Abstract
DNA double-strand breaks (DSBs) contribute to genome instability, a key feature of cancer. DSBs are mainly repaired by homologous recombination (HR) and non-homologous end-joining (NHEJ). We investigated the role of an isoform of the multifunctional cyclin-dependent kinase 9, CDK9-55, in DNA repair, by generating CDK9-55-knockout HeLa clones (through CRISPR-Cas9), which showed potential HR dysfunction. A phosphoproteomic screening in these clones treated with camptothecin revealed that CDC23 (cell division cycle 23), a component of the E3-ubiquitin ligase APC/C (anaphase-promoting complex/cyclosome), is a new substrate of CDK9-55, with S588 being its putative phosphorylation site. Mutated non-phosphorylatable CDC23(S588A) affected the repair pathway choice by impairing HR and favouring error-prone NHEJ. This CDK9 role should be considered when designing CDK-inhibitor-based cancer therapies.
Similar content being viewed by others
Introduction
Genome instability is a key feature of cancer since it generates the genetic diversity that enables the acquisition of hallmarks capabilities during tumour development and progression [1]. DNA double-strand breaks (DSBs) are among the most toxic DNA lesions threatening genome stability. To tackle this threat, DSBs are repaired by two principal processes: homologous recombination (HR) and non-homologous end-joining (NHEJ) [2]. HR uses the sister chromatid as a template for faithful repair and requires DNA end-resection to generate long 3’ single-stranded DNA (ssDNA) to invade this template; conversely, NHEJ joins DNA ends irrespective of their original sequence and, thus, is error-prone. NHEJ can act throughout the cell cycle, whereas HR can only occur in the S/G2 phase when sister chromatids are available. The mechanisms underlying the choice between HR and NHEJ pathways in S/G2 have extensively been studied. Although the DNA end structure, repair timing, DNA end-resection, and chromatin context have been identified as major factors affecting this choice, some determinants still need to be defined [3, 4].
Cyclin-dependent kinases (CDKs), which are serine/threonine kinases whose function depends on the interaction with cyclin regulatory subunits, were first identified in yeast as drivers of cell cycle progression [5]. However, growing evidence indicates that CDKs are also involved in other cell processes, such as transcription, DNA damage response (DDR), cell death, differentiation, immune response, and metabolism [6]. Among the CDKs performing these non-canonical functions, CDK9, which was identified about three decades ago by our group [7, 8], is a multifunctional kinase involved in the regulation of transcription as part of the positive transcription elongation factor b complex (P-TEFb) [9], HIV gene expression [10] and also in replication stress response and DDR [11,12,13,14].
CDK9 exists in two isoforms, CDK9-42 (42 kDa) and CDK9-55 (55 kDa), which originate from different promoters separated by around 500 bp, and with CDK9-55 having 117 additional amino acids in its N-terminal portion [15]. Here, through a global phosphoproteomic screening in camptothecin (CPT)-treated HeLa cells knocked out for CDK9-55, we identified cell division cycle 23/anaphase-promoting complex subunit 8 (CDC23/APC8) as a possible new CDK9 substrate regulating the activity of the E3-ubiquitin ligase Anaphase Promoting Complex (also called cyclosome or APC/C) for the correct DNA repair pathway choice.
Results
Differential expression of CDK9 isoforms modulates DNA damage response
CDK9 protein was recently described to be involved in DDR [11,12,13,14] and the discrimination between its two protein isoforms in the DDR was not already performed. We silenced CDK9 proteins through siRNAs (siCDK9) followed by differential expression of CDK9 42 or 55 (Fig. 1A and Fig. S1A, S1B) to characterise the CPT response. First of all, silencing of CDK9 reduces foci number of phosphorylated Replication Protein A 32Kda (pRPA32) and BRCA1, confirming its role regulation of DNA repair end-resection dependent (Fig. 1B, C); the overexpression of 42 or 55 isoforms recovers pRPA32 foci in a similar manner but not for BRCA1 (Fig. 1D–E). To check if CDK9 silencing could affect the HR repair favouring the NHEJ, we performed a Replication timing regulatory factor 1 (RIF1) immunofluorescence [16]. As reported in Fig. 1F–G, siCDK9 increased the number RIF1 foci in response to CPT in HeLa cells, followed by their reduction with the differential overexpression of CDK9 isoforms. Over all these data showed a possible involvement of both CDK9 protein isoforms in the DDR.
A Western blot analysis of CDK9 in HeLa cells transfected with siRNA CDK9 (siCDK9), or control (siCTR), and after 24 h transfected with the single CDK9 isoforms (pCEFLHA CDK9 55 kDa and pCEFL HA CDK9 42 kDa) or pCEFL HA control plasmid as indicated by + or −. Upon transfection HeLa cells were treated with 1 μM camptothecin (CPT), or not treated, for additional 2 h. RPA32 was used as a DNA damage control and Lamin A/C as a protein loading control. Red and black arrows indicate CDK9 endogenous and overexpressed protein isoforms, respectively. B Immunofluorescence analysis of HeLa cells transfected and treated as indicated in A; pRPA32 S4/8 foci number was measured by Fiji software. More than 30 cells were analysed for each condition in 3 independent experiments; data represent means ± standard deviation. C Representative immunofluorescence image of pRPA32, from HeLa cells transfected as in B followed by incubation with CPT for two hours. D BRCA1 foci number of HeLa cells, transfected with siCDK9 for 24 h followed by transfection with indicated coding plasmid DNA, was measured by Fiji software. E Representative immunofluorescence image of BRCA1 of HeLa cells transfected and treated as in B. F RIF1 foci number was measured by Fiji software from HeLa cells transfected and treated as in B. For all immunofluorescence analyses more than 30 cells were analysed for each condition in 3 independent experiments. G Representative immunofluorescence image of RIF1, from HeLa cells silenced with CDK9 for 24 h followed by differential overexpression of CDK9 42 or 55 kDa and treated with CPT for two hours. All graphs represent means ± standard deviation. Data were subjected to one-way repeated measures ANOVA with Kruskal-Wallis post-test, to compare all groups. Statistically significant differences are indicated with: **P-value < 0.01 and ***P-value < 0.001.
CDK9-55KO HeLa impaired DNA end-resection in response to CPT and sensitised cells to PARP inhibition
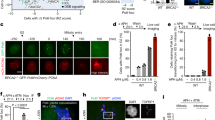
To best described the possible role of CDK9 isoforms in the HR, we used the CRISPR-Cas9-D10A-nickase system to separately knockout each isoform in HeLa cells. We generated clones knockout for CDK9-55 (55KO cl2, cl3, and cl4) without affecting CDK9-42 levels (Fig. 2A), whereas we failed in generating viable CDK9-42 knockout clones. We then treated wt HeLa cells and 55KO clones with CPT analysing the expression of pRPA32 S4/8 [17], and observed its impaired phosphorylation in 55KO clones compared with control cells after 2 h of treatment (Fig. 2B). The ectopic expression of CDK9 55 HA tag protein, in 55KO cl2 cells, rescued the pRPA S4/8 protein levels (Fig. S1C). Moreover, cell cycle analysis of CDK9 55KO clones reveals no cell distribution alteration confirming that the reduction of RPA32 phosphorylation was due a DNA repair defect (Fig. S2A). Consistently, Single-Molecule Analysis of Resection Tracks assay [18, 19] (SMART) showed a marked reduction of resected DNA length in 55KO cl2 compared with wt HeLa cells upon CPT treatment (Fig. 2C). Finally, we analysed a chromatin loading of RAD51 recombinase (RAD51) showing an impaired loading in response to CPT (Fig. 2D). These data suggest that CDK9-55 knockout affects the resection dependent double strand breaks DNA repair. CDK9-55 interacts with the X-ray repair cross complementing 6 (KU70) [20], a DDR protein; we checked if the 55KO could be able to modify the KU70 chromatin loading ability in response to CPT. In Fig. S2B we showed that, CPT reduced chromatin loading respect to the untreated condition, in HeLa cells; conversely, the cl2 55KO cells showed a similar KU70 chromatin loading (as reported in P fraction) with respect to control. Moreover, we performed cell survival and clonogenic assays with different doses of either Olaparib [21] or CPT, showing an increased sensitivity to both drugs in 55KO clones compared with control cells (Fig. 2E, F and Fig. S2C, S2D). Finally, we checked if 55KO cell clones could be sensible to another DSB-inducing agent as the Etoposide, a topoisomerase II inhibitor [22], showing an increased sensitivity of CDK9 KO clones to etoposide treatment, with respect to control (Fig. S3A e, S3B).
A Western blotting analysis of CDK9 in wild-type (wt) HeLa cells and CDK9-55KO clones. GAPDH was used as protein loading control. B Western blot analysis of CDK9 and phosphorylated RPA 32 (pRPA32 S4/8) in wt HeLa cells and CDK9-55KO clones (cl2 and cl4) treated with 1 μM CPT for 2 h or not treated (NT). The black arrow indicates the phosphorylated RPA32 bands. Total RPA 32 and lamin A/C were used as DNA damage and a loading control, respectively. C SMART (single-molecule analysis of resection tracks) assay of wt HeLa and cl2 cells pulse-labelled with BrdU for 24 h and treated with 1 μM CPT, as above. The graph represent means ± standard deviation of more than 200 fibres for each cell type in 3 independent experiments. Statistically significant differences were evaluated by multiple comparisons with Mann-Whitney to compare 2 groups and indicated as follows: ***P-value < 0.001. D The soluble (S) and chromatin-enriched cell fractions (P) of HeLa and CDK9 55KO cells treated, or not (NT), with 1 μM of CTP for 2 h were analyzed by western blotting through the indicated antibodies. Total RPA32 was used as DNA damage control and lamin A/C as a loading control. Under RAD51 blot was represented a densitometric values of protein normalised with lamin A/C by using Fiji software. E Percentage of colonies generated through clonogenic assay of wt HeLa cells and CDK9-55KO clones treated with Olaparib at crescent concentrations (1.5-2-2.5 μM). The graphs represent the means ± standard deviation of 3 independent experiments. In the lower panel significant differences, obtained from one-way ANOVA with Dunnett’s post-test to compare all groups versus control (wt), are indicated with *P-value < 0.05, **P-value < 0.01. F Clonogenic assay of wt HeLa cells and CDK9-55KO clones treated with CTP at crescent concentrations (2.5-5-7 nM). All colonies were stained 10 days after treatment (Olaparib or CTP) with crystal violet. The graphs represent means ± standard deviation of 3 independent experiments. Statistically analysis was performed by using one-way repeated measures ANOVA with Dunnett’s post-test to compare all groups versus control (wt) and significant differences are indicated with: *P-value < 0.05, **P-value < 0.01, in the lower panel.
Global phosphoproteomic screening revealed new potential substrates of CDK9-55 involved in DNA repair
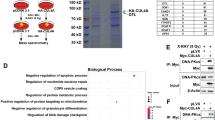
Phosphorylation is one of the principal PTM regulating the DDR through the action of CDKs or other kinases [23, 24]. We performed a global phosphoproteomic screening in CPT-treated and untreated wt HeLa and 55KO cl2 to identify possible new CDK9-55 substrates involved in DDR (Fig. 3A). We detected, through the phospho-signal intensity of each serine/threonine/tyrosine (STY), around 7700 proteins differentially phosphorylated in HeLa and cl2 (Table S1). We selected only peptides with fold-changes (treated/untreated) > 2 in both wt HeLa and cl2. Thus, we identified 944 unique peptides exceeding the fold-change threshold in wt HeLa and not in 55KO cells and corresponding to 646 unique proteins, which are possible CDK9 substrates (Table S2). Gene ontology analysis (http://geneontology.org/) showed enrichment of 2.43-fold in the “DNA damage response” (GO:0006974) (Fig. 3B and Table S3). In particular, we identified 23 proteins involved in the regulation of DNA damage response (Table S4), whose further analysis revealed a > 100-fold enrichment in the “double-strand break repair via break induced replication” term (GO:0000727), supporting the possible role of CDK9-55 in this pathway (Table S5). To verify the reproducibility of our screening, we performed a GeneVenn analysis between the 529 putative CDK9 substrates, out of the 646 proteins identified in our screening - since they mapped into the Uniprot database - and 169 proteins involved in response to CPT, detected in genetic screening by Olivieri et al. [25]. This analysis showed 12 proteins identified in both screenings (Fig. 3C and Table S6). We used the phospho (STY) probability score to indicate the probability that the STY in each peptide could be phosphorylated, with the best score set as 1 (Table S1). The amino acid with the best phospho (STY) score in the peptide sequence was designated as position P0, and the flanking amino acids were indicated from P-9 to P + 9 (Table S7). The resulting 19-aa peptides from all the 646 potential CDK9-55 substrates were analysed by using the IceLogo software (https://iomics.ugent.be/icelogoserver/). This analysis showed a prevalence of phosphorylated serines followed by proline at P + 1 and preceded by arginine in P-3, thus outlining the putative consensus sequence of CDK9-55 substrates as RxxSP (Fig. S3C). This consensus sequence was found in 48 peptides corresponding to 43 unique putative substrates with different functions (Table S8 and Fig. S3D); in particular, functional analysis with the DAVID bioinformatics software (https://david.ncifcrf.gov/) showed 4 proteins with ubiquitin-protein transferase activity (Table S9). We used PhosphoSite Plus (https://www.phosphosite.org/homeAction) to analyse the frequency of Rxx(phospho)SP in the 4 ubiquitin transferases, as assessed in high-throughput screenings (HTSs) reported in the database. All steps from phosphoproteomic screening all summarised in a flow chart (Fig. S4). CDC23 phosphorylated at S588 (581-NTPTRRVSPLNLSSV-595) and Damage Specific DNA Binding Protein 2 (DDB2) phosphorylated at S26 were identified in all the HTSs (Table S9). To explore the possible role of the RxxSP motif in cancer biology, we searched the catalogue of somatic mutations in cancers (COSMIC, https://cancer.sanger.ac.uk/cosmic) and found a deletion in the CDC23 consensus sequence in large intestine cancer samples, and no mutation in DDB2. Moreover, protein sequence analysis showed that the RxxSP motif of CDC23, but not of DDB2, was conserved in Xenopus laevis and Gallus gallus, suggesting a possible important role of this motif through the species (Fig. S5A). In order to validate the phosphoproteomic data, we performed a protein kinase assay to check if CDK9-55 specifically phosphorylates CDC23 protein at S588; to this aim, we purified, from E.coli, GST-CDK9-42, GST-CDK9-55, GST-cyclin T1, GST-CDC23 wt, GST-CDC23 S588A and RNAPII-CTD [26] wt, as a positive CDK9 substrate control [27]. As reported in Fig. S5B, CDK9-42 was able to phosphorylate CDC23 independently by the mutated Serine at position 588. Surprisingly, CDK9-55 phosphorylate CDC23 wt protein in a more specific manner respect to the short CDK9 protein isoform. Finally, the amino acid point mutation S588A reduces the phosphorylation ability of CDK9-55 protein, revealing CDC23 as its new possible substrate. CDC23 (also known as APC8) is a subunit of the E3-ubiquitin ligase APC/C, a 1.5-MDa protein complex consisting of four domains: the scaffolding platform, Tetratricopeptide repeat (TPR) lobe, catalytic core, and co-activators [28]. This complex, besides playing a role in cell cycle control by mediating the timely degradation of components of the cell cycle machinery [29], has recently been implicated in DNA repair [30,31,32,33]. Overall, given that CDC23 is part of a complex involved in DDR, considering the conservation of its RxxSP motif among species and its mutation in cancer, we decided to further investigate the role of CDK9 55 kDa phosphorylation on CDC23 S588 in DNA repair.
A Graphical representation of the global phosphoproteomic assay. B Gene ontology analysis of the 646 putative CDK9-55 substrates identified, showing enrichment in the gene ontology terms “DNA damage response” (GO:0006974). C GeneVenn analysis of the putative CDK9-55 substrates and the DDR proteins identified in screening by Olivieri et al. [65].
pCDC23 S588 is not involved in the regulation of the APC/C complex assembly
We generated HeLa cells expressing a mutated non-phosphorylatable CDC23, in which S588 was substituted with alanine (S588A) by using the CRISPR-Cas9-D10A system obtaining two cell clones carrying heterozygous substitution (Fig. S5C). Given the central role of APC/C in cell cycle regulation [29], we analysed the cell cycle profile of both wt HeLa cells and CDC23(S588A)-mutated clones (cl8 and cl9) and observed no meaningful difference in cell distribution (Fig. S5D). First of all, we analysed, by western blot and immunofluorescence, the activation of pRPA32 S4/8 upon treatment with CPT for two hours, showing the same pRPA32 activation levels in cl8 and cl9 respect to HeLa control cells (Fig. 4A–C). Moreover, we performed a reciprocal endogenous co-immunoprecipitation of CDC23 and APC1, a platform component [28], in wt HeLa and cl9 treated with CPT observing similar assembly ability (Fig. 4D and Fig. S6A). To further confirm these observations, we performed reciprocal co-immunoprecipitation of CDC23 and APC1 also in CDK9-55KO cl2 and observed no meaningful difference compared with wt HeLa cells (Fig. 4E and Fig. S6B).
A Western blotting analysis of CDC23 and pRPA32 (pRPA32 S4/8) in wild type (wt) HeLa and CDC23 (S588A)-mutated clones (cl8 and cl9) (treated or not with 1 μM CPT for 2 h). The black arrow indicates the phosphorylated RPA32 bands. Total RPA32 and lamin A/C used as a DNA damage and loading control, respectively. B pRPA32 S4/8 foci intensity, measured by Fiji software, of HeLa and CDC23 S588A cl8 and cl9 treated with CPT at 1 μM for 2 h. More than 30 cells were analysed for each condition in 3 independent experiments; data represent means ± standard deviation. C Representative immunofluorescence images of pRPA32 S4/8, from HeLa wt and CDC23 S588A cells treated as in B. D Immunoprecipitation assay of HeLa cells and CDC23(S588A)-mutated cl9, with CDC23 antibody, or IgG control. All cells were treated with 1 μM CPT for 2 h or not treated (NT). Total RPA32 was used as DNA damage and loading control. E Immunoprecipitation assay, with CDC23 antibody, performed in wt HeLa cells and CDK9-55KO cl2 treated or not with 1 μM CPT. Total RPA32 antibody was used as in D. The black arrow indicates the phosphorylated RPA32 bands. An Anti-CDK9 antibody was used to verify CDK9 55KO in cl2 cells respect to HeLa wt and the black arrows indicate the corresponding protein kinases.
CDC23(S588A) impairs HR
We analysed the effect of CDC23 silencing on DDR in Hela cells through a small interfering RNA against CDC23 (siCDC23). We monitored the phosphorylation of RPA32 at S4/8 (pRPA32 S4/8) and checkpoint kinase 1 (CHK1) at S345 (pCHK1 S345), as a readout of DNA end-resection and [17, 34, 35], upon CPT treatment, showing an impaired phosphorylation of both factors in CDC23-silenced cells (Fig. 5A and Fig. S6C), consistent with the recently described role of APC/C in DNA repair [30,31,32,33]. To analyse the role of Serine 588 phosphorylation in CDC23 protein, we cloned a CDC23 coding sequence in a pFLAG vector to generate non-phosphorylable S588A and phosphomimetic S588D mutants. First of all, CDC23 silencing reduced the pRPA32 foci intensity which was rescued by the wt protein but also by the two S588 mutants in a similar manner (Fig. 5B, C). In Fig. S6D and S6E, we demonstrated that the CDC23 silencing reduced the BRCA1 foci intensity, rescued by the expression of wt protein but not by the overexpression of CDC23 S588A. Conversely, overexpression of the CDC23 phosphomimetic mutant increases the BRCA1 signal in a similar manner to wt protein (Fig. S6D and S6E). Consistently, transfection with siCDC23, markedly reduced the HR frequency, measured by Direct Repeat-Green Fluorescent Protein (DRGFP)-expressing HeLa cells, [36] (Fig. 5D); moreover, re-expression of wt CDC23 led to HR recovery. Consistently with the BRCA1 foci (Fig. S6D and S6E), the complementation of HeLa-DRGFP siCDC23 cells with the phosphomimetic mutant rescued the HR ability in a similar manner to wt CDC23 protein, with respect to S588A mutant (Fig. 5D). We performed a SMART assay, after treatment with CPT, showing a reduced resected DNA length in CDC23(S588A)-mutated cl9 cells compared with the control (Fig. 5E). Moreover, FACS analysis of Bromodeoxyuridine (BrdU) incorporation in non-denaturing conditions [37], confirmed an impaired formation of resected ssDNA (Fig. 5F) [27]. One of the important steps of the HR is the strand invasion which is regulated by RAD51 [38]; the analysis of RAD51 foci, by immunofluorescence, showed a reduced signal in cl9 (Fig. S7A). Consistently, chromatin loading of RAD51 in HeLa CDC23 S588A cell clones was reduced with respect to the control in response to CPT treatment (Fig. S7B). Recently, APC/C was described to regulate CtIP protein stability through a phosphorylation mechanism for correct DNA end resection [31, 39]. We checked if 55Kda knockout could be involved in the regulation of CtIP activity treating HeLa and cl2 cells with CPT followed by drug washout to monitor CtIP protein degradation. As reported in Fig. S7C, two hours of CPT treatment induced a phosphorylation of CtIP protein, indicated by the presence a higher MW protein band (black arrow). Moreover, we observed a reduction of CtIP phosphorylation in cl2 followed by unaltered protein degradation upon CPT washout, respect to HeLa control cells, consistent with a reduction of DNA end resection described in Fig. 2. In order to analyse the effect of CDC23 S588A onto CtIP, we carried out the same experiment in clone 9 showing non-significant differences in CtIP phosphorylation and degradation with respect to control cells at any time points (Fig. S7D) suggesting a different inhibition mechanism onto HR. Finally, APC/C was described to be involved in the regulation of DNA damage response through polyubiquitination [30] of Ubiquitin specific peptidase 1 (USP1) favouring the HR; we showed in Fig. S7E, a reduced USP1 polyubiquitination signal in cl9 respect to HeLa cells consistent with an impaired HR. As reported in literature, Poly [ADP-ribose] polymerase 1 (PARP1) inhibitors [40] showed a synthetic lethal phenomenon with an impaired HR defective tumour; to check if HeLa cells, carrying CDC23 S588A point mutation, could be synthetic lethal with Olaparib treatment, we performed cell survival and clonogenic assays, showing an increased sensitivity to Olaparib in CDC23 S588A cell clones compared with control cells (Fig. S8A and S8B). Moreover, we analysed cell survival and clonogenic assays of CDC23 S588A cell clones in response to CPT treatment showed reduced viability with respect to HeLa wt cells (Fig. S8C and S8D). To analyse the possible effect of another DNA damaging agent onto the cell viability, we performed cell survival and clonogenic assays, in response to Etoposide treatment, showing an increased sensitivity of cl8 and cl9 cell lines, respect to control (Fig. S9A and S9B). Furthermore, we analysed the possible involvement of CDC23 S588 phosphorylation in CDK9 55 DNA damage signalling, we transfected CDC23 coding plasmids in HeLa cells CDK9 55KO cl2 cells followed by CPT treatment; as reported in supplementary Fig. S9C, overexpression of CDC23 S588A and S588D induces an increased foci intensity of pRPA32 S4/8 respect to the control. Conversely, only the overexpression of CDC23 phosphomimetic mutant (S588D) was able to recover the RAD51 foci intensity (Fig. S9D). Finally, to check if the overexpression of CDC23 mutants could negatively dominate the checkpoint activation in response to camptothecin we monitored pCHK1 S345 and pCHK2 T68 activations [41]. As reported in Fig. S10A and S10B, CDC23 protein mutants do not affect CHK1 and CHK2 phosphorylation ability in response to camptothecin; interestingly, CDC23 S588D was able to induce the activation of pCHK1 and pCHK2 in untreated conditions.
A Western blot analysis of pRPA32 S4/8 and CDC23 of HeLa cells transfected with a small interfering RNA (siRNA) CDC23 (siCDC23) or with a non-targeting control siRNA (siCTR) and treated, after 72 h, with 1 μM of CPT treatment for 2 h. Total RPA32 and Lamin A/C total were used as DNA damage and loading controls, respectively. B The plot represents the pRPA32 S4/8 foci intensity analysis of HeLa cells transfected with siCDC23 or siCTR for 24 h and transfected also with the indicated coding plasmids for additional 48 h. All cells were treated with 1 μM CPT. The analysis of data was carried out with Fiji software. Plot represent means ± standard of more than 30 cells for each condition of 3 independent experiments. Moreover, data were subjected to one-way repeated measures ANOVA with Kruskal-Wallis post-test, to compare all groups. Statistically significant differences are indicated with *P-value < 0.05, **P-value < 0.01, and ***P-value < 0.001. C Representative immunofluorescence images of pRPA32 S4/8 of HeLa cells transfected and treated as in B. D HeLa cells, stably expressing pDR-GFP vector, were transfected with plasmids (pFLAG-CMV1) expressing siRNA-resistant mRNAs encoding either wt CDC23, mutated CDC23(S588A) or CDC23 (S588D). The empty vector was used as a control. GFP levels were measured by FACS. Homologous recombination (HR) frequency is expressed as a percentage relative to siCTR (set as 100%). Data represent means ± standard deviation (n = 3 independent experiments) and they were subjected to ANOVA with multiple comparison post-test to compare all groups. Statistically significant differences are indicated with: ***P-value < 0.001. E SMART (single-molecule analysis of resection tracks) assay in wt HeLa cells and CDC23(S588A)-mutated cl9 pulse-labelled with BrdU for 24 h and then treated with 1 μM camptothecin (CPT) for 2 h. More than 200 fibres were analysed for each cell type in 3 independent experiments; data represent means ± standard deviation. Data were subjected to an unpaired Student t-test. Statistically significant differences are indicated with ***P-value < 0.001. F FACS analysis of BrdU incorporation in non-denaturing conditions in wt HeLa and cl9, treated or not treated (NT) with 1 μM camptothecin (CPT) for 2 h.
HeLa CDC23 S588A cells showed a NHEJ repair in response to CPT treatment
Considering that APC/C has been recently implicated in choosing the DSB repair pathways [30], and our data obtained from the complementation assay with the CDC23 mutants (Fig. 5), we explored the role of CDC23 phosphorylation at S588 in the regulation of the NHEJ. First of all, we performed a RIF1 immunofluorescence analysis in HeLa cells silenced for CDC23 protein and complemented with its different mutants. As reported in Fig. 6A, B, CDC23 silencing does not change the RIF1 foci intensity; conversely, complementation with wt or S588D proteins reduces immunofluorescence signals compared to siCDC23 pFLAG cells. Interestingly, the transfection with S588A mutant increases a RIF1 foci intensity (Fig. 6A, B). To confirm these data, we used HeLa cells stably expressing pimEJ5GFP, [36] a reporter system to measure NHEJ [42]. We observed similar NHEJ efficiency in cells transfected with siCDC23 and siCTR but also after re-expression of a siRes wt CDC23 (Fig. 6C). The expression of S588D does not affect NHEJ repair ability contrarily to S588A mutants which increases its repair frequency. These results suggest that CDC23 S588A mutant has a dominant negative effect on the endogenous wt protein favouring NHEJ repair (Fig. 6A, C and Fig. S6D). Moreover, we analysed the possible activation of NHEJ, in response to CPT induction, in Hela wt and CDC23 S588A cell clones. To this aim, we performed a plasmid DNA random integration assay [43] and observed an increased plasmid integration in cl9 compared with wt HeLa cells (Fig. 6D). Furthermore, in wt HeLa cells treated with CPT, the loading of 53BP1 onto chromatin was reduced upon 60 and 120 min of CPT washout, consistent with repair DBSs through HR (Fig. 6E); conversely, 53BP1 chromatin loading persisted in cl9 at all time points (Fig. 6E). Consistently, 53BP1 foci decreased in wt HeLa cells upon CPT washout, whereas remained unchanged in cl9 (Fig. S10C and S10D). Finally, we observed an increase of RIF1 foci in cl9 compared with wt HeLa cells, upon CPT treatment (Fig. S10E). Overall, these data showed an impairment of the HR favouring the repair of the CPT-induced DNA damage via NHEJ for the CDC23(S588A)-mutated cells.
A Immunofluorescence analysis of HeLa cells, transfected with siCDC23 or siCTR for 24 h followed by second transfection with indicated coding plasmids for additional 48 h. All cells were treated with 1 μM CPT and RIF1 foci intensity was analysed with Fiji image analysis software. The graph shows the means ± standard deviation of 3 independent experiments and more than 30 cells for each condition. Statistically differences were analysed one-way repeated measures ANOVA with Kruskal-Wallis post-test, to compare all groups and indicated with: *P-value < 0.05 and ***P-value < 0.001. B Representative immunofluorescence images of RIF1 of HeLa cells transfected and treated as in A. C HeLa cells, stably expressing pimEJ5GFP, were transfected with siCDC23 or siCTR and then co-transfected with either wt CDC23, mutated CDC23(S588A) and CDC23(S588D). The empty vector was used as a control. GFP levels were measured by FACS analysis. NHEJ frequency is expressed as a percentage relative to siCTR (set as 100%). Data represent means ± standard deviation (n = 3 independent experiments). Data were subjected to multiple to one-way repeated measures ANOVA with multiple comparison post-test to compare all groups. Statistically significant differences are indicated with: ***P-value < 0.001. D The graph shows the number of clones expressed as a percentage relative to control (set as 100%) of HeLa and cl9 cells co-transfected with a small interfering RNA (siRNA) 53BP1 (si53BP1) and selected by hygromycin B. Data represent means ± standard deviation (n = 3 independent experiments). Statistically differences were evaluated by one-way Anova with Tukey post-test to compare all pairs of data and indicated with: ***P-value < 0.001. E Western blotting of 53BP1 loading onto chromatin obtained from wt HeLa and CDC23(S588A)-mutated cl9, treated or not (NT) with 1 μM camptothecin (CPT) for 2 h followed by CPT washout for 60- and 120-min. CSK purification was carried out as in material and methods to obtain a soluble (S) and a chromatin-enriched (P, as pellet) fraction. Total RPA32 and Lamin A/C were used as controls of the supernatant or the chromatin-enriched fraction, respectively. The black arrow indicates the phosphorylated RPA32 bands. Densitometric analysis of 53BP1 was carried out by using Fiji.
Discussion
Previous studies implicated CDK9 in DDR and replication stress response, thus pointing to this multifunctional kinase as a new possible regulator of DNA repair [11,12,13,14]. CDK9 has two isoforms, CDK9-42 and CDK9-55; most previous studies exploring CDK9 functions used siRNAs targeting the common portion of these two proteins [44, 45]. First of all, we silenced CDK9 though siRNA, followed by differential overexpression of CDK9 isoforms showing an involvement of 42 and 55 in DDR. We set out to study the role of the CDK9 isoforms in DDR through the CRISPR-Cas9-D10A nickase system to obtain a complete knockout of each isoform separately in HeLa cells. We generated 55KO clones without affecting CDK9-42 levels. Conversely, we failed in generating viable CDK9-42KO clones probably related to its central role of this isoform in the regulation of RNA polymerase II activity [46,47,48]. We demonstrated a marked impairment of DNA end-resection, which is fundamental for HR, upon CPT treatment in 55KO clones compared with the control cells, as revealed by both pRPA32 S4/8 expression and SMART assay. Consistently, CtIP phosphorylation and degradation were impaired in CDK9 55 KO clone, upon DNA damage induction, confirming a reduction of DNA repair mechanism resection dependent. Moreover, 55KO clones were more sensitive to PARP inhibition, which previously proved to be synthetic lethal with HR defects [21], thus, suggesting possible HR dysfunction in 55KO cells. Among the PTMs, phosphorylation plays a crucial role in replication stress response [49] and DSB repair [23, 24, 50]; we performed a global phosphoproteomic screening in wt HeLa and 55KO cells, treated or not with CPT, to identify potential CDK9-55 substrates involved in DDR. We identified 7700 proteins deregulated, confirming the high impact of phosphorylation in DDR. Proteomic data analysis revealed possible roles of CDK9-55 mainly in regulating ubiquitination, DNA replication, and DSBs repair. We compared our results with those obtained in genetic screening by Olivieri et al. [25] to verify the reproducibility of our screening for the identification of potential DDR players. We detected 12 proteins common to both screenings. CDKs phosphorylate their substrates in consensus sequences of the target proteins, with some differences among family members [51]. Generally, CDKs show preferences for the phosphorylable amino acids serine and threonine, followed by proline in the +1 position and a basic residue at the +3 position [52]. As previously reported, CDK9 seems to lose its dependency on the basic residue at P + 3, as in the case of the RNA polymerase II, phosphorylated by CDK9 at the C-terminal domain (YSPTSPS) [9]; moreover, CDK9 phosphorylation sites also seem not to depend on the proline at P + 1 [53]. Through the analysis of the phospho (STY) probabilities of the peptides detected by the phosphoproteomic screening, we identified a new CDK9-55 consensus sequence, RxxSP. The search for proteins containing this motif, among the putative CDK9-55 substrates, led to the selection of CDC23 as a possible target of CDK9-55 involved in DDR, with S588 being the putative phosphorylation site. We focused on this protein because it is part of the E3-ubiquitin ligase complex APC/C involved in DDR, and its RxxSP motif is conserved among species and mutated in cancer. APC/C regulation by PTMs is crucial for controlled target degradation [54, 55]; in particular, the APC/C binding with the two co-activators CDC20 and CDH1 during mitosis is regulated by CDK1 [56] and polo-like kinase PLK1 [57]-mediated phosphorylation. CDC23 is involved in substrate recognition and APC/C activity through the binding with the CDC20 protein during mitosis [58]. We generated a CDC23 FLAG tag non-phosphorylable (S588A) and phosphomimetic (S588D) coding plasmids showing the inability of S588A to activate the HR, through the DR-GFP reporter plasmid, leading to the activation of the NHEJ in response to CPT treatment. Conversely, CDC23 phosphomimetic protein was able to rescued the HR ability in similar manner to CDC23 wt protein. Interestingly, CDC23 S588D could be able to induce the activation of checkpoint control proteins in absence of DNA damage mimicking a DDR activation. Through the CRISPR-Cas9-D10A system, we generated HeLa cells stably expressing the non-phosphorylable CDC23(S588A) mutant. These cells showed similar APC/C complex formation ability compared with wt cells during DDR. Moreover, we observed the interaction of CDC23 with the platform protein APC1, identifying most of the APC/C components by LC/MS analysis of the proteins co-immunoprecipitated with either wt CDC23 or CDC23(S588A), but not the CDC20 and CDH1 co-activators. We also observed no effects of CDC23 mutation (S588A) on cell cycle profile. Consistently with the experiments in HeLa cells transfected with the CDC23 S588A coding plasmid, this mutation impacted the DNA repair pathway choice, impairing HR and favouring NHEJ, as detected by different methods in HeLa cells. Moreover, we observed no NHEJ activity increase in HeLa siCDC23 cells probably dependent by a reduced APC/C complex assembly. However, the end-resection marker pRPA32 S4/8 [17, 37] showed similar levels between wt HeLa and CDC23(S588A)-mutated cells; this phenomenon could be in agreement with a possible negative role of RPA32 S4/8 phosphorylation [59] onto the DNA end-resection process. Although further studies are needed to clarify the role of RPA32 phosphorylation in the contest of DNA repair pathway choice. Recently, APC/C has also been implicated in choosing the DNA repair pathway by mediating the polyubiquitination of the USP1 and, thus, preventing it to destroy the K63-linked poly-ubiquitin chains on histones, which are necessary for the proper recruitment of the HR factor BRCA1 [30]. Moreover, APC/C inhibition caused USP1 persistence and increased 53BP1 and RIF1 foci, leading to NHEJ [30]. Consistently, we observed increased ubiquitination of USP1 in HeLa cells, treated with CPT, as pro-HR mechanism; conversely, in HeLa CDC23 S588A cell clone we observed reduced USP1 ubiquitination as a pro-NHEJ mechanism. Similarly, we observed persistence of 53BP1 and RIF1 foci upon CPT treatment in CDC23(S588A)-mutated cells. The possible crosstalk between USP1 and CDK9 deserves future investigation. Overall, we found a new CDK9-55 consensus sequence, which led to the identification of the APC/C subunit CDC23 as a new CDK9-55 substrate guiding the DNA repair pathway choice. Considering the recently established role of CDKs in DDR, their inhibition has been proposed to sensitise cancers to DNA-damaging chemotherapeutics or PARP inhibitors [23]. CDK9 could be a valuable target, especially in tumours with dysregulated transcriptional programs [60]. Our data add a new understanding to CDK9 functions, which should be considered when designing CDK-inhibition-based therapies.
Material and methods
Cell culture, DNA constructs, and transfection
The HeLa cell line was obtained by the American Type Culture Collection (ATCC, CCL-2). Cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 (Thermo Fisher Scientific) supplemented with 10% foetal bovine serum (FBS, Thermo Fisher Scientific), penicillin (100 U/ml), streptomycin (100 µg/ml), and 2 mM glutamine at 37 °C in a humified atmosphere containing 5% CO2. The plasmid encoding wt CDC23 was constructed by cDNA amplification form HeLa cells, followed by digestion with the EcoRI and BamHI restriction enzymes and ligation in the pFLAG-CMV1 vector. We generated the CDC23(S588A) and siCDC23 resistant plasmids through the QuikChange II Site-Directed Mutagenesis Kit (Agilent) according to the manufacturer instructions with the primers indicated in Supplementary Table S10. For silencing experiments, HeLa cells were transfected with 50 nM siCDC23 (5′-GAAAUUAAAUCCUCGGUAUUU-3′), [58] si53BP1 (5’-GAAGGACGGAGUACUAAUAdTdT-3’) [30], siCDK9 (Table S10) or siCTR using Dharmafect 1 (Horizon Discovery) according to the manufacturer instructions. Complementation experiments were carried out by co-transfecting siRNA resistant plasmids and siRNAs targeting protein by using the Lipofectamine 3000 following the manufacturer instructions. CDK9-55KO and CDC23(S588A)-mutated HeLa cells were generated through the CRISPR-Cas9-D10A system. Briefly, cells were transfected with lenti dCAS-VP64_Blast (gift from Feng Zhang; Addgene plasmid #61425; http://n2t.net/addgene:61425; RRID: Addgene_61425) and selected with Blasticidin; CRISPR-Cas9 expression was verified by western blotting; HeLa dCAS9 cells were transfected with the pGL3-U6-sgRNA-PGK-puromycin plasmids (gift from Xingxu Huang; Addgene plasmid #51133; http://n2t.net/addgene:51133; RRID: Addgene_51133) containing guide RNAs targeting CDK9-55 (G1: 5′-GGTGGCGCGTCCCGCTGCAT-3′ and G2: 5’-ATGCAGCGGGACGCGCCACC-3’) to edit ATG for CDK9 55 (NC_000009.12:127785798-127790792 Homo sapiens chromosome 9) or targeting CDC23 for S588A generation (G1: 5’-CGGTGGTAGGAGTCTCGCCT-3’ and G2: 5’-GTCTTCTGTCACGCCATAGT-3’) to edit TCT for CDC23 S588A (NC_000005.10:c138189010-138187650 Homo sapiens chromosome 5), in the presence of the corresponding replacement DNA for HR (Table S10), followed by selection with 5 μg/ml of puromycin. The generated cell clones were analysed by western blotting and sequenced to verify the knock-out/knock-in. The pCBASceI [61] plasmid was a gift from Maria Jasin (Addgene plasmid #26477; http://n2t.net/addgene:26477; RRID: Addgene_26477), pDRGFP [62] plasmid was a gift from Maria Jasin (Addgene plasmid #26475; http://n2t.net/addgene:26475; RRID: Addgene_26475), pEJ5GFP [42] plasmid was a gift from Jeremy Stark (Addgene plasmid # 44026; http://n2t.net/addgene:44026; RRID: Addgene_44026). For pCEFL HA CDK9 42 Kda we designed a oligonucleotides sequence, indicated in the supplementary table S10, followed by PCR using a cDNA and cloning. Finally, to clone CDK9 55 Kda into pCEFL HA vector we used the oligonucleotides (Table S10) for PCR reaction using a pGEM-T CDK9 55 Kda as DNA.
Antibodies and western blotting
The following antibodies were used: CDK9 D7 (1:500, sc-13130, Santa Cruz Biotechnology), RPA32 (1:5000, A300–244 A, Bethyl Laboratories), RPA32 S4/S8 (1:2000, A300–245 A, Bethyl Laboratories), Lamin A/C (1:1000, #4777, Cell Signalling), CDC23 (1:1000, ab182003, Abcam), APC1 (1:1000; A301-653A, Bethyl Laboratories), 53BP1 (1:1000, NB100-304, Novus Biological), HA-tag (1:500, sc-805, Santa Cruz Biotechnology), CHK1 S345 (1:1000, #2348, Cell Signalling), CHK1 (1:1000, #2360, Cell Signalling), GAPDH (1:1000, sc-25778, Santa Cruz Biotechnology), phospho-CHK2 T68 (1:1000, #2197, Cell Signalling), USP1 (1:1000, 14346, Proteintech), RAD51 (1:1000, NB100-148, NovusBiological), KU70 (1:1000, sc-1486, Santa Cruz Biotechnology). For total protein extraction, cells were lysed at 4 °C in 50 mM HEPES pH7.5, 1% Triton X-100, 150 mM NaCl, 5 mM EGTA, supplemented with a protease and phosphatase inhibitor cocktail (Roche Applied Science). Lysates were clarified by centrifugation at 10.000 × g for 20 min at 4 °C. Lysates containing equal amounts of proteins, as estimated through the Bradford assay (BioRad), were subjected to SDS-PAGE. Chemiluminescent images were obtained using the ImageQuant LAS 500 (GE Healthcare). Densitometric analysis of phosphorylated protein was carried out in the following way: first of all, total protein was normalised with the loading control followed by normalisation of phosphorylated protein to the corresponding protein.
Immunoprecipitation
For protein co-immunoprecipitation, HeLa cells were lysed in protein extraction buffer as for western blot. The protein lysate was quantified, and 2 mg, for each condition, were pre-cleared with protein G plus agarose (22851, ThermoFischer Scientific) 45 min at 4 °C on rocking. Immunoprecipitation was carried out at 4 °C on rocking overnight with either CDC23 (1 μg Ab to 1 mg of proteins, ab182003, Abcam), APC1 (1 μg Ab to 1 mg of proteins, A301-653A, Bethyl Laboratories), anti-FLAG M2 (1 μg Ab to 1 mg of proteins,F1804, Sigma Aldrich), IgG isotype mouse control (1 μg Ab to 1 mg of proteins, #31903, ThermoFisher Scientific), IgG isotype rabbit control (1 μg Ab to 1 mg of proteins, #02-6102, ThermoFisher Scientific), USP1 (1 μg Ab to 1 mg of proteins, 14,346, Proteintech).
Cell fractionation
Cell fractionation was performed as previously described with minor modifications [63]. Briefly, 3 × 106 cells, per condition, were collected and re-suspended in 200 μl of CSK buffer (10 mM PIPES pH 6.8, 100 mM NaCl, 300 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.1% Triton X-100, 0.34 M sucrose) supplemented with protease and phosphatase inhibitors and kept 5 min on ice. The soluble cytoplasmic fraction (S) was separated from nuclei (P) by 4 min centrifugation at 1300×g at 4 °C. The P fraction was washed with CSK then re-suspended in 200 μl of western blot buffer, sonicated and centrifuged for 30 min at 4 °C at 10.000 × g. Samples were subjected to SDS-PAGE and analysed by western blot with the indicated antibodies.
Immunofluorescence
HeLa cells, grown on glass coverslips, were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. Samples were blocked 10 min in 1% BSA at RT and incubated 1 h with anti-RAD51 (1:200, sc8349, Santa Cruz Biotecnology), anti-53BP1 (1:300, NB100-304, Novus Biological), anti-RIF1 (1:300, A300-567A Bethyl Laboratories), anti-RPA32 S4/8 (1:200, A300-245A Bethyl Laboratories), anti-BRCA1 (1:200, A300-000A Bethyl Laboratories) at 37 °C. After washing, samples were incubated 45 min at 37 °C with AlexaFluor 594-conjugated chicken anti-rabbit (ThermoFischer Scientific) and analysed with a Zeiss LSM900 confocal microscope. The foci intensity and foci per cell values were calculated by Fiji software.
ssDNA formation assay
The ssDNA formation assay was performed as reported previously [64] with some modifications. Briefly, wt HeLa and CDC23(S588A)-mutated cells were pulse-labelled for 24 h with 10 μM BrdU and treated with 1 μM CPT (Santa Cruz Biotechnology) for 2 h. In order to quantify the amount of resected ssDNA we worked in non-denaturing conditions. Cells were harvested by trypsinization. After washing with 1X PBS, 1 × 106 cells for each experimental point were fixed with 4% paraformaldehyde in 1X PBS for 15 min at RT followed by permeabilisation with 1X PBS containing 0.1% Triton X-100 for 30 min at RT. Cells were washed in 1X PBS twice and re-suspended in 100 μl of incubation buffer (1X PBS + 0.5% Saponin) with the anti-BrdU antibody (1:100, clone B44, 347580, BD Biosciences) for 1 h at RT or just incubation buffer, as a control. Cells were washed with incubation buffer and re-suspended with 100 μl of 488-conjugated rabbit anti-mouse (Thermo Fisher Scientific) for 45 min at RT. The percentage of BrdU positive cells was determined using the CellQuest Software (Becton Dickinson). The threshold level identifying FITC positivity was set following comparison with cells incubated with only the secondary antibody.
Single-Molecule Analysis of Resection Tracks (SMART)
SMART was carried out as previously described [18] with some modifications [19]. Briefly, wt HeLa, CDK9-55KO cl2, and CDC23(S588A)-mutated cl9 were pulse-labelled with 10 μM IdU and incubated at 37 °C in a standard 5% CO2 incubator for 24 h, followed by treatment with 1 μM CPT for 2 h. HeLa cells were resuspended in ice-cold 1X PBS at the concentration of 2.5 × 105 cells/ml and diluted in a 1:8 proportion with unlabelled cells at the same concentration; 2.5 μl of cells were mixed with 7.5 μl of spreading buffer (0.5% SDS in 200 mM Tris-HCl, pH7.4, 50 mM EDTA) directly on the upper part of the slides and incubated at room temperature (RT) for 10 min. The slides were tilted to 15°, and the DNA spreads were air-dried for 10 min. The DNA fibres were fixed in 3:1 methanol/acetic acid at −20 °C for 15 min; the slides were washed in 1X PBS twice and incubated in 70% ethanol and 30% water overnight at 4 °C. The slides were then washed twice in 1X PBS, blocked with 5% BSA in 1X PBS for 30 min at RT, and incubated with 1:300 Anti-BrdU clone BU1/75 (ICR1) for one hour at 37 °C, followed by incubation with 1:400 Alexa Fluor 488-conjugated chicken anti-rat antibody. The DNA fibres were washed with 1X PBS twice and the slides were mounted with ProLong Gold Antifade Reagent (Life Technologies).
Phosphoproteome analysis
Proteins were extracted from cells using a lysis buffer consisting of 9 M urea, 50 mM Tris-Cl, pH 7.5, 1 mM EDTA, and phosphatase inhibitors. 1.5 mg of each sample was reduced with 5 mM dithiothreitol, alkylated with 10 mM iodoacetamide, diluted to 2 M urea, and digested with trypsin at 1:50 enzyme: protein ratio. Peptides were purified using C18 SepPak column (Waters) and phosphopeptides were enriched using Titansphere Phos-TiO (GL Sciences Inc.). Liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis was performed by the Proteomics and Metabolomics Facility at the Wistar Institute using a Q Exactive Plus mass spectrometer (ThermoFisher Scientific) coupled with a Nano-ACQUITY UPLC system (Waters). Samples were injected onto a UPLC Symmetry trap column (180 µm i.d. x 2 cm packed with 5 µm C18 resin; Waters). Peptides were separated by reversed phase HPLC on a BEH C18 nanocapillary analytical column (75 µm i.d. x 25 cm, 1.7 µm particle size; Waters) using a 2-h gradient formed by solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile). Eluted peptides were analyzed by the mass spectrometer set to repetitively scan m/z from 400 to 2000 in positive ion mode. The full MS scan was collected at 70,000 resolutions followed by data-dependent MS/MS scans at 17,500 resolutions on the 20 most abundant ions exceeding a minimum threshold of 20,000. Peptide match was set as preferred, exclude isotopes option and charge-state screening were enabled to reject singly and unassigned charged ions. Peptide sequences were identified using MaxQuant v1.5.2.8 [65]. MS/MS spectra were searched against a UniProt human protein database (9/12/2016) using full tryptic specificity with up to two missed cleavages, static carboxamidomethylation of Cys, and variable oxidation of Met, phosphorylation on Ser, Thr and Tyr, and protein N-terminal acetylation. Consensus identification lists were generated with false discovery rates of 1% at protein, peptide, and site levels. A minimum localisation probability of 0.75 was applied for phosphosites identification.
Clonogenic assay
300 cells were seeded in each well of 24-wells plates and either treated with the indicated doses of CPT, Olaparib (Selleckchem), Etoposide (Sellckchem) or not treated. Cells were incubated for 10 days. Colonies were counted after fixation with methanol and staining with crystal violet.
Cell viability assay
HeLa cells were seeded in triplicates in 96-well plates at a density of 1500 cells/well for and allowed to adhere for 24 h. Cells were treated at the indicated drug concentrations and incubated for additional 72 h. At the end of the treatment cells were fixed with 50% v/v trichloroacetic acid and stained with 0.4% w/v sulforhodamine B (SRB) in 1% v/v acetic acid. The percentage of cell viability after treatment was calculated assuming as 100% the number of untreated cells.
HR and NHEJ reporter assays
HeLa-pDRGFP cells [36], stably expressing the reporter plasmid DR-GFP, were co-transfected with a plasmid coding for the ISceI endonuclease and the indicated siRNAs and vectors. After 48 h of incubation, we analysed the GFP values (as a readout of HR frequency) through FACS analysis. HeLa cells stably expressing the reporter plasmid pimEJ5GFPwere co-transfected with the plasmid coding for the endonuclease ISceI and the indicated siRNAs and vectors. After 48 h of incubation, we analysed the GFP values (as a readout of NHEJ frequency) through FACS analysis.
Random Integration assay
The random Integration assay was carried out as described previously [43]. Briefly, wt HeLa and CDC23(S588A)-mutated cl9 were co-transfected with the siCTR or si53BP1 and the pCMV3-C-His, previously linearised by ApaI restriction enzyme. After 24 h from transfection, we seeded 50.000 cells for each point in 60 mm dishes. Cells were incubated for additional 24 h and then subjected to hygromycin B selection for 15 days at the 800 μg/ml working concentration. Cell colonies were counted after fixation with methanol and staining with crystal violet.
Cell cycle analysis
For DNA content analysis, cells were fixed in 70% ice-cold ethanol at –20 °C. Cells were incubated with 5 mg/ml propidium iodide and 0.25 mg/ml RNase A (Sigma-Aldrich). At least 10.000 cells were analysed by FACS (Becton Dickinson). Data were analysed through the CellQuest Software (Becton Dickinson).
Ubiquitination assay
The ubiquitination assay was performed as described previously [66]. We pre-treated HeLa cells with 30 μM MG132 (sc-201270, Santa Cruz Biotechnology), followed by incubation with 1 μM CPT. For the ubiquitination assay performed in cl9 with the proTAME inhibitor (I-440, R&D System), we pre-treated cells for 1 h with 30 μM MG132, alone or in combination with 20 μM proTAME, followed by incubation for additional 2 h with 1 μM CPT.
Recombinant GST protein production
GST-CDK9-42, GST-CDK9-55, GST-cyclin T1, GST-CDC23 wt, GST-CDC23 S588A were cloned in the pGEX2T vector by using the primers listed in Table S10; pGEX-2T-RNAPII-CTD wt #14-HA was a gift from Jeffry Corden (Addgene plasmid # 160688; http://n2t.net/addgene:160688; RRID: Addgene_160688) [26]. Cloned vector was transformed in BL21 DE3 (Rosetta) followed by induction with 300 μM IPTG for 4 h. The bacterial pellet was resuspended in lysis buffer (20 mM Tris-HCL ph 7.5, 250 mM, 2 mM EGTA, 2 mM EDTA, 1% Triton and protease inhibitors) followed by sonication. Cleared bacterial protein extracts were incubated for two hours at 4 °C with Glutatione Sepharose 4 Fast Flow resin (Cytiva). At the end of incubation time, the resin was washed six times with lysis buffer without Triton but with 0.03% NP40 followed by elution in wash buffer and 20 mM Glutathione ph 8. The dialysis was carried out over night at 4 °C with 40 mM Tris-HCL ph 7.5, 20 mM MgCl2 and 15% glicerol.
Kinase assay
CDK9 kinase assay was carried out with ADP-Glo Kinase assay as described in the manufactured instructions. Briefly, we incubated kinase and substrates on ice for 30 min in kinase buffer (400 mM Tris-HCL ph 7.5, 200 mM MgCl2, 1 mg/ml BSA, 0,5 mM DTT, 5 mM ATP) followed by incubation at 30 °C for 30 min.
Statistical analysis
Statistical analysis was performed using the GraphPad Prism Software, version 9 for Mac. To evaluate differences between the means of two groups, we used two-sided Student t test, whereas to analyse differences among the means of multiple groups we used one-way Anova with either Kruskal-Wallis post-test or Tukey post-test to compare all pairs of data; Dunnett’s multiple comparison to compares all groups versus control. To evaluate to groups unpaired, we used unpaired t-test with Mann-Whitney post-test. P < 0.05 was considered to be statistically significant. The number of independent experiments and P-values are reported in the figure legends.
Data availability
The mass spectrometry proteomics data have been deposited into the MassIVE (http://massive.ucsd.edu) with the accession number MSV000087856.
References
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Mao Z, Bozzella M, Seluanov A, Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair. 2008;7:1765–71.
Scully R, Panday A, Elango R, Willis NA. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol. 2019;20:698–714.
Ceccaldi R, Rondinelli B, D’Andrea A. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26. https://doi.org/10.1016/J.TCB.2015.07.009.
Russell P, Nurse P. Schizosaccharomyces pombe and Saccharomyces cerevisiae: a look at yeasts divided. Cell. 1986;45:781–2.
Hydbring P, Malumbres M, Sicinski P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat Rev Mol Cell Biol. 2016;17:280–92.
De Falco G, Giordano A. CDK9 (PITALRE): a multifunctional cdc2-related kinase. J Cell Physiol. 1998;177:501–6.
Graña X, De Luca A, Sang N, Fu Y, Claudio PP, Rosenblatt J, et al. PITALRE, a nuclear CDC2-related protein kinase that phosphorylates the retinoblastoma protein in vitro. Proc Natl Acad Sci USA. 1994;91:3834–8.
Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305.
Khan SZ, Mitra D. Cyclin K inhibits HIV-1 gene expression and replication by interfering with cyclin-dependent kinase 9 (CDK9)-cyclin T1 interaction in Nef-dependent manner. J Biol Chem. 2011;286:22943–54.
Yu DS, Zhao R, Hsu EL, Cayer J, Ye F, Guo Y, et al. Cyclin-dependent kinase 9-cyclin K functions in the replication stress response. EMBO Rep. 2010;11:876–82.
Zhang H, Park S-H, Pantazides BG, Karpiuk O, Warren MD, Hardy CW, et al. SIRT2 directs the replication stress response through CDK9 deacetylation. Proc Natl Acad Sci USA. 2013;110:13546–51.
Nepomuceno TC, Fernandes VC, Gomes TT, Carvalho RS, Suarez-Kurtz G, Monteiro AN, et al. BRCA1 recruitment to damaged DNA sites is dependent on CDK9. Cell Cycle. 2017;16:665–72.
Storch K, Cordes N. The impact of CDK9 on radiosensitivity, DNA damage repair and cell cycling of HNSCC cancer cells. Int J Oncol. 2016;48:191–8.
Shore SM, Byers SA, Maury W, Price DH. Identification of a novel isoform of Cdk9. Gene. 2003;307:175–82.
Zimmermann M, de Lange T. 53BP1: pro choice in DNA repair. Trends Cell Biol. 2014;24:108–17.
Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284:9558–65.
Cruz-García A, López-Saavedra A, Huertas P. BRCA1 accelerates CtIP-ediated DNA-end resection. Cell Rep. 2014;9:451–9.
Altieri A, Dell’Aquila M, Pentimalli F, Giordano A, Alfano L. SMART (single molecule analysis of resection tracks) technique for assessing DNA end-resection in response to DNA damage. Bio Protoc. 2020;10:e3701.
Liu H, Herrmann CH, Chiang K, Sung T-L, Moon S-H, Donehower LA, et al. 55K isoform of CDK9 associates with Ku70 and is involved in DNA repair. Biochem Biophys Res Commun. 2010;397:245–50.
Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21.
Montecucco A, Zanetta F, Biamonti G. Molecular mechanisms of etoposide. EXCLI J. 2015;14:95.
Johnson N, Shapiro GI. Cyclin-dependent kinases (cdks) and the DNA damage response: rationale for cdk inhibitor-chemotherapy combinations as an anticancer strategy for solid tumors. Expert Opin Ther Targets. 2010;14:1199–212.
Bai M, Ti D, Mei Q, Liu J, Yan X, Chen D, et al. The role of posttranslational modifications in DNA repair. Biomed Res Int. 2020;2020:1–13.
Olivieri M, Cho T, Álvarez-Quilón A, Li K, Schellenberg MJ, Zimmermann M, et al. A genetic map of the response to DNA damage in human cells. Cell. 2020;182:481–96.e21.
Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, et al. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–94.
Kim YK, Bourgeois CF, Isel C, Churcher MJ, Karn J. Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 Is directly responsible for human immunodeficiency virus type 1 tat-activated transcriptional elongation. Mol Cell Biol. 2002;22:4622.
Peters J-M. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–56.
Castro A, Bernis C, Vigneron S, Labbé J-C, Lorca T. The anaphase-promoting complex: a key factor in the regulation of cell cycle. Oncogene. 2005;24:314–25.
Ha K, Ma C, Lin H, Tang L, Lian Z, Zhao F, et al. The anaphase promoting complex impacts repair choice by protecting ubiquitin signalling at DNA damage sites. Nat Commun. 2017;8:15751.
Lafranchi L, Boer HR, Vries EG, Ong S, Sartori AA, Vugt MA. APC / C C dh1 controls Ct IP stability during the cell cycle and in response to DNA damage. EMBO J. 2014;33:2860–79.
de Boer HR, Guerrero Llobet S, van Vugt MATM. Controlling the response to DNA damage by the APC/C-Cdh1. Cell Mol Life Sci. 2016;73:949–60.
Cotto-Rios XM, Jones MJK, Busino L, Pagano M, Huang TT. APC/CCdh1-dependent proteolysis of USP1 regulates the response to UV-mediated DNA damage. J Cell Biol. 2011;194:177–86.
Liu S, Opiyo SO, Manthey K, Glanzer JG, Ashley AK, Amerin C, et al. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res. 2012;40:10780–94.
Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011. https://doi.org/10.1101/gad.2003811.
Alfano L, Caporaso A, Altieri A, Dell’Aquila M, Landi C, Bini L, et al. Depletion of the RNA binding protein HNRNPD impairs homologous recombination by inhibiting DNA-end resection and inducing R-loop accumulation. Nucleic Acids Res. 2019;47. https://doi.org/10.1093/nar/gkz076.
Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–14.
Cruz C, Castroviejo-Bermejo M, Gutiérrez-Enríquez S, Llop-Guevara A, Ibrahim YH, Gris-Oliver A, et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann Oncol. 2018;29:1203–10.
Anand R, Ranjha L, Cannavo E, Cejka P. Phosphorylated CtIP functions as a Co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol Cell. 2016;64:940–50.
Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–8.
Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–9.
Bennardo N, Cheng B, Huang N, Stark J. Alternative-NHEJ Is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4. https://doi.org/10.1371/JOURNAL.PGEN.1000110.
Nishi R, Wijnhoven P, le Sage C, Tjeertes J, Galanty Y, Forment JV, et al. Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat Cell Biol. 2014;16:1016–26. 1–8
Garriga J, Graña X. CDK9 inhibition strategy defines distinct sets of target genes. BMC Res Notes. 2014;7:301.
Ou J, Li H, Qiu P, Li Q, Chang H-C, Tang Y-C. CDK9 modulates circadian clock by attenuating REV-ERBα activity. Biochem Biophys Res Commun. 2019;513:967–73.
Zhou Q, Chen D, Pierstorff E, Luo K. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 1998;17:3681–91.
Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–22.
Yik JHN, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–82.
Li Z, Xu X. Post-translational modifications of the mini-chromosome maintenance proteins in DNA replication. Genes. 2019;10:331.
Summers KC, Shen F, Sierra Potchanant EA, Phipps EA, Hickey RJ, Malkas LH. Phosphorylation: the molecular switch of double-strand break repair. Int J Proteom. 2011;2011:1–8.
Brown NR, Lowe ED, Petri E, Skamnaki V, Antrobus R, Johnson L. Cyclin B and Cyclin A confer different substrate recognition properties on CDK2. Cell Cycle. 2007;6:1350–9.
Brown NR, Noble MEM, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol. 1999;1:438–43.
Baumli S, Lolli G, Lowe ED, Troiani S, Rusconi L, Bullock AN, et al. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008;27:1907–18.
Zhang S, Chang L, Alfieri C, Zhang Z, Yang J, Maslen S, et al. Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature. 2016;533:260–4.
Lee CC, Li B, Yu H, Matunis MJ. Sumoylation promotes optimal APC/C activation and timely anaphase. Elife. 2018;7. https://doi.org/10.7554/eLife.29539.
Blanco MA, Sánchez-Díaz A, de Prada JM, Moreno S. APC(ste9/srw1) promotes degradation of mitotic cyclins in G(1) and is inhibited by cdc2 phosphorylation. EMBO J. 2000;19:3945–55.
Golan A, Yudkovsky Y, Hershko A. The cyclin-ubiquitin ligase activity of cyclosome/APC is jointly activated by protein kinases Cdk1-Cyclin B and Plk. J Biol Chem. 2002;277:15552–7.
Izawa D, Pines J. How APC/C-Cdc20 changes its substrate specificity in mitosis. Nat Cell Biol. 2011;13:223–33.
Soniat MM, Myler LR, Kuo HC, Paull TT, Finkelstein IJ. RPA phosphorylation inhibits DNA resection. Mol Cell. 2019;75:145–53.e5.
Cassandri M, Fioravanti R, Pomella S, Valente S, Rotili D, Del Baldo G, et al. CDK9 as a valuable target in cancer: from natural compounds inhibitors to current treatment in pediatric soft tissue sarcomas. Front Pharmacol. 2020;11. https://doi.org/10.3389/FPHAR.2020.01230.
Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–42.
Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–8.
Ishii T, Shiomi Y, Takami T, Murakami Y, Ohnishi N, Nishitani H. Proliferating cell nuclear antigen-dependent rapid recruitment of Cdt1 and CRL4Cdt2 at DNA-damaged sites after UV Irradiation in HeLa cells. J Biol Chem. 2010;285:41993–42000.
Forment JV, Walker RV, Jackson SP. A high-throughput, flow cytometry-based method to quantify DNA-end resection in mammalian cells. Cytom Part A. 2012;81A:922–8.
Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72.
Singh AN, Oehler J, Torrecilla I, Kilgas S, Li S, Vaz B, et al. The p97-Ataxin 3 complex regulates homeostasis of the DNA damage response E3 ubiquitin ligase RNF8. EMBO J. 2019;38:e102361.
Acknowledgements
We are thankful to the Sbarro Health Research Organization (http://www.shro.org) and Italian Ministry of Health Ricerca Corrente 2022 Grant L3/1. This work was supported by Sbarro Health Research Organization and by the Italian Ministry of Health Ricerca Corrente 2022 Grant L3/1. We are grateful to Professor David H. Price, University of Iowa, for the pGEM-T CDK9 55Kda used in pCEFL HA CDK9 55Kda cloning.
Author information
Authors and Affiliations
Contributions
AL contributed to conceptualization, investigation, development of methodology, writing- original draft, funding acquisition; MDA, AA, AC, GM, CR, SB, CAI, IMF, DB, MCR, MC, LM, contributed to investigation, development of methodology, validation; PI contributed to writing - review & editing; AG contributed to conceptualization, supervision, funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Alfano, L., Iannuzzi, C.A., Barone, D. et al. CDK9-55 guides the anaphase-promoting complex/cyclosome (APC/C) in choosing the DNA repair pathway choice. Oncogene 43, 1263–1273 (2024). https://doi.org/10.1038/s41388-024-02982-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-024-02982-w