model for definitive radiation of localized prostate cancer. Int J Radiat Oncol Biol Phys. 2022;114:e105–e6.
Bhattacharya T, Brettin T, Doroshow JH, Evrard YA, Greenspan EJ, Gryshuk AL, et al. AI meets exascale computing: advancing cancer research with large-scale high performance computing. Front Oncol. 2019;9:984.
Davenport T, Kalakota R. The potential for artificial intelligence in healthcare. Future Healthc J. 2019;6:94.
Hines PA, Herold R, Pinheiro L, Frias Z. Artificial intelligence in European medicines regulation. Nat Rev Drug Discovery. 2023;22:81–2.
Harvey HB, Gowda V. How the FDA regulates AI. Acad Radiol. 2020;27:58–61.
Sankaran S, Zhang C, Aarts H, Markopoulos P. Exploring peoples’ perception of autonomy and reactance in everyday AI interactions. Front Psychol. 2021;12:713074.
Author information
Authors and Affiliations
Contributions
Conceptualization: AB and DES. Writing: AB and DES. Reviewing & Editing: AB, AYJ, NGZ, RK, SR, JS, RAV, LKB, RZ, AP, THA, and DES. All authors approved the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
AB: None. AYJ: Personal Fees from Myovant. NGZ: Support by the National Institutes of Health Grant LRP 1 L30 CA231572-01 (2018-2022) and by the American Cancer Society – Tri State CEOs Against Cancer Clinician Scientist Development Grant, CSDG-20-013-01-CCE (2020-). Remuneration from Springer Nature for his textbook, Absolute Clinical Radiation Oncology Review (2019). Remuneration from the American College of Radiation Oncology for chart review and accreditation of radiation oncology facilities nationally (2020-). RK: None. SR: Personal speaking fees from Seagen. JS: Grant support from BMS Foundation, Damon Runyon Cancer Research Foundation. Key opinon leader for Fortec Medical. RAV: None. LKB: None. RZ: None. AP: Honoraria with both ViewRay and SunNuclear. THA: None. DES: Personal fees from AstraZeneca, Blue Earth, Bayer, Boston Scientific, Janssen, Novartis, Myovant, Pfizer, Varian, Elekta, Gamma Tile.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Baydoun, A., Jia, A.Y., Zaorsky, N.G. et al. Artificial intelligence applications in prostate cancer. Prostate Cancer Prostatic Dis 27, 37–45 (2024). https://doi.org/10.1038/s41391-023-00684-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41391-023-00684-0
This article is cited by
-
Prostate cancer detection and complications of MRI-targeted prostate biopsy using cognitive registration, software-assisted image fusion or in-bore guidance: a systematic review and meta-analysis of comparative studies
Prostate Cancer and Prostatic Diseases (2024)
-
Developers-Doctor-patients: the artificial intelligence’s trifecta
Prostate Cancer and Prostatic Diseases (2024)
-
Quality of information and appropriateness of Open AI outputs for prostate cancer
Prostate Cancer and Prostatic Diseases (2024)
-
Expected impact of MRI-targeted biopsy interreader variability among uropathologists on ProScreen prostate cancer screening trial: a pre-trial validation study
World Journal of Urology (2024)
-
Bioinformatics in urology — molecular characterization of pathophysiology and response to treatment
Nature Reviews Urology (2023)